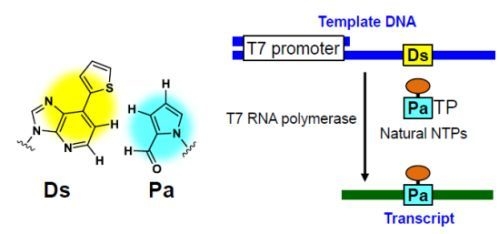
Site-Specific Incorporation of Functional Components into RNA by an Unnatural Base Pair Transcription System
Abstract
:1. Introduction
2. Results and Discussion
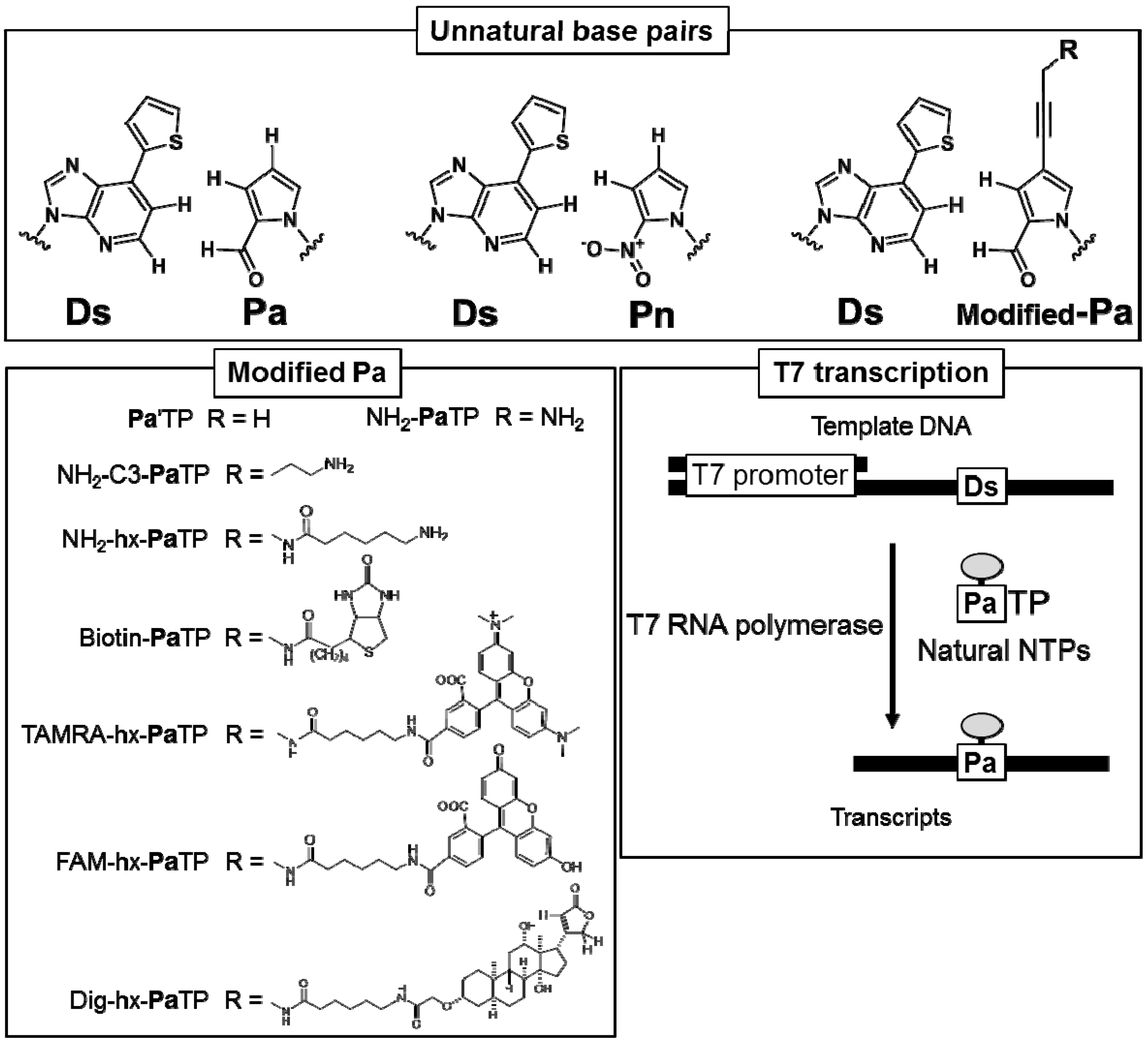
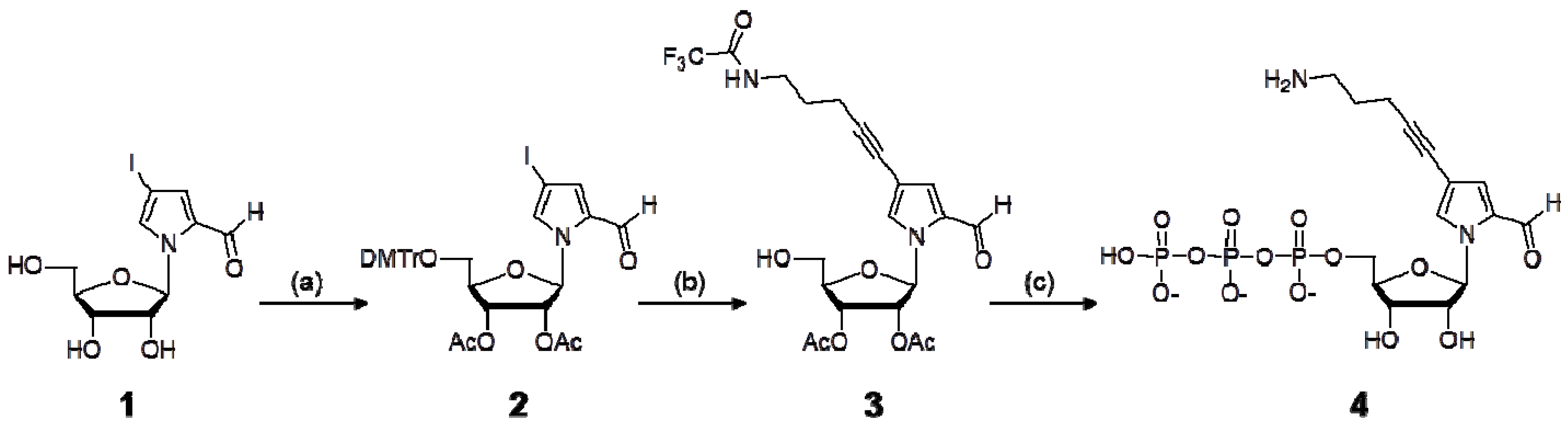
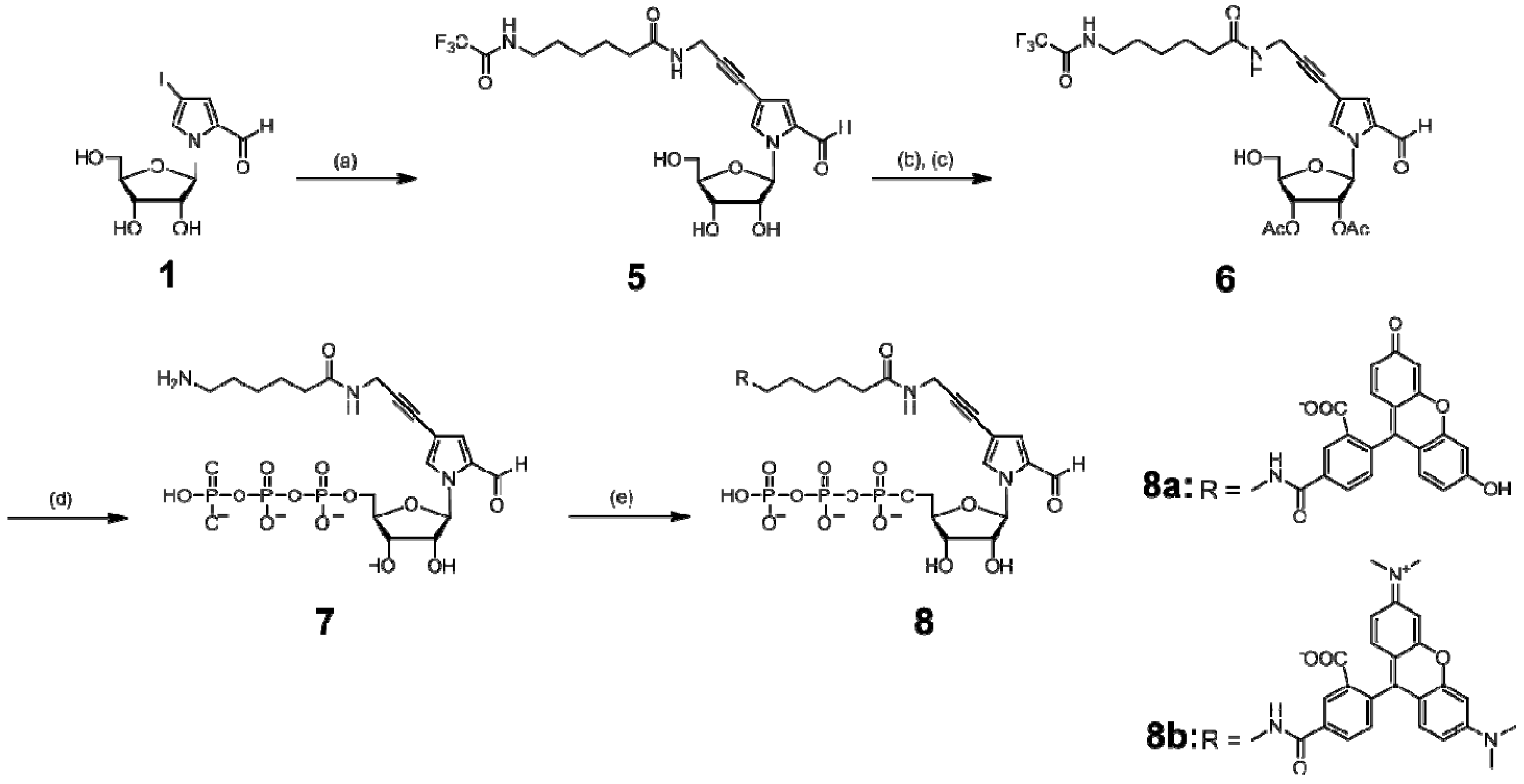
2.1. Chemical Syntheses of Modified-Pa Triphosphates (Modified-PaTPs) and Ds-Containing DNA Templates
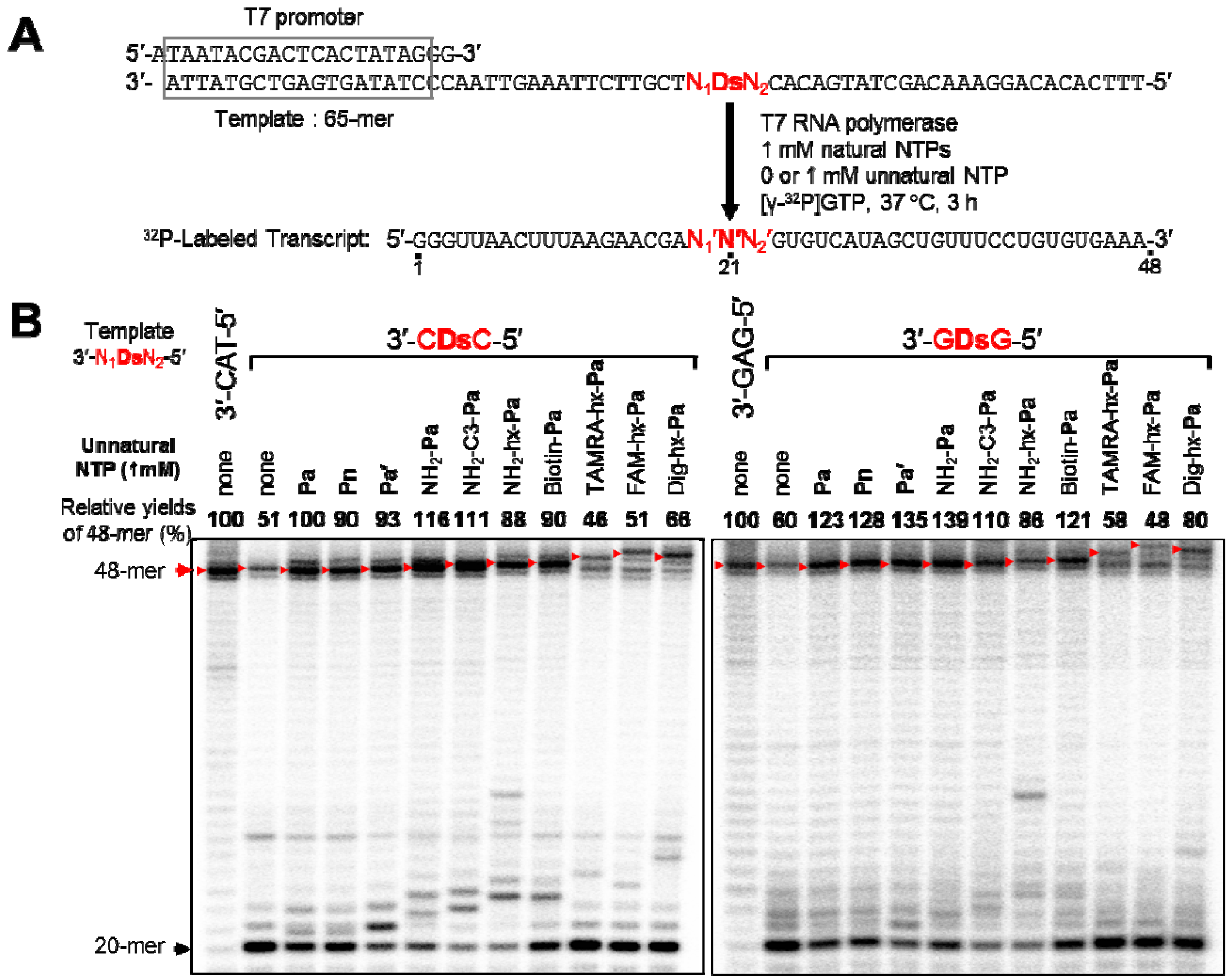
2.2. T7 Transcription for 48-Mer Transcripts Containing Modified-Pa at Internal Positions
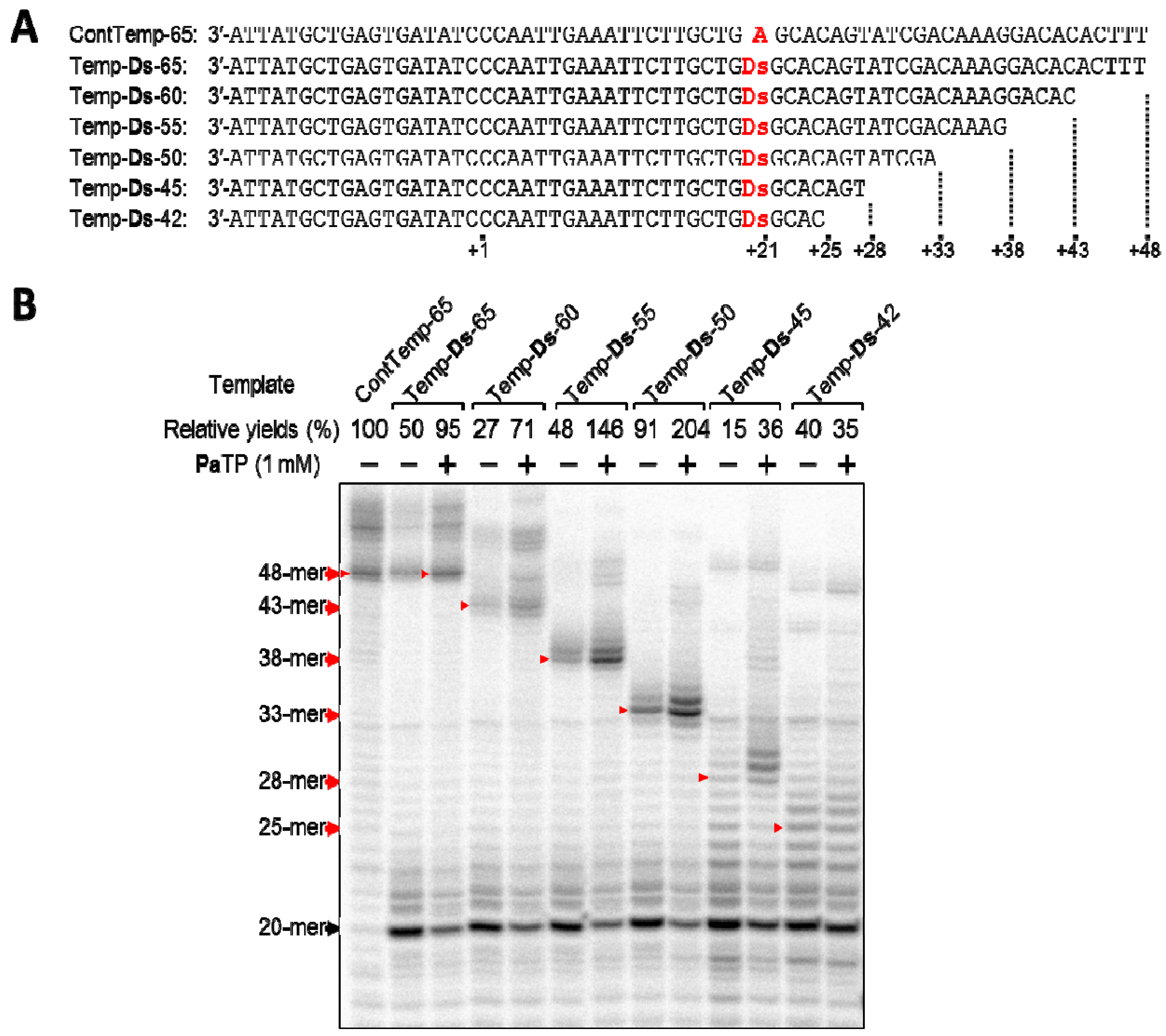
2.3. T7 Transcription for Transcripts Containing Pa at Different Positions from the 3′-Terminus
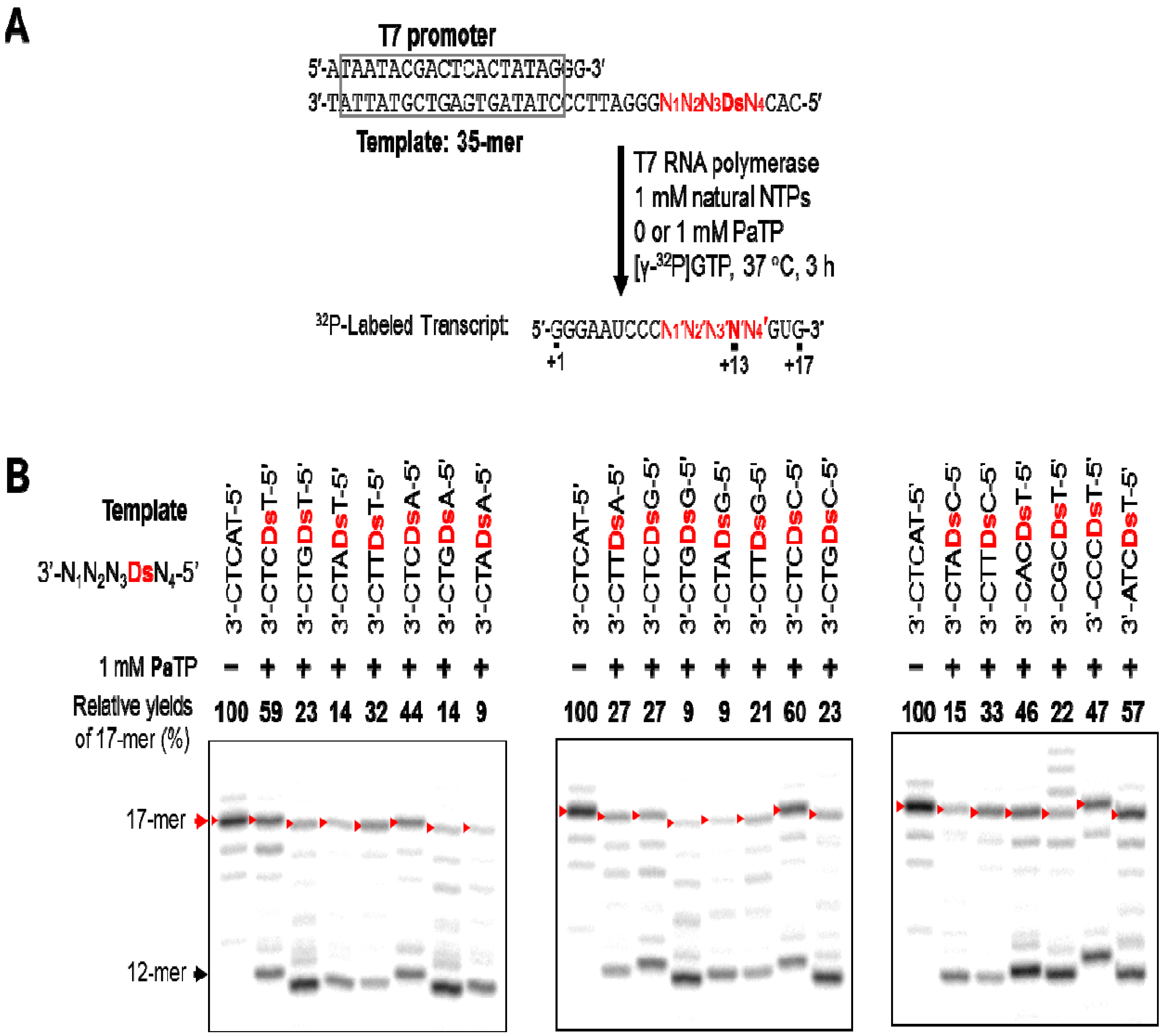
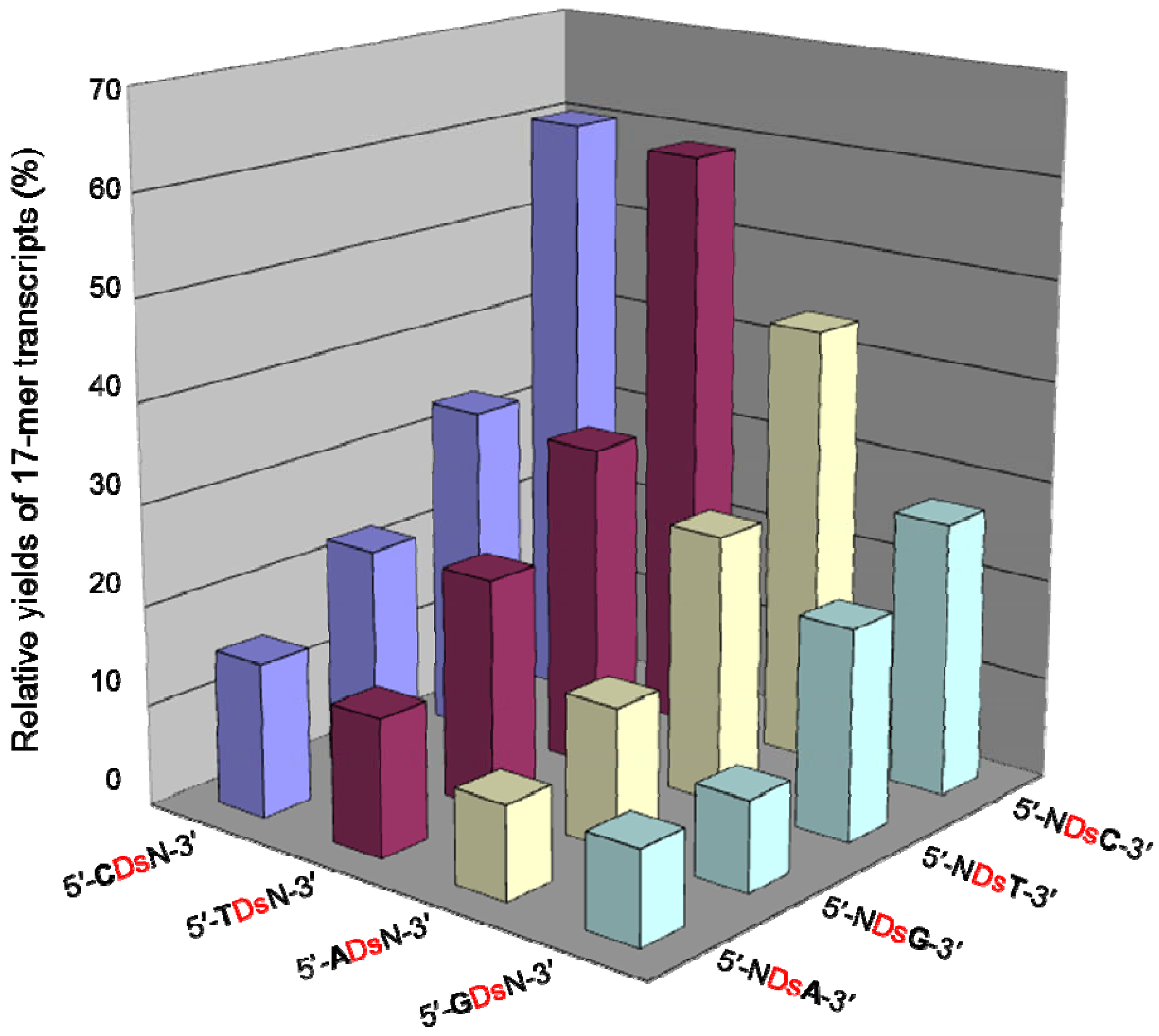
2.4. Pa Incorporation into the 3′-Region of Transcripts with Different Sequence Contexts around the Unnatural Base
2.5. DNA Template Preparation for the Ds–Pa Transcription System
3. Experimental
3.1. General
3.2. Synthesis of NH2-C3-PaTP (4)
3.3. Synthesis of NH2-hx-PaTP (7), FAM-hx-PaTP (8a), and TAMRA-hx-PaTP (8b)
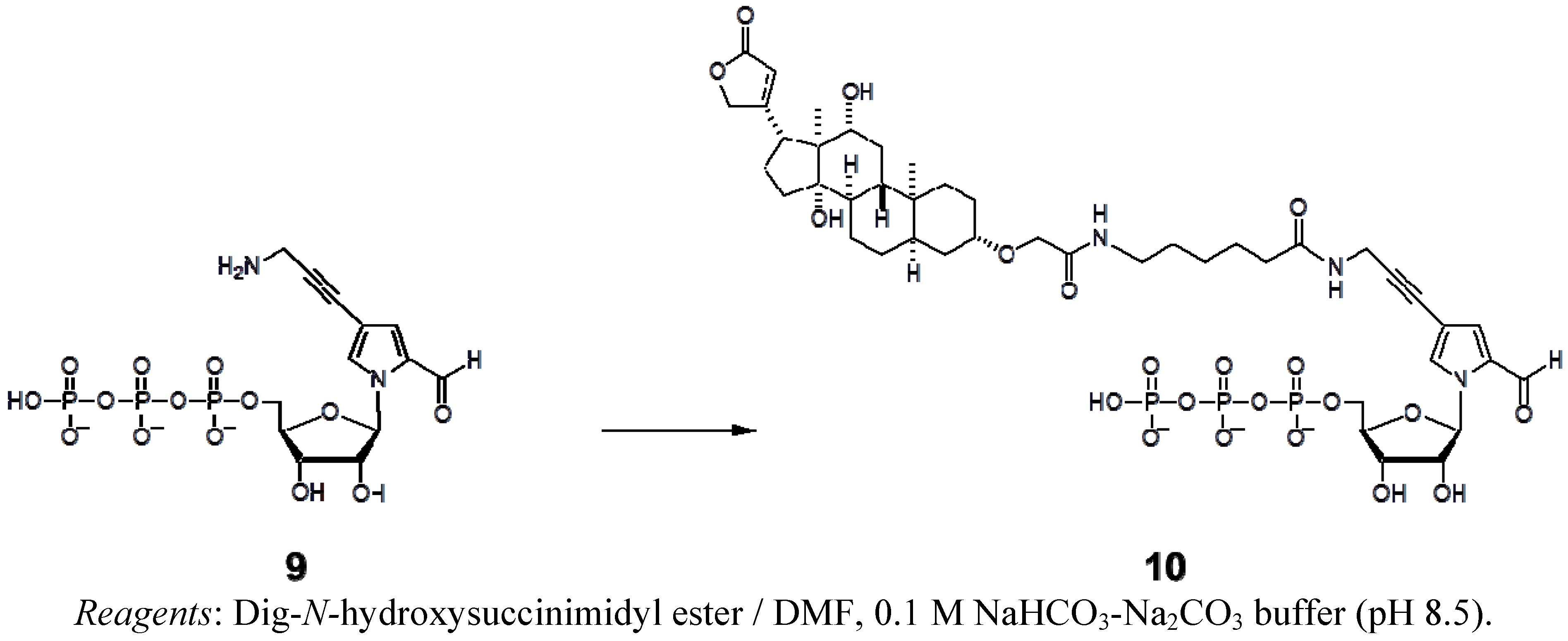
3.4. Synthesis of Dig-hx-PaTP (10)
3.5. DNA Templates for T7 Transcription
3.6. Transcription
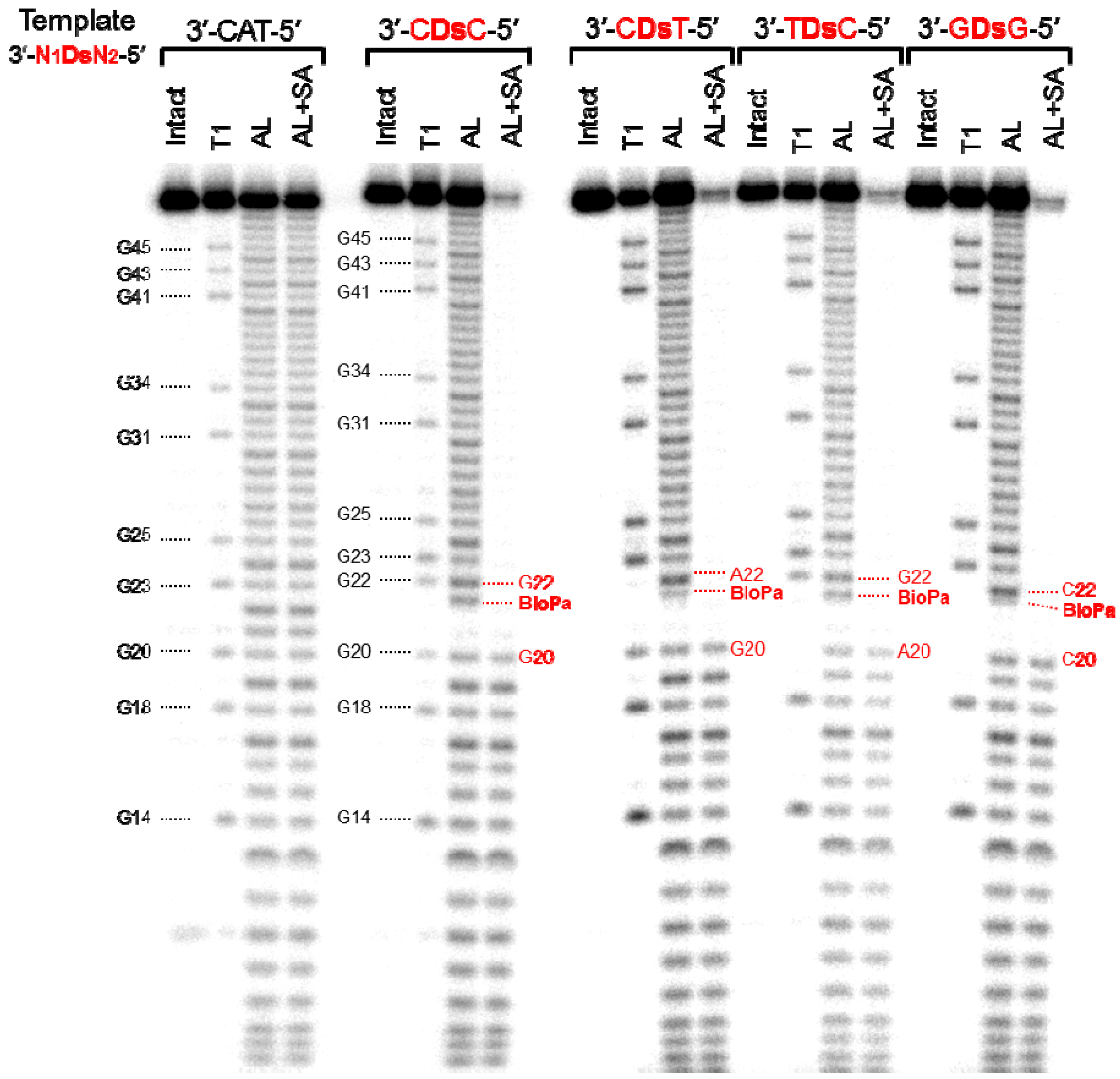
3.7. Analysis of the Insertion Position of Biotin-Pa in the 48-Mer Transcripts
4. Conclusions
Acknowledgements
References and Notes
- Benner, S.A. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 2004, 37, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Henry, A.A.; Romesberg, F.E. Beyond A, C, G and T: Augmenting nature’s alphabet. Curr. Opin. Chem. Biol. 2003, 7, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.T.; Kool, E.T. Redesigning the architecture of the base pair: Toward biochemical and biological function of new genetic sets. Chem. Biol. 2009, 16, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006, 10, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Cox, R.S., 3rd.; Hirao, I. Unnatural base pair systems for sensing and diagnostic applications. Expert Rev. Mol. Diagn. 2011, 11, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Kimoto, M.; Mitsui, T.; Fujiwara, T.; Kawai, R.; Sato, A.; Harada, Y.; Yokoyama, S. An unnatural hydrophobic base pair system: Site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods 2006, 3, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Kawai, R.; Mitsui, T.; Yokoyama, S.; Hirao, I. An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 2009, 37, e14. [Google Scholar] [CrossRef] [PubMed]
- Yamashige, R.; Kimoto, M.; Takezawa, Y.; Sato, A.; Mitsui, T.; Yokoyama, S.; Hirao, I. Highly specific unnatural base pair systems as a third base pair for PCR amplification. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, D.A.; Seo, Y.J.; Ordoukhanian, P.; Romesberg, F.E. PCR with an expanded genetic alphabet. J. Am. Chem. Soc. 2009, 131, 14620–14621. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, D.A.; Pfaff, D.A.; Ippoliti, S.I.; Hwang, G.T.; Dwyer, T.J.; Romesberg, F.E. Solution structure, mechanism of replication, and optimization of an unnatural base pair. Chem. Eur. J. 2010, 16, 12650–12659. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sismour, A.M.; Sheng, P.; Puskar, N.L.; Benner, S.A. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007, 35, 4238–4249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, F.; Alvarado, J.B.; Benner, S.A. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 2011, 133, 15105–15112. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Ohtsuki, T.; Fujiwara, T.; Mitsui, T.; Yokogawa, T.; Okuni, T.; Nakayama, H.; Takio, K.; Yabuki, T.; Kigawa, T.; Kodama, K.; Nishikawa, K.; Yokoyama, S. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002, 20, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hikida, Y.; Kimoto, M.; Yokoyama, S.; Hirao, I. Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis. Nat. Protoc. 2010, 5, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Hirao, I. Site-specific incorporation of extra components into RNA by transcription using unnatural base pair systems. Methods Mol. Biol. 2010, 634, 355–369. [Google Scholar] [PubMed]
- Seo, Y.J.; Malyshev, D.A.; Lavergne, T.; Ordoukhanian, P.; Romesberg, F.E. Site-specific labeling of DNA and RNA using an efficiently replicated and transcribed class of unnatural base pairs. J. Am. Chem. Soc. 2011, 133, 19878–19888. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Matsuda, S.; Romesberg, F.E. Transcription of an expanded genetic alphabet. J. Am. Chem. Soc. 2009, 131, 5046–5047. [Google Scholar] [CrossRef] [PubMed]
- Tor, Y.; Dervan, P.B. Site-specific enzymatic incorporation of an unnatural base, N6-(6-aminohexyl)isoguanosine, into RNA. J. Am. Chem. Soc. 1993, 115, 4461–4467. [Google Scholar] [CrossRef]
- Kimoto, M.; Mitsui, T.; Yamashige, R.; Sato, A.; Yokoyama, S.; Hirao, I. A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J. Am. Chem. Soc. 2010, 132, 15418–15426. [Google Scholar] [CrossRef] [PubMed]
- Kawai, R.; Kimoto, M.; Ikeda, S.; Mitsui, T.; Endo, M.; Yokoyama, S.; Hirao, I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005, 127, 17286–17295. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Endo, M.; Mitsui, T.; Okuni, T.; Hirao, I.; Yokoyama, S. Site-specific incorporation of a photo-crosslinking component into RNA by T7 transcription mediated by unnatural base pairs. Chem. Biol. 2004, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, K.; Kimoto, M.; Mitsui, T.; Yokoyama, S.; Hirao, I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005, 33, e129. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Mitsui, T.; Kimoto, M.; Yokoyama, S. An efficient unnatural base pair for PCR amplification. J. Am. Chem. Soc. 2007, 129, 15549–15555. [Google Scholar] [CrossRef] [PubMed]
- Nacheva, G.A.; Berzal-Herranz, A. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur. J. Biochem. 2003, 270, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Yamashige, R.; Kimoto, M.; Mitsui, T.; Yokoyama, S.; Hirao, I. Monitoring the site-specific incorporation of dual fluorophore-quencher base analogues for target DNA detection by an unnatural base pair system. Org. Biomol. Chem. 2011, 9, 7504–7509. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of three triphosphate derivatives (NH2-hx-PaTP, Biotin-PaTP, FAM-PaTP) are available from the authors. |



© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Morohashi, N.; Kimoto, M.; Sato, A.; Kawai, R.; Hirao, I. Site-Specific Incorporation of Functional Components into RNA by an Unnatural Base Pair Transcription System. Molecules 2012, 17, 2855-2876. https://doi.org/10.3390/molecules17032855
Morohashi N, Kimoto M, Sato A, Kawai R, Hirao I. Site-Specific Incorporation of Functional Components into RNA by an Unnatural Base Pair Transcription System. Molecules. 2012; 17(3):2855-2876. https://doi.org/10.3390/molecules17032855
Chicago/Turabian StyleMorohashi, Nobuyuki, Michiko Kimoto, Akira Sato, Rie Kawai, and Ichiro Hirao. 2012. "Site-Specific Incorporation of Functional Components into RNA by an Unnatural Base Pair Transcription System" Molecules 17, no. 3: 2855-2876. https://doi.org/10.3390/molecules17032855