Abstract
Unlike the secondary structure elements that connect in protein structures, loop fragments in protein chains are often highly mobile even in generally stable proteins. The structural variability of loops is often at the center of a protein’s stability, folding, and even biological function. Loops are found to mediate important biological processes, such as signaling, protein-ligand binding, and protein-protein interactions. Modeling conformations of a loop under physiological conditions remains an open problem in computational biology. This article reviews computational research in loop modeling, highlighting progress and challenges. Important insight is obtained on potential directions for future research.
1. Introduction
Virtually all biological mechanisms in the living cell involve protein molecules. Proteins are central components of cellular organization and function. The mechanistic view that shape, also referred to as structure, governs biological function in proteins has been confirmed in wet laboratory experiments [1]. The unique set of atoms that make up a protein molecule determines to a great extent the spatial arrangement or conformation assumed by these atoms for biological function. Experiment, theory, and computation, however, show that proteins are not rigid molecules but employ internal motions to populate different structures or conformations through which they tune their biological function [2,3,4]. Elucidating the role of motion in protein function is now at the forefront of protein research [5].
The state in which a protein carries its biological activity is also referred to as the protein native state. Microscopically, this macrostate is an ensemble of native conformations also referred to as the native state ensemble. Even for a protein where the native state ensemble is largely the result of local fluctuations around an average structure, certain fragments of the protein chain are more mobile than others. For instance, unlike secondary structure elements, such as α-helices and β-sheets (see Figure 1), loop fragments in protein chains are often highly mobile even in generally stable proteins.
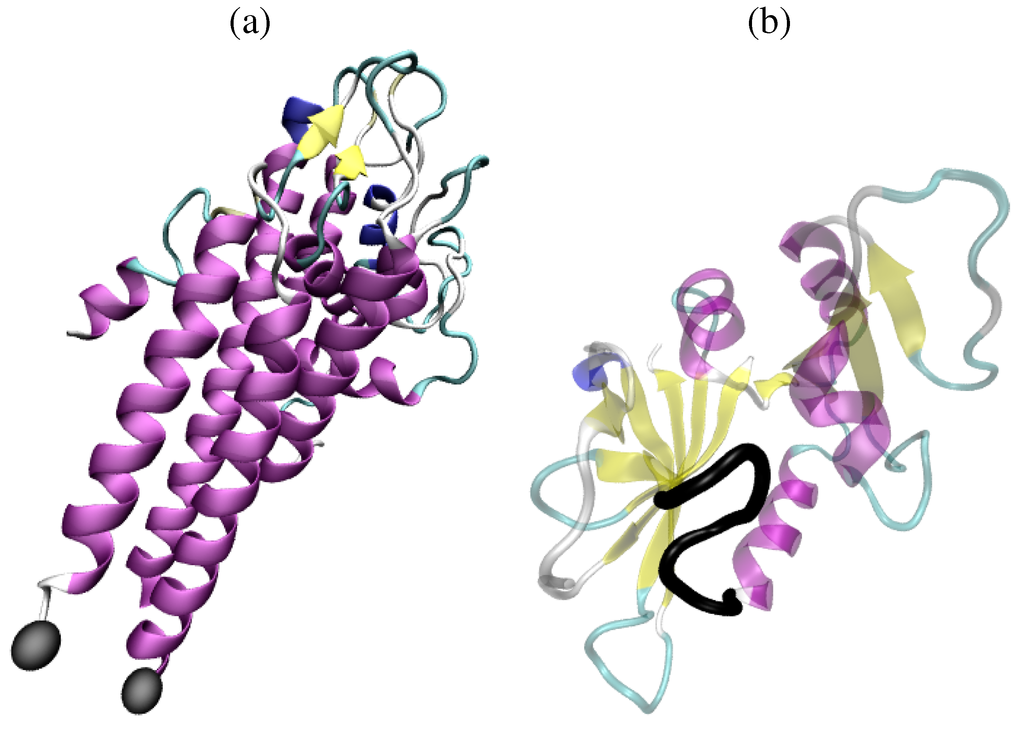
Figure 1.
(a) The crystal structure of the variable surface antigen (PDB ID 1LW8) is partially resolved, with a loop of 20 amino acids missing. The van der Waals spheres of the atoms of amino acids at both ends of the missing loop are drawn in gray. (b) Three loops surround the active site of dihydrofolate reductase. The Met20 loop, drawn in thick black and spanning amino acids 9 to 24, undergoes a significant conformational change in the enzyme’s four distinct functional states. The structure drawn in transparent is the X-ray structure under PDB ID 3DFR. (a)–(b) Secondary structure elements are visible in the protein structures drawn here. α-helices are drawn as purple helices, and β-sheets are drawn in yellow as thick pointed arrows. With the exception of the Met20 loop highlighted in black, other loop fragments are drawn as coils in green or white. All protein structures here are drawn with the Visual Molecular Dynamics Software (VMD) [6].
Loops are fragments of a protein chain that are generally void of secondary structure. In addition to connecting secondary structure elements in protein structures, loops play an important role in protein folding and stability. Additionally, loops often determine the functional specificity of a protein molecule. Loops mediate important biological processes. They are found on active and binding sites to mediate binding of antigens to immunoglobulins [7], toxins to protein receptors [8], metal ions to proteins [9], DNA to DNA-binding proteins [10], and protein substrates to serine proteases [11]. Due to their role in protein function, loops are also an important consideration in protein engineering [12].
Loops often lie on protein surfaces and so are exposed to solvent. This allows them more structural variability, which is a primary reason why loops are not easy to characterize through wet-laboratory techniques, such as NMR and X-ray crystallography. It is often the case that protein structures resolved in the wet laboratory are incomplete. In particular, mobility introduces significant disorder in a protein crystal. In such cases, partially-resolved protein structures are reported, with the loop missing. Figure 1a shows the partially-resolved X-ray structure of the variable surface antigen obtained from the Protein Data Bank (PDB) [13]. A loop of 20 amino acids is missing from the crystal structure of this protein [14].
Modeling a missing loop is important in completing a protein’s native structure, particularly when the loop may mediate the biological activity of the protein. Guessing coordinates for a loop fragment is not an easy task. Because loops are often on the surface of protein structures, they are susceptible to insertions and deletions of amino acids. This sequence variability limits the application of comparative modeling techniques in extracting the conformation for a loop at hand from a template structure.
In many cases, proposing a single loop conformation may not address the mobility of the loop under native conditions. In light of the high structural variability of loops in proteins, one or a few loop conformations may not adequately capture the structural diversity in the ensemble of conformations assumed by a mobile missing loop. Figure 1b shows the native structure of dihydrofolate reductase, an enzyme that employs distinct backbone conformations of the M20 loop to regulate its substrate binding [15].
The above examples illustrate that understanding protein function may depend on modeling the equilibrium mobility or flexibility of a loop fragment. Modeling the equilibrium flexibility of a loop can be addressed in different ways. A qualitative description of flexibility can be obtained without explicitly computing conformations populated at equilibrium. For instance, given a loop conformation in a complete native protein structure, rigidity-based analysis of the network of bonds in the protein structure can be conducted to locate flexible and rigid regions [16,17,18]. Even though this information does not readily yield equilibrium conformations, elucidating which regions of a loop are more flexible than others can improve understanding of the functional role of the loop. It is worth pointing out that rigidity-based analysis can be employed to offer alternative conformations, but there are currently no direct applications on loops. Instead of focusing on a qualitative description of flexibility, this review concerns itself with methods that explicitly show the motions available at equilibrium by computing conformations for the given loop.
Finding a physically-relevant conformation for a (missing or not) loop in a given protein structure is known as the loop modeling problem. Due to the role of loops in protein function, loop modeling is an important problem in computational biology. Because the problem has been studied in various forms by different communities, it has alternative names, such as loop/fragment completion, gap completion, loop closure, and fragment fitting. Loop modeling is important to address not only in automated crystallographic protein structure determination, but also in comparative modeling and ab-initio structure prediction, where incomplete protein structures with missing loops are often obtained from protein structure prediction protocols [19,20]. In fact, loop modeling is often regarded as a somewhat easier version of the ab-initio structure prediction problem for two main reasons. First, loops are shorter than protein chains. Second, the presence of the protein structure at either end of the loop poses constraints that can be exploited to obtain conformations for a loop more efficiently than conformations for an entire protein chain.
Loop modeling involves fitting a generated loop conformation with a given protein structure so the loop connects with the rest of the protein structure and completes it. When the loop is missing, the only pieces of information about the loop and the protein at hand are the loop’s amino-acid sequence and coordinates for the atoms in the rest of the protein structure. Figure 2 shows a loop conformation that fits and so connects with the rest of the protein structure. The amino acids that precede and follow the loop are referred to as stems. The given protein structure provides coordinates for the stem amino acids. Figure 2a shows the protein structure at either end of the loop and draws the planes defined by the three main-chain atoms of the stems.
In loop modeling, the loop is often defined as the fragment of the protein chain that includes the stem amino acids (shown in Figure 2b). It is important to note that two sets of coordinates are available for these amino acids. One set is available from the given protein structure. The other is obtained from a generated loop conformation. Fitting a generated loop conformation with a given protein structure involves modifying the conformation so that the coordinates of the stem amino acids in this conformation superimpose with those of the stem amino acids in the given protein structure. The distinction between the stem amino acids in the loop versus those in the given protein structure is made clearer by borrowing robotics terminology. In robotics-inspired approaches to loop modeling that exploit analogies between protein chains and articulated robotic mechanisms, the stems in the protein structure are referred to as stationary anchors, whereas the stems in the loop are referred to as mobile anchors.
Borrowing further terminology used in robotics, the mobile anchors are constrained to assume the poses (position and orientation) of their corresponding stationary anchors. Figure 2b shows that the loop conformation goes through the planes defined by the main-chain atoms of the stationary anchors in the given protein structure. Since the stationary anchors provide geometric constraints for the mobile anchors, loop modeling can be viewed as a constraint satisfaction problem. In a treatment of loop modeling where a loop conformation is first generated and then modified to fit with the rest of the protein structure, the geometric constraints need to be satisfied only on one of the loop’s two mobile anchors. Rigid-body transformations can be employed to translate and rotate a selected mobile anchor with its stationary counterpart. Modifying the resulting loop conformation in order to place the remaining mobile anchor in the target pose provided by its corresponding stationary anchor is non-trivial, and many methods have been proposed to address this problem.
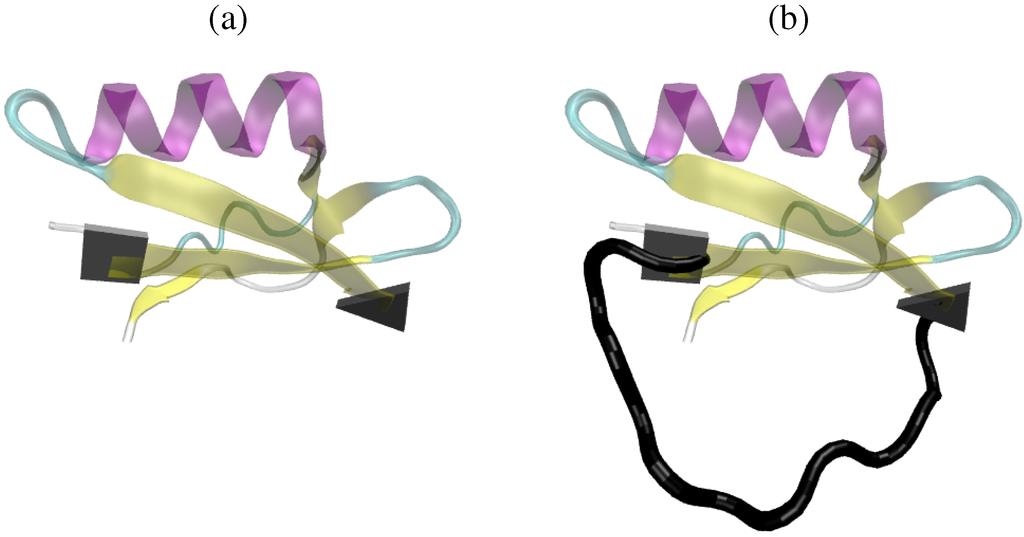
Figure 2.
(a) The X-ray structure of chymotrypsin inhibitor 2 (CI2), PDB ID 1COA, is drawn in transparent with a loop of 12 amino acids (amino acids 53-64) removed. The planes defined by the three main-chain atoms of the stationary anchors (amino acids 52 and 65) are drawn in opaque. (b) The complete structure is now drawn, with the loop conformation present. The loop, drawn in black, connects with the rest of the CI2 structure. The loop’s mobile anchors overlap with the stationary anchors.
The loop conformation where the geometric (also referred to as kinematic) constraints are satisfied are also referred to as closed conformations, as opposed to open loop conformations where the constraints are not satisfied. The conformational space that contains closed loop conformations is also referred to as the closure space of the loop. It is important to note that this space is a superset of the set of physically-relevant loop conformations. Considerations of energetic interactions between the loop’s atoms and between the loop and the rest of the protein structure allow discriminating against energetically unfavorable loop conformations in the closure space. In fact, an understanding and treatment of proteins that goes beyond geometry and includes physics-based interactions is crucial to successfully address loop modeling. Section 2 elaborates the role of energy in protein modeling.
The review in this article summarizes loop modeling methods often contributed from diverse communities of researchers, such as computational biologists, chemists, computer scientists, and roboticists. The review tracks the great progress that has been made by loop modeling methods, while highlighting remaining challenges. Particular attention is paid to methods that are able to compute not just one conformation to model a given loop but can reveal more about the mobility of the loop by elucidating its conformational ensemble under native conditions. This ensemble view is more suitable to shed light on the potential biological role of a loop, as the gamut of conformations available to the loop under native conditions may elucidate the ability of the loop to take part in diverse molecular interactions.
The article is organized as follows. Section 2 provides a brief introduction into protein geometry and modeling. Section 3, Section 4 and Section 5 then describe loop modeling methods, categorizing them according to representative approaches. Section 6 describes in greater detail selected methods that are representative of successful approaches. The article concludes in Section 7 with a summary of current progress, remaining challenges, and potential directions for future research.
2. Short Primer on Protein Modeling
Blocks of atoms known as amino acids connect in a series-like fashion to form a protein chain. Each amino acid contains an alpha carbon () atom connected to a hydrogen atom, an amino group, a carboxylic group, and a group of atoms known as a side chain. Side-chain atoms and the connectivity among them confer to amino acids unique physico-chemical properties and result in 20 naturally-occurring amino acids. The amino nitrogen of one amino acid connects through a peptide bond with the carboxylic carbon of another amino acid to form a dipeptide. Consecutive peptide bonds link amino acids in a protein or polypeptide chain, as shown in Figure 3a. Amino acids are numbered from the N- to the C-terminus, which refer to the amino and carboxyl groups not involved in peptide bonds. The backbone is what remains of the polypeptide chain after stripping off all side chains.
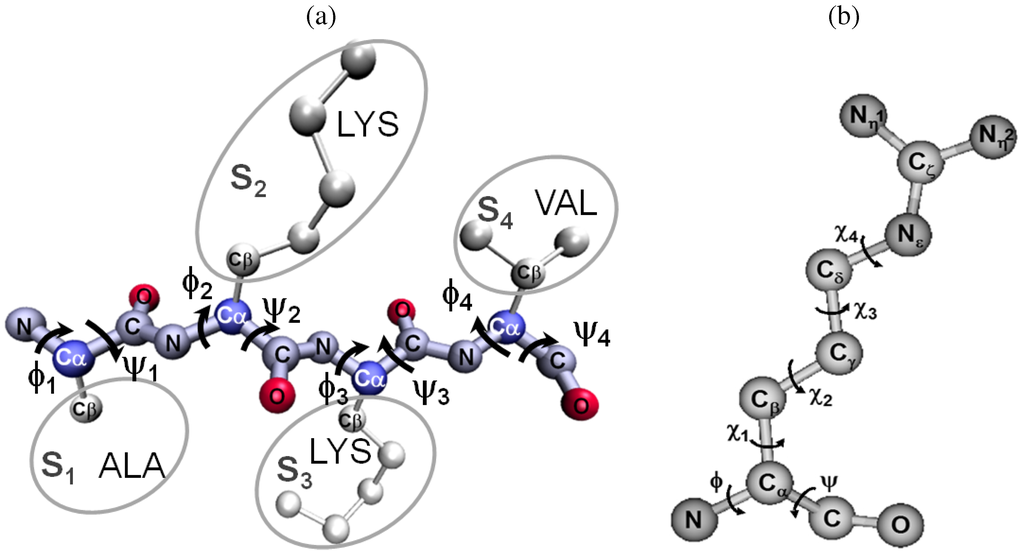
Figure 3.
(a) A chain of four amino acids is shown. The N, , C, and O backbone atoms are labeled for each amino acid. Main-chain atoms refer to all backbone atoms but the oxygen atoms. A peptide bond between the backbone N and C atoms links two amino acids together. The termini atoms in this short chain are the N and C backbone atoms not involved in peptide bonds. There are two backbone () angles per amino acid, as labeled here. Atoms in white are labeled S for side chain. There are 20 distinct side chains in naturally-occurring proteins. Side-chains dangle off the backbone. (b) There are at most four side-chain dihedral angles per amino acid in a protein chain, as shown here in detail for the long arginine amino acid.
The spatial arrangement of atoms in a protein chain is referred to as a conformation. There are different representations of a protein chain, which result in different degrees of detail in computed conformations. A conformation C of a protein chain comprised of N atoms may be represented as a vector , where are coordinates of atom . The atom coordinates maintained to represent C are often referred to as degrees of freedom (dofs).
The Cartesian representation of a protein chain is often viewed as redundant. An internal representation reduces the number of dofs by recording only bond lengths, bond angles (the angle between two consecutive bonds), and dihedral angles (the angle that can be defined on the second bond of a series of three consecutive bonds). The internal representation allows recovering atomic coordinates through forward kinematics: essentially, rotations about bonds are propagated down the protein chain to update atomic positions [21,22,23].
Analysis of protein structures deposited in structure databases reveals that bond lengths and bond angles are constrained to characteristic values [24]. This observation, a consequence of energetic constraints on native conformations, is exploited to idealize the protein geometry employed in modeling and remove bond lengths and bond angles as dofs. The resulting idealized geometry representation contains only dihedral angles defined over three consecutive bonds as dofs, as illustrated in Figure 3b. Idealizing protein geometry is appealing, as it reduces the total number of dofs to an average of dofs for a protein chain of N atoms [25]. Idealizing the protein geometry relevant for computing native conformations excludes from consideration improbable but not impossible deviations of bond lengths and bond angles from equilibrium characteristic values. These deviations are strongly disfavored due to energetic constraints in native conformations.
Idealizing protein geometry reveals mechanistic analogies between protein chains and robot kinematic chains with revolute joints. As a joint rotation changes the positions of following links, so does rotation about a bond change the positions of following atoms [21]. These analogies have long been employed by robotics researchers to apply algorithms that plan motions for kinematic chains with revolute dofs to the study of protein conformations [26,27,28,29,30,31,32,33,34,35,36,37]. Unlike typical articulated mechanisms, protein chains have a high number of dofs. A short backbone chain of 15 amino acids has 30 dofs.
It is important to note that protein chains are more than kinematic chains. The backbone dihedral angles in conformations that the protein chain assumes to carry out a biological function, also known as native conformations, do not populate the entire but are limited to specific regions in amino-acid dependent (Ramachandran) maps [38]. These regions are associated with local secondary structures in native conformations, such as α-helices, β-sheets, and coils. Additionally, a limited set of rotamer configurations are observed for side chains in native conformations [39].
Significant efforts in protein modeling go towards finding representations that reduce the number of dofs and yet allow capturing important physical properties. Typical representations range from fine-grained all-atom, which model all atoms, to coarse-grained, which model only a subset of the atoms. Coarse-grained representations range from traces, where only the central atom is modeled in an amino acid, to backbone representations which model only backbone atoms. We refer the reader to [40] for a review of current state-of-the-art representations employed by computational methods. Choosing a representation for a protein chain is an important decision in a computational method, as the representation affects not only the feasibility of the method, but also its accuracy. A coarse-grained representation changes both the conformational space and the effective energy surface underlying the conformational space.
Conformational changes in proteins are the result of favorable and unfavorable interactions among the atoms in a conformation and with the surrounding solvent. The totality of these interactions results in a potential energy value that can be associated with a protein conformation. Organizing the multitude of conformations of a protein chain by their potential energies elucidates a multi-dimensional funnel-like energy surface [41,42,43]. The vertical axis of this surface records the potential energy (or the “internal free energy”) of a conformation. The lateral axes represent the many underlying dimensions, and the width of this multi-dimensional surface denotes the entropy a protein system [41]. Entropy essentially measures the degree of conformational redundancy that allows a protein chain to flex while maintaining the same potential energy. Stable conformations are a compromise between low potential energy and high entropy, a quantity captured by the notion of free energy as in F = E − TS (F is free energy, E is potential energy, T is temperature, and S is entropy). Due to evolutionary bias, the most stable or native state in naturally-occurring proteins is also the one with the minimum free energy [1]. We refer the reader to [41,43,44] for detailed reviews on proteins and free energy.
Free energy is difficult to measure in silico. Measuring entropy is the main challenge, as it requires computing the range of values of underlying dofs corresponding to different conformations with the same potential energy. Since introducing the effects of entropy requires extensive free energy sampling, many methods forego entropy considerations and focus instead on probing the energy surface essentially one potential energy at a time. Most loop modeling methods summarized in this review fall in this category. Since the steepness of the energy surface is due to the potential energy, the goal is often to obtain low-energy conformations and then to select from among them the one(s) reproducing the sought protein native state.
Measuring potential energy is also non-trivial. All current energy functions, even state-of-the-art ones, are approximations that allow probing not the true protein energy surface but an effective energy surface [40]. For instance, all-atom energy functions that essentially sum interatomic interactions into a potential energy value sacrifice the electronic dofs. The functional formula of physics-based energy functions is often limited to pairwise interactions. This is another approximation necessitated by the computational cost of summing over multi-body interactions and the difficulty in designing physically robust multi-body energy functions. Energy functions that calculate multi-body interactions do exist and often outperform pairwise-based functions in reproducing experimental kinetic data [45].
Coarse-grained energy functions sacrifice even more dofs and introduce more approximations. Their role, however, is not limited to mainly offering a computationally expedient alternative to the costly all-atom energy functions. In fact, coarse-grained energy functions (see [40] for a detailed review on them) provide a smoother energy surface compared to all-atom energy functions. The smoothness is due to removal of some structural frustration in proteins that results in energetic barriers between two conformations corresponding to local minima in the energy surface. A less rugged energy surface helps search algorithms to more feasibly locate the global minimum. Coarse-grained energy functions additionally incorporate some configurational entropy due to the dofs that are integrated out in these functions. Moreover, when employed to elucidate the folding mechanism of given proteins, effective coarse-grained energy functions provide important insights into the minimal set of dofs that are most relevant to folding [40]. Many loop modeling methods employ coarse-grained representations and energy functions. These methods benefit from the feasibility afforded by coarse-grained representations but incorporate potential errors inherent to coarse-grained energy functions. In fact, many loop modeling studies show that both coarse- and fine-grained energy functions suffer from inaccuracies or insensitivities [46,47,48,49].
3. Inverse Kinematics Methods
Fitting a loop conformation with a given protein structure so that the mobile anchor assumes the target pose in the corresponding stationary anchor naturally lends itself to analogies with controlling motions of a robot arm so that the hand/gripper assumes a given pose [26]. One end of the loop, a mobile anchor that is trivially overlapped with its corresponding stationary anchor through rigid-body transformations can be treated as the base of a kinematic chain. The other end (the remaining mobile anchor) can be treated as an end effector that needs to reach a target pose (that of the stationary counterpart) to connect with the rest of the protein and so complete the protein structure. This is known as the Inverse Kinematics (IK) problem originally formulated for robotic manipulators [21]: solve for the chain dofs so the resulting configuration places the end effector in the target pose (the term configuration is employed in robotics instead of conformation). In the context of loop modeling under idealized geometry, angles are sought for the dihedral dofs so the resulting loop conformation places the mobile anchor in the pose of its stationary counterpart. When formulated as an IK problem, loop modeling can be addressed with a rich set of IK techniques originally developed in the robotics and computer graphics communities [50].
3.1. Exact IK Techniques
Exact or analytic IK techniques employ mathematical formulations of the given geometric constraints and seek exact solutions to the resulting algebraic equations. For manipulators with no more than 6 dofs, the number of solutions is finite [21]. There is, however, no analytical technique able to find these solutions for all types of manipulators. On kinematic chains with 6 revolute dofs (6R mechanisms), the number of constraints is identical to the number of dofs. Hence, configurations can be found as discrete solutions to polynomial equations that formulate the constraints. A tight upper bound of 16 solutions has been established for the IK problem for 6R kinematic chains operating in a 3D workspace [51].
Since protein fragments with idealized geometry and 3 amino acids are analogous to 6R manipulators, the solutions can be enumerated. An efficient technique able to enumerate all solutions was proposed in [52] and later applied to short molecular chains in [26,53]. Separate from progress in the robotics community on the IK problem, specialized solutions in biology appeared as early as the 70s [54]. Conformations for protein fragments of up to 6 dofs were predicted by solving a set of polynomial equations representing geometric transformations. These equations were originally applied to build tripeptide loops under the ideal geometry assumption [54]. Later work offered efficient analytical solutions for three consecutive amino acids through spherical geometry and polynomial equations [53,55,56,57].
Later work in [58] generalizes the approach to protein fragments of an arbitrary number of amino acids by solving for any 6 not-necessarily-consecutive dofs separated by any number of rigid segments of the protein. Essentially, the sought configurations are found as real solutions of a -degree polynomial equation in one variable. Later work in [59] pushes the dimensionality limit from 6 to 9 dofs through an efficient subdivision of the solution space. IK techniques based on curve approximation are proposed in [60] for the inverse kinematics of hyper-redundant chains with a very large number of regularly-distributed joints. Other exact IK techniques that deal with molecular chains include [53,54,57,58,59]. Bounding inverse kinematics solutions for chains with no more than 6 dofs within small intervals is applied in the context of drug design in [23,61].
3.2. Optimization-Based IK Techniques
Optimization-based or numerical techniques can address the IK problem for kinematic chains with an arbitrary number of dofs. Techniques like random tweak [62,63] and cyclic coordinate descent (CCD) [64] are representative of optimization-based IK techniques that iteratively solve a system of equations until the kinematic constraints are satisfied.
Random tweak defines the kinematic constraints of interest somewhat differently, choosing to focus on satisfying distance constraints between atoms of the mobile anchors in the loop rather than between atoms of mobile anchor in the loop and its corresponding stationary anchor in the given protein structure. A Jacobian matrix is employed to maintain the first derivatives of these distances with respect to the dihedral angles. Minimization of these distances relies on inversion of the Jacobian, but Lagrange multipliers are used to minimize changes in the dihedral angles [63]. Essentially, starting from a random loop conformation, all dihedral angles are modified at once. This modification is repeated for a number of iterations, until the distance constraints are satisfied or deemed infeasible. Random tweak remains popular and has been incorporated in many loop modeling packages, such as Biopolymer by Tripos, Inc. St. Louis, MO, USA, Drawbridge [65] and Loopy [48].
Because random tweak relies on the inversion of the Jacobian, its computations are demanding and even numerically unstable, as the matrix may lose rank. In addition to suffering from singularities, random tweak does not allow placing additional constraints on individual amino acids of the loop, because modifications to dihedral angles are introduced all at once, with a strong dependence of each proposed dihedral change on all the others. Additional constraints on the dihedrals may result in unpredictable motions of the atoms away from rather than toward their target positions.
Unlike random tweak, CCD does not compute an inverse or pseudoinverse of a Jacobian matrix, but solves instead a 1-dof IK problem. Given a loop conformation, the dihedral angles are modified one at a time to minimize the distance of the end effector/mobile anchor in the ensuing conformation to the target pose. The process is illustrated in Figure 4a, where a selected atom in a current position M needs to reach a target position F. The optimal rotation of the dihedral bond between the and C atoms, shown by α, is the one that positions the atom on the projection of the target position F on the circle defined by the -C rotation axis. Figure 4b shows the ensuing conformations as CCD iterates over the rotatable bonds of a short chain of 7 bonds and rotates them by the optimal angles needed for the selected end atom to reach its target pose. In applications of CCD to loop modeling, α is the solution to the equation that minimizes the distance between the current positions of the main-chain atoms in the mobile anchor and the target positions of these atoms in the stationary anchor.
By avoiding the use of a Jacobian, CCD is computationally inexpensive, numerically stable, and free of singularities. The technique avoids the inter-dependence of dihedrals by adjusting one at a time. CCD was first introduced in the context of non-linear programming and was originally applied to robotics [64]. Its linear time complexity on the number of dofs make it very appealing for computational biology applications that need to model and close loops of arbitrary length [66,67,68,69]. Unlike random tweak, CCD-obtained solutions can be modified to incorporate additional constraints with a predictable motion of atoms toward target positions. This is exploited in [66] to steer CCD-obtained angle values towards the closest occupied region in the Ramachandran map. CCD has been incorporated into the Rosetta ab-initio structure prediction package to model loops in computed structures [20].
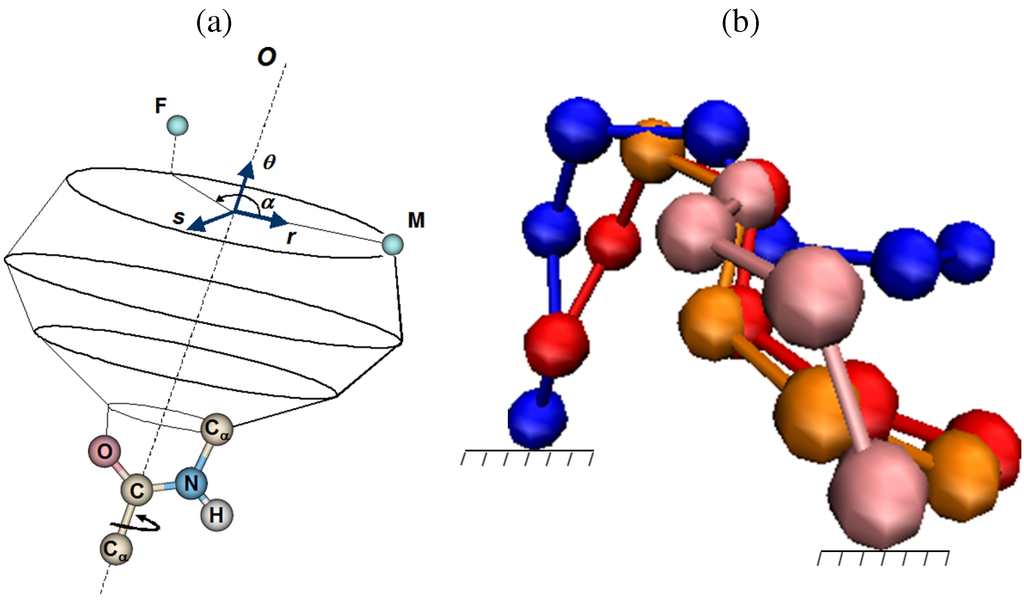
Figure 4.
(a) In this simple illustration of how CCD can be employed, the atom at the end of the chain in the current position M needs to reach its target position F. The dihedral bond between the and C atoms shown can be modified by the angle α in order to place the atom as close to F as possible. The angle α minimizes the distance between the position of the atom in the ensuing conformation of the chain and F, as it places the atom on the projection of F on the circle around the -C rotation axis. (b) The initial conformation of a chain of 7 bonds is shown in blue. In this illustration, CCD modifies each of the dihedral angles of the rotatable bonds one at a time so the end atom approaches the target position. The conformations in red, orange, and pink are the intermediate conformations resulting during the modification of the dihedral angles. The target position is reached in the pink conformation.
3.3. Remaining Challenges Related to IK Techniques
IK techniques can be incorporated in a search algorithm to compute different loop conformations that satisfy the given constraints. For instance, CCD has been employed to map sampled open loop conformations to closed conformations to explore the closure space of the loop [70]. Work in [70,71] shows that incorporating CCD in a probabilistic search algorithm allows obtaining an ensemble of closed conformations for loops of different lengths and modeling equilibrium loop flexibility [70,71].
An important consideration when employing IK techniques in search-based methods to sample the closure space of a loop is the coverage of this space. Analysis in [71] has shown that IK techniques, such as CCD, can recover all known exact solutions of 6R kinematic chains when employed to map random open conformations to closed conformations where the constraints are satisfied. A full understanding of the ability of optimization-based IK techniques to cover constrained conformational spaces remains elusive. Some efforts are made in [71] to understand the nature of the constrained conformational space into which CCD maps random open conformations that lie in a narrow neighborhood. The analysis shows that CCD maps neighbor conformations into distant regions in the constrained conformational space, potentially allowing to sample diverse geometrically-constrained loop conformations when applied to an ensemble of randomly sampled open loop conformations. A better understanding is needed of the potential of optimization-based IK techniques to allow obtaining a broad view of the constrained conformational space relevant for the equilibrium flexibility of a loop.
It is also worth noting that IK techniques only address the geometric aspect of the loop modeling problem. For instance, even if all 16 solutions are enumerated for a loop of 6 dihedral dofs, not all solutions are feasible. Many of them may contain steric clashes between atoms of the loop itself or between atoms of the loop and the rest of the protein structure. Even optimization-based IK techniques suffer from this shortcoming. Moreover, in addition to potential steric clashes, the local geometry of obtained loop conformations may not be protein-like. Obtained dihedral angles may lie outside of preferred values in protein native structures. Statistical analysis has revealed that the backbone dihedral angles in native structures do not populate the entire range but are limited to specific regions in amino-acid dependent (Ramachandran) maps [38]. Current IK techniques do not readily allow to limit sought dof values to specific ranges, such as those obtained from Ramachandran maps.
Due to their ability to model only the geometry of loop conformations in loop modeling, IK techniques are often employed only as one of the components of a multi-stage method. Post-processing steps are often taken to remove steric clashes and refine local geometries in loop conformations obtained from IK techniques. These steps often employ an energy minimization protocol with a given energy function. The purpose of the energy function is to remove unfavorable interactions (such as steric clashes) in the loop itself and between the loop and the rest of the protein structure (see Section 2). Additional components in the energy function may allow modeling other important effects of the environment on the loop under consideration. The environment can be solvent, membrane, or crystal [72,73].
4. Database Methods
Database methods were some of the earliest to model loops and obtain loop conformations that satisfy given geometric constraints [74,75,76,77,78,79,80,81,82]. These methods were first proposed in the context of electron density fitting [83]. They operate on the assumption that loops in protein structures deposited in protein structure databases provide natural examples to model unknown loops.
In essence, database methods search for loop conformations in databases of native protein structures, such as the Protein Data Bank (PDB) [13]. Suitable loop conformations are selected by how well they satisfy constraints on length (number of amino acids) and geometry (constraints on the mobile loop termini). Selected loop conformations that satisfy these constraints very well are of high quality and in naturally-occurring geometries. For instance, database methods have had great success in modeling antibody hyper-variable loops that form very specific folds based on few key amino acids [74,81,84,85].
Database methods have the advantage of producing loop conformations very fast, as the selection process involves few computations. However, the structural diversity of the database affects their general ability to find constraint-satisfying conformations of a loop of a given length. From an historical perspective, database methods have often struggled with the quality of obtained loop conformations. For instance, early work in [77] demonstrated that loop conformations selected from structure databases did not overlap satisfactorily with given termini in all instances. Additionally, large-scale studies in the late 1990s showed that database methods were severely limited by the lengths of the loops they could model and had practical limitations of 4 amino acids [86]. Later work in [87] demonstrated that loop conformations obtained from databases were good candidates to model loops up to 9 amino acids but with extensive restrained energy minimizations of extracted conformations.
Significant efforts have been made since then to address the main limitations of database methods. For instance, optimization protocols have been proposed to improve the overlap of the mobile anchor in a loop conformation selected from the database with the stationary anchor in the loop to be modeled [87,88]. Briefly, the energy function can be enhanced with pseudo-energy terms that penalize poor overlap. The measurement of overlap can take into account either only Euclidean distances among main-chain atoms of the mobile and stationary anchor or incorporate additional constraints on orientation. A simple energy minimization protocol can then ensure that improvement of overlap is part of the solutions that minimize the given energy function. Selection of candidate loop conformations from a database, subsequent optimization of these conformations, and then ranking according to lowest energy values achieved can result in physically-reasonable conformations for a loop under investigation [88].
The growing structural diversity of protein structure databases has recently allowed database methods to model loops of up to 15 amino acids [89], but the quality of conformations obtained for loops of more than 12 amino acids (in terms of root-mean-squred-deviation, RMSD, to the known native structure of the loop) is lower than that obtained with ab-initio methods (the summary below shows that some ab-initio methods can achieve impressing sub-angstrom RMSDs to the known loop native structure). Longer loops can be addressed if the process of selecting candidate conformations from a database is encapsulated in more powerful algorithmic frameworks. For instance, the divide-and-conquer approach in [90] provides a template on how to do so. The method in [90] constructs conformations of a given long loop by putting together configurations of shorter fragments. While in [90] these configurations are analytic solutions to geometric constraints, the process illustrates how one can address limitations of loop length in database methods. For instance, the fragment configurations can be excised from protein structures deposited in databases, and the assembly process can be encapsulated in a search framework that addresses both the geometric constraints of the loop and the energetic feasibility of the resulting conformation. Indeed, this process has recently been employed in [91], but the resulting method bears little resemblance with database methods.
The method in [91] provides a template on how databases can be employed to help a search-based algorithm sample physically-realistic loop conformations. Methods that implement such templates fall in the category of knowledge-based methods and can be employed to obtain not just one physically-realistic loop conformation but an entire ensemble of loop conformations and so better capture the structural variability of a loop. It is worth emphasizing that, while in database methods sampling is strictly limited to the discrete space encoded in the library of structures, piecing together loop conformations with shorter pieces obtained from these libraries allows sampling a larger space. There are currently no applications of knowledge-based methods to sample the constrained conformational space of a loop for the purpose of modeling the loop’s equilibrium flexibility. Most existing methods choose to focus instead on producing one accurate loop conformation in a given native protein structure.
5. Search-Based Methods
Many methods employ search algorithms to explore the constrained conformational space of a loop instead of relying on databases to readily obtain loop conformations. Ab-initio methods aim to compute closed loop conformations from physico-chemical principles. However, many of these methods make use of information extracted from structural databases either in their modeling of protein chains or in the employed energy function. While the boundary between ab-initio and knowledge-based methods is getting murkier, it is common practice to reserve the knowledge-based designation for methods that use database-extracted configurations of small fragments to assemble a physically-realistic open loop conformation or refine a closed, distorted loop conformation.
Search-based methods can use different representations of a protein chain, different energy functions to obtain physically-realistic conformations, and different search algorithms to obtain loop conformations. To ensure that loop conformations fit with a given protein structure, one of two approaches is usually followed: (i) open conformations that do not satisfy the constraints are first sampled for the loop at hand, and then geometry- or energy-based adjustments are carried out on open conformations to close them so they fit with the given protein structure; (ii) closed, possibly physically unrealistic, conformations are directly obtained, followed by geometry- or energy-based adjustments to correct local geometry and energetic interactions of the closed loop conformation with itself and the rest of the protein structure.
5.1. Generate-and-Close Methods
Generate-and-close methods sample open loop conformations with search algorithms commonly used in computational biology for protein conformational search. They include Molecular Dynamics (MD), Monte Carlo (MC), or other more powerful search methods that enhance the sampling capability of the classic MD and MC frameworks. Briefly, in MD simulations, atomic coordinates are updated to obtain new conformations by numerically solving Newton’s equations of motions (the reader is referred to [2,92] for detailed reviews on the topic). In contrast, MC employs a set of available moves to modify a conformation and obtain a new one, resulting in a biased probabilistic walk in conformational space that has often higher sampling capability than an MD trajectory (cf. to [93,94] for a detailed review on the topic).
Generate-and-close methods rely on energy- or geometry-based treatments to close open loop conformations. The following summary categorizes these methods according to these two subcategories.
5.1.1. Energy-Based Approaches
Energy-based approaches to closing sampled open loop conformations rely on the fact that the distance between a sampled conformation’s mobile anchor and the corresponding stationary anchor can be captured in a term that can be added to a physics-based energy function. The resulting pseudo-energy function penalizes both unfavorable atomic interactions (through its original terms) and long distances between the mobile and stationary anchor (through the newly added term). At the very least, the energy function can be employed to identify and discard sampled loop conformations that do not fit with the given protein structure, as done in very early work on loop modeling (described below). The lowest-energy conformation among those remaining can be offered as the one that closes the loop. Additionally, optimization protocols can be pursued. Through the minimization of the pseudo-energy function, these protocols close the computed open loop conformations and correct unfavorable interactions of the loop with itself and the rest of the protein structure.
Some of the earliest methods in this category employ systematic search to thoroughly explore the conformational space of short loops [86,88,95,96,97,98,99]. Essentially, an exhaustive combinatorial search is conducted by systematically rotating dihedral bonds in a loop fragment with discrete angle increments. Loop conformations that achieve lowest energies according to a designed pseudo-energy function that evaluates each loop conformation in the context of the given protein structure are deemed closed. It is worth noting that these methods were among the first to employ energy functions to either select closed conformations among those sampled or modify sampled conformations to close them.
The potential combinatorial explosion of conformations enumerated in systematic search is controlled in different ways. For instance, work in [88] focuses on short loops of up to 6 amino acids and biases dihedral angles by their distribution in known protein structures. The method in [99] samples from a discretized solution space by biasing the search toward more populated regions of the Ramachandran maps. Subsequent work in [100] employs finer-grained amino acid-specific state sets or angle pairs with as many as 722 states per amino acid. In contrast from methods that simplify the search space through discretizations, the method in [96] analytically solves for a selected short fragment in a given loop, while enumerating conformations for the remaining part of the loop. Computed loop conformations are then optimized through energetic minimization protocols like Metropolis MC Simulated Annealing [98] or high-temperature MD [101]. Systematic or exhaustive methods are limited by the lengths of loops they can consider, but their performance in the early 1990s was superior to database methods [86].
Other methods address length limitations by employing more powerful conformational search algorithms. Over the years, the list of these algorithms has grown very diverse. Just to mention a few, conformational search algorithms employed in energy-based approaches include importance sampling with local minimization of randomly generated conformations [102,103,104], global energy minimization by mapping a trajectory of local minima [105,106], MD simulations [101,107,108,109], genetic algorithms [65,110,111], dynamic programming in a discretized space [112,113], biased probability MC searches [12,114,115], MC combined with MD [116], MC Simulated Annealing [117,118,119,120,121], multi-copy searches [122,123,124], extended scaled collective variable MC [125], self-consistent mean field optimization [126], or enumeration based on graph theory [127].
The method in [46] follows a slightly different approach in sampling loop conformations and is worth describing in some detail. Instead of modifying angle values from the N- to the C-termini of the loop, the loop is divided into two equal fragments that duplicate the middle amino acid of the loop (referred to as the closure amino acid). Each of the fragments is sampled independently, modifying angles starting from the loop stem to the middle. Obtained fragment conformations where the carboxyl carbon atoms of the end amino acids are within Å of each other are retained for further structural and energetic refinement. The position of this atom that is essentially duplicated in the two fragments is averaged over the values provided by each fragment in order to obtain a connected loop conformation. An extensive optimization protocol is then conducted on a resulting loop conformation.
The method in [46] represents one of the most successful methods that employ an energy-based approach. For instance, average prediction accuracies of and Å in backbone RMSD from the crystal structure are reported for loops of 8 and 11 amino acids, respectively. A more powerful sampling algorithm and a more accurate energy function with a novel hydrophobic term in the employed SGB solvation model have further improved the performance of the method on loops longer than 10 amino acids [72]. Median backbone RMSDs of , , and Å, are reported between native loop conformations and lowest-RMSD conformations on loops of length 11, 12, and 13 amino acids, respectively. Later extensions of the method in [73] improve modeling of loops in inexact environments, such as those in incomplete native protein structures obtained from comparative modeling techniques. In these structures, side chains surrounding the loop also move in order to accommodate a closed loop conformation. The method in [46] and its extensions and improvements in [72,73] are available as part of the Protein Local Optimization Program (PLOP) [46].
While most methods in loop modeling focus on potential energies, the suite of methods in [46,72,73] are unique in their addition of a novel term to the potential energy function to approximate introducing the effects of entropy without resorting to free energy sampling. The new term relies on clustering of the sampled loop conformations and effectively lowers the overall energy of loop conformations that are close (in terms of low RMSD) to other conformations. The resulting overall energy, referred to as “colony energy”, effectively tends to promote conformations that are located in broad energy basins and is successful in the ab-initio prediction of native loop conformations. Employment of the colony energy has been shown to significantly improve the ability to predict the native loop conformation within of the actual native loop structure without prior knowledge of this structure for loops up to 8 amino acids long [48].
The employment of a term that mimics entropy in the colony energy showcases the importance of ab-initio prediction; that is, predicting the native loop conformation(s) from among the ones generated through energetic criteria. While most loop modeling methods illustrate their accuracy by showing that they can generate conformations with low RMSD to the known native loop conformation, they cannot guarantee that the low-RMSD conformations will also be among the ones with lowest energies. Phrased otherwise, these methods cannot guarantee that conformations with low potential energy will also have low RMSD from the known native conformation. This limitation is a consequence of employing potential rather than free energy, as laid out in our short protein primer in Section 2. In tandem with efforts to incorporate an approximation of entropy, other efforts in loop modeling focus on employing more detailed/fine-grained and expensive energy functions on a few loop conformations that are of similar low energy according to a coarse-grained energy function employed during the generation procedure [70]. The hope is that the fine-grained energy function will allow better discriminating against non-native loop conformations. It is worth noting that the employment of energetic criteria to select native loop conformations is not limited to energy-based approaches. While the geometry-based approaches summarized below focus on treating the geometry aspects of loop modeling in order to efficiently generate closed loop conformations, energy is an important component of the loop modeling problem in order to select native loop conformations among those generated.
5.1.2. Geometry-Based Approaches
Geometry-based methods do not rely on an energy function and optimization to obtain closed loop conformations. These methods can handle longer loops, achieve higher success rates in loop closure, and do so in less time than energy-based methods. While energy-based methods like the one in [46] take hours to days to accurately model a loop of 8 amino acids, the Loopy method in [48], which employs random tweak to close sampled open conformations, can model loops of similar lengths in minutes on a MHz processor. The method in [66], which applies CCD to close open loop conformations sampled uniformly at random, is similarly efficient. A comparative analysis on a single-dual 1.4 GHz Xeon processor in [128], which measures the time required for the generation of closed loop conformations free of steric clashes, shows that CCD requires minutes for loops of 8 amino acids. The time demands jump to minutes for loops of 12 amino acids. The actual time demands of CCD are expected to be seven times less than those reported in the comparison study in [128] (the closure of a conformation in [66] is obtained on average seven times faster than in the CCD implementation in [128]).
Geometry-based methods also employ energy functions but do so only to improve the physical relevance of computed closed conformations. Often, the energy function is used primarily to identify closed loop conformations with severe steric clashes in the loop itself and between the loop and the rest of the given protein structure. While optimization protocols may also be employed to improve the accuracy of the obtained closed loop conformations, the focus in most geometry-based methods is on improving time demands rather than in reproducing the native loop conformation with high fidelity. Nonetheless, due to the ability of geometry-based methods to compute many different closed loop conformations in a reasonable amount of time, high-quality loop conformations are often reported by these methods. For instance, the method in [66] achieves minimum backbone RMSDs of , , and Å for loops of length 8, 11, and 12 amino acids, respectively.
Many geometry-based methods are contributed from robotics researchers due to the similarity of the loop modeling problem to the problem of controlling motions of a robotic manipulator. Early robotics-inspired approaches like the one proposed in [129] sample chain conformations ignoring the constraints, while later address the constraints with gradient descent. Other methods subject open loop conformations to attractive mechanical forces formulated to pull the mobile anchor to its target pose in the stationary anchor [33]. Yet other methods incorporate IK techniques in a probabilistic sampling framework to close open loop conformations [32,68,69,70,130,131,132,133,134,135]. In [130], for instance, the loop is broken into an active part, for which open conformations are generated disregarding the constraints, and a passive part of exactly three amino acids that is closed through exact IK methods [130]. An efficient extension of the above method for longer chains is later provided in [131]. Sampling the active part of the chain one dof at a time ensuring that the active part’s endpoints are always reachable by the passive part has been pursued as a natural extension of this line of work in [132]. The resulting Random Loop Generator (RLG) algorithm has been embedded in sampling-based path planning methods to efficiently obtain closed conformations for long protein loops [134].
The group of methods that rely on the active and passive designation of fragments in a given loop have a very high failure ratio as the loop grows in length [130,131,132]. Conformations of long loops that are free of steric clashes are typically found in very narrow regions of the closure space. Therefore, naive methods that do not consider steric constraints in the process but keep sampling closed loop conformations until they find clash-free ones have a very low success rate. The method proposed in [136] aims to address this problem through a two stage exploration. In stage 1, closed loop conformations are obtained with a seed sampling technique that samples broadly from the closure space. The chain of a loop is divided into three fragments (beginning, middle, and end), as one follows the N- to C-termini of the loop. The middle fragment is chosen to be at most half the loop’s length in terms of number of amino acids. Values for the dihedral angles in the front and end fragments are sampled while avoiding steric clashes. Values for the dihedral angles in the middle fragment are then obtained through the IK technique proposed in [58].
Conformations obtained in stage 1 of the method in [136] are employed as seeds and refined in stage 2 through a deformation sampling technique. The technique explores the conformational space around a seed conformation at a finer-grained level of detail by modifying the dofs on a seed conformation without breaking closure. Motions in the self-motion manifold are employed to move towards a local minimum of the energy function while keeping closure (this approach was originally employed in [68,69] to obtain a closed loop conformation that also fit with the electron density map of the protein crystal structure). Applications on loops of length up to 25 amino acids show that the method is able to efficiently obtain diverse closed conformations of long loops. The average reported time to obtain one closed clash-free conformation on a 3 GHZ Intel Pentium processor with 1 GB RAM is seconds for a loop of 25 amino acids. This is impressive compared to more than 800 seconds that would be needed by a naive approach that keeps sampling closed conformations until it finds a clash-free one. The method is available in the LoopTK toolkit [69].
Geometry-based methods are actually capable of modeling loops with very high accuracy due to their extensive conformational sampling of the closure space. The method in [91] employs a knowledge-based approach to obtain physically-realistic open loop conformations. Open loop conformations are assembled with configurations of short fragments extracted from protein structures, a process known as fragment-based assembly that is very popular in protein structure prediction [20]. The fragment configurations are employed as moves in an MC framework. Open conformations obtained this way are subjected to the IK technique proposed in [58] to close and fit them with the given protein structure. The method is capable of recovering the known native loop conformation in loops of lengths 4 to 12 amino acids. The lowest-RMSD conformations obtained by this method for loops of length 4 to 12 amino acids range from to Å in backbone RMSD from the corresponding known native conformation, respectively. The method is available through the FALC-Loop webserver [137].
In an impressive result in loop modeling, the method in [138] reports a sub-angstrom backbone accuracy in reconstruction of 25 different loops of 12 amino acids in protein structures obtained with the Rosetta structure prediction package [20]. The approach in [138], which builds over the IK technique proposed in [58], is to obtain all kinematically accessible conformations for 6 not necessarily consecutive dihedral angles of the loop, while simultaneously sampling the remaining dihedral angles using polynomial resultants [139].
A further extension of the work in [140] incorporates the loop closure algorithm in [58] within an MC framework to obtain an ensemble of low-energy loop conformations. Qualitative comparison with NMR data show the method is promising for additionally modeling loop flexibility.
5.2. Close-and-Relax Methods
In close-and-relax methods, closed loop conformations are obtained directly, at the expense of correct local geometries and energetic interactions of the loop. The bond scaling with relaxation method in [141,142] was the first to construct a closed loop conformation by directly placing both mobile anchors of the loop on the stationary anchors (essentially copying the stationary anchors’ coordinates). The size of a loop conformation obtained from a protein structure database is scaled in order to fit the anchors. The conformation is later returned to ideal bond lengths through an energy minimization or a short MD simulation. The method in [47] follows a similar approach, but the loop conformation is not obtained from a database. Instead, after placing both mobile anchors of the loop on the stationary anchors, the rest of the loop’s main-chain atoms are positioned with uniform spacing on the line connecting the backbone N and C atoms of the loop’s mobile anchors.
In [47], a set of different closed loop conformations are obtained by adding to the coordinates of all the loop’s atoms’ (excluding the stems) a number distributed uniformly at random in Å. An optimization protocol employing a specially-designed energy function is then applied to each closed conformation to improve local geometry and energetically refine each conformation. The protocol consists of a short conjugate gradient minimization, followed by a short MD simulation with simulated annealing, and concluded with another short conjugate gradient minimization. The method in [47] can obtain conformations that reproduce the native loop conformation with high fidelity in terms of main-chain RMSD; of loops of 8 amino acids are predicted within Å of the native structure. Some of the best cases for loops of 4 and 12 amino acids reach the native structure within and Å, respectively. The method is available as the ModLoop web server [47].
Recent work in [143] employs structural information extracted from protein structure databases in order to improve the quality of closed conformations. The terminal atoms of the loop are placed in their target positions. Instead of placing the atoms of the loop in a straight line, the self-organizing method in [143] places the remaining atoms of the loop in random positions in the vicinity of the terminal atoms. An iterative procedure then refines the positions of these remaining atoms with information derived from ideal structural templates obtained for the fragments from a precomputed library. The resulting method reproduces native structures of loops of 4, 8, and 12 amino acids with backbone RMSDs no higher than , , and Å, respectively. Extensive analysis in [143] shows that the method is both more accurate and more efficient than CCD.
5.3. Accuracy and Time Demands
The above summary highlights that loop modeling methods are quite diverse and are characterized by different results in terms of accuracy and time demands. For instance, while database methods are fast and can obtain physically-relevant loop conformations, they are often limited to applications on short loops. More powerful methods are often needed to handle longer loops. These methods either implement search algorithms over a core process that still makes use of a structure database, like the divide-and-conquer method in [90], or attempt to reconstruct closed loop conformations from first principles, ab initio. Due to their reliance on an energy function and an optimization protocol to search the loop closure space, ab-initio energy-based methods are less efficient and have practical limitations of loop length and success rates. In contrast, geometry-based methods that address primarily the geometric constraints posed by the closure of the loop can compute closed loop conformations more efficiently. These methods still have to rely on an energy function and an optimization protocol to incorporate energetic considerations and improve the relevance of computed loop conformations.
Whether relying on the generate-and-close or the close-and-relax paradigm, steering the loop’s ends, its middle amino acid, or designating active and passive fragments in the loop chain, current search-based methods are able to model loops as long as 12 amino acids within at most 3 Å of the known loop native structure. Impressive cases exist when these methods report closed conformations that reproduce the known native loop structure with less than 1 Å backbone RMSD for loops of 12 amino acids and more [73,138]. Indicatively, as far as running times are concerned, comparative studies in a dual-core 2 GHZ Intel processor with 1.96 GB 667 MHz DRAM report that search-based methods that employ CCD to close open loop conformations are quite efficient, obtaining a closed conformation of a loop of 12 amino acids in about seconds. The self-organizing method in [143] reports further savings over employing CCD, but does not improve time demands over the IK-based method in [58]. Work in [136] shows that the consideration of steric constraints increases these time demands to seconds to obtain a closed conformation that is also free of steric clashes for a loop of 25 amino acids.
6. Highlights of Selected Representative Methods
Four methods are chosen to highlight current progress in loop modeling in greater detail. The first method is based on evolutionary search and is selected here due to its unique approach and employment of energy functions for improving accuracy in loop modeling. The second method represents the state of the art in close-and-relax methods. The third and fourth methods are representative of geometry-based generate-and-relax methods that incorporate IK techniques in probabilistic sampling frameworks. The fourth one, in particular, has been applied to model equilibrium loop flexibility.
6.1. Pareto Optimal Sampling Method
The method in [144] addresses the fact that energy functions for loop modeling, whether coarse- or fine-grained, suffer from inaccuracies or insensitivities, as demonstrated by many loop modeling studies [46,47,48,49]. A Pareto Optimal Sampling (POS) method is proposed in [144] to address this issue and improve accuracy in loop modeling. The essential idea is to sample an ensemble of diverse loop conformations that belong to the Pareto optimal solution set. This set contains conformations u that are not dominated by others; that is, no other conformation v has lower energy than a conformation u in the set according to many different energy functions. Three different popular energy functions, Rosetta [20], DFIRE [145], and Triplet [146] are selected for this purpose.
A novel population-based sampling algorithm is proposed to cover the Pareto optimal front with diverse conformations. The algorithm is inspired from evolutionary search and evolves populations of conformations toward the Pareto optimal front. Only the top-ranked conformations of a population (according to a designed fitness function) are allowed to “breed” and reproduce conformations for a new population. The first population consists of N random open loop conformations (the backbone dihedral angles are the only dofs). Conformations of a new population are obtained through differential evolution (DE) crossover [147]. The Metropolis criterion determines whether a newly-generated conformation is accepted or not in the population.
The fitness function is based on the strength of each non-dominated conformation C, which is defined as the proportion of conformations in the population that are dominated by C. Specific schemes are designed to maintain diversity while seeking high fitness scores. The DE mechanism, illustrated in Figure 5a, combines fragments of conformations selected at random over a population to obtain a new loop conformation. CCD is employed to restore closure for the new loop conformation. Given that conformations are modeled at a backbone level of detail, selected conformations are later refined in atomistic detail (details in [144]).
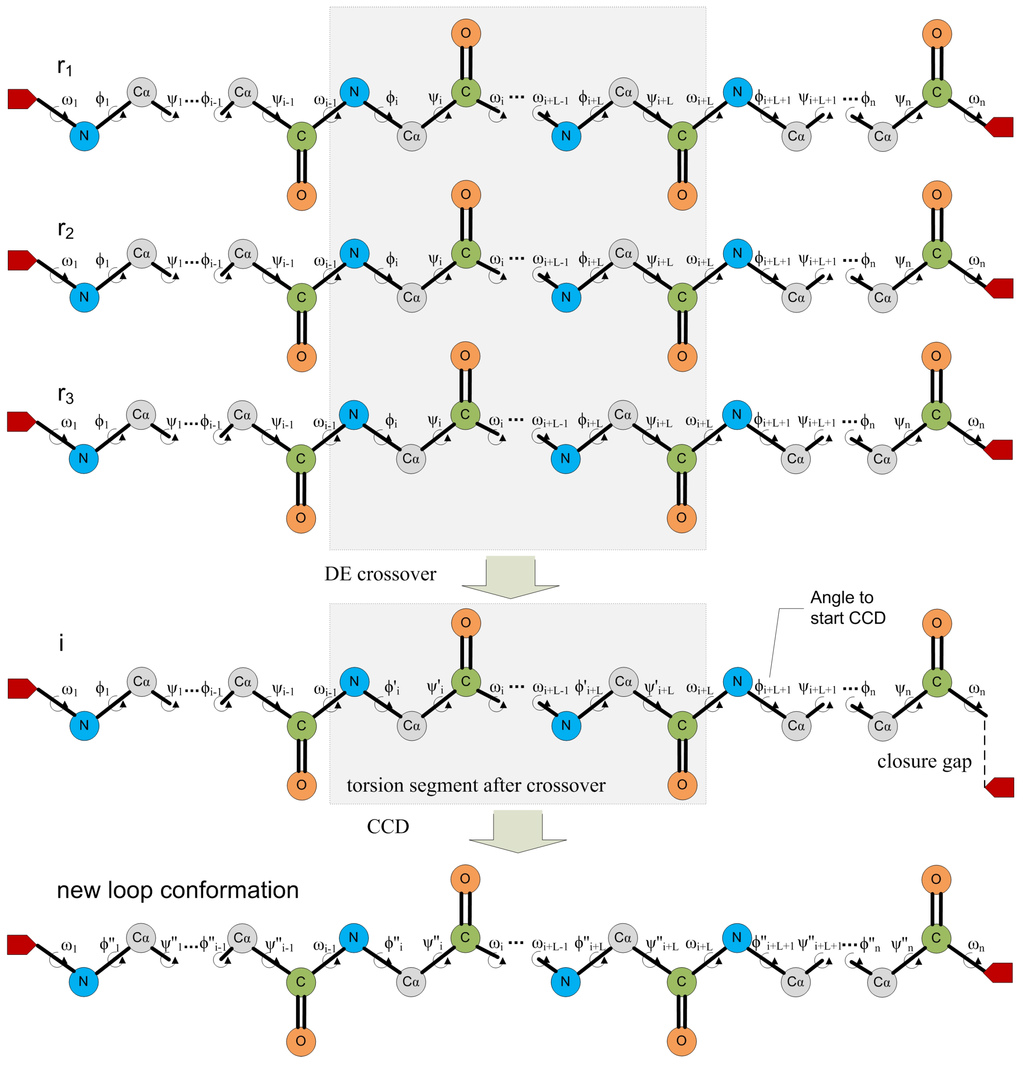
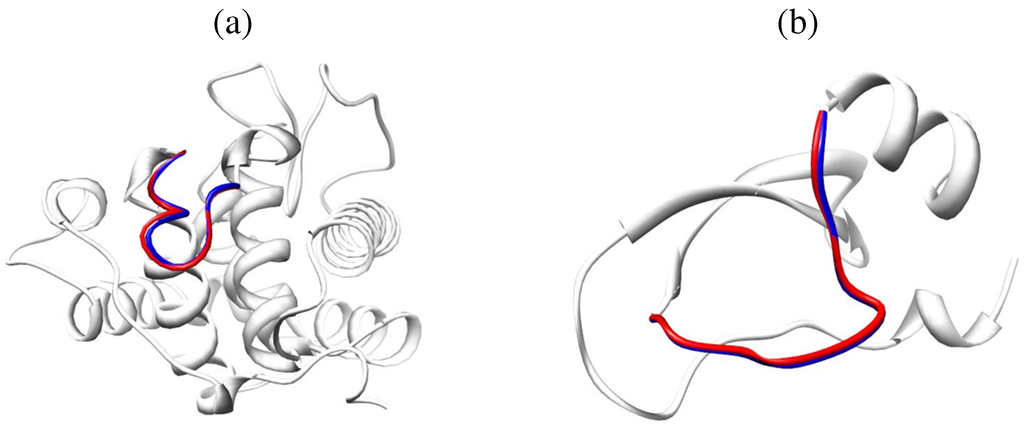
Figure 5.
Top: Schematic illustration of the process employed in POS to generate closed loop conformations in a population. Fragments of conformations selected at random from a population are combined to obtain a new open loop conformations. CCD is employed to close the new loop conformation. Reprinted with permission from [144]. Copyright 2011 American Chemical Society. Bottom: Predicted loop conformations in red are superimposed over the native loop structures in blue. The rest of the protein structures are drawn in white. The shown protein structures are for the goose lysozome protein in (a), PDB ID 153l, and the bovine pancreatic trypsin inhibitor in (b), PDB ID 5 pti. The selected loops span amino acids 36–47 in (a) and 98–109 in (b). The RMSD between the predicted and computed loop conformations are and in (a) and (b), respectively. Contributed by Yaohang Li.
Application of POS on the Jacobson decoy set [46] reveals interesting results. On short loops of 4–6 amino acids, around of the POS top-ranked conformations approach the native structure of the loop within 1 Å backbone RMSD. On medium loops of 7–9 amino acids, around of POS top-ranked conformations lie within 1 Å of the native structure. This result changes to for long loops of 10–12 amino acids. Figure 5a,b show cases two of the best results obtained by POS for loops of 12 amino acids, where the backbone RMSDs from the known native structures of the loops are below Å.
6.2. Self-Organizing Method
The self-organizing method proposed in [143] is one of the most recent close-and-relax approaches to loop modeling. The method aims to simultaneously address the satisfaction of geometric, steric, planarity, and chirality constraints. The closure constraint is trivially satisfied by placing the loop’s mobile anchors over the stationary anchors. The method then proceeds to determine physically-relevant coordinates for the remaining atoms of the loop.
The initial coordinates of the loop’s atoms (excluding the stems) are sampled uniformly at random in the vicinity of the stationary anchors. The method then modifies these coordinates in iterations, until certain distance constraints resulting from the loop’s covalent structure, planarity, and chirality constraints are satisfied. Each iteration consists of pairwise distance adjustments and superimposition of structural templates to gradually refine and obtain a physically-relevant closed loop conformation.
What makes the method unique and extremely efficient is its employment of template structures for short rigid fragments of the loop chain. The method consists of two main stages, initialization and embedding. In initialization, the loop is decomposed into rigid fragments. Peptide bonds and bonds on side-chain rings are considered rigid. The remaining rotatable bonds define excision points to decompose a chain into rigid fragments. The ideal conformational template for each fragment is extracted from a library of pre-computed templates (obtained, for instance, from the PDB). Upper and lower interatomic distance bounds are then constructed. Lower bounds follow from standard covalent geometry for bonded atoms. The sum of van der Waals radii defines the lower bound for nonbonded atoms. Upper bounds are set to the sum of bond lengths along the shortest path connecting two atoms and obtained through the Floyd-Warshall algorithm.
In the second embedding stage, a series of pairwise distance adjustments and template fittings are performed in order to obtain loop conformations consistent with the distance bounds. Each iteration adjusts each defined fragment for a set number of cycles. In each cycle, two random atoms are selected from the fragment for adjustment. The atoms’ coordinates are adjusted so that their distance lies between the lower and upper distance bounds. Afterwards, the template for the selected fragment is superimposed over the configuration of the fragment. The coordinates of all atoms in the fragment are replaced with those of the superimposed template (see Figure 6a for an illustration). This process of iteratively adjusting atomic coordinates and fitting templates results in a closed loop conformation that is free of steric clashes and satisfies planarity and chirality constraints.
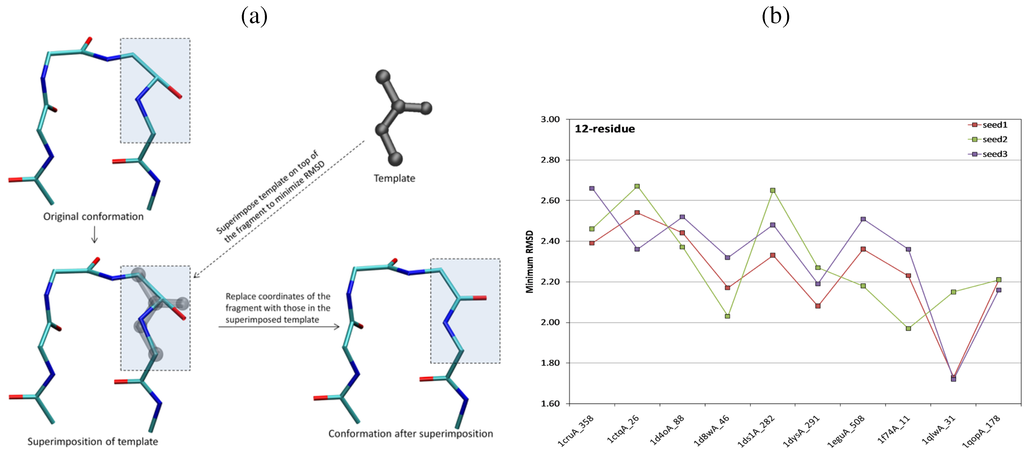
Figure 6.
(a) Illustration of the superimposition operation for a randomly selected amide fragment (highlighted with the rectangular box) in a loop of 4 amino acids. Coordinates of the fragment are replaced with those of the superimposed template (drawn in grey). This ensures correct geometry for the fragment (bond lengths, angles, and planarity). (b) Plot shows the lowest achieved RMSD to the known native loop structure for loops of 12 amino acids. The three different lines, drawn in red, blue, and green, show the values obtained from three independent runs of the method. (a),(b) are reprinted with permission from [143].
The method in [143] is numerically stable and computationally efficient. More importantly, additional distance constraints, such as those obtained from NMR, can be directly incorporated into the method. Comparisons of the method with the CCD algorithm and the IK technique in [58] show that the method achieves similar or better prediction accuracies on diverse loops of 4, 8, and 12 amino acids. Native structures of loops of 4, 8, and 12 amino acids are reproduced with backbone RMSDs no higher than , , and Å, respectively, as shown in Figure 6b. Extensive analysis shows that the method does not improve over the IK technique in [58] but is more accurate and efficient than CCD.
6.3. A Robotics-Inspired Method for Sampling the Loop Closure Space
The method proposed in [32] is representative of robotics-inspired approaches originally developed for the exploration of robot configurational spaces that have been adapted to study protein conformations. The method in [32], based on similar previous work by the authors [134], employs a tree-based search to efficiently explore the closure space of a loop and model its flexibility. The RLG algorithm is embedded in the search in order to efficiently obtain closed loop conformations.
The tree-based search proposed in [32] is an adaptation of a path planning algorithm employed in the robot motion planning community to find feasible paths that take a robot from an initial to a goal state while satisfying specified constraints. The algorithm, the rapidly-exploring random tree (RRT) [148], is a tree-based variant of the Probabilistic RoadMap (PRM) framework [149]. It is worth noting that the introduction of the PRM method in robotics enabled the efficient exploration of constrained high-dimensional search spaces. Essentially, a computed connectivity graph (the roadmap) encodes feasible paths in the space. Vertices in the roadmap are randomly sampled configurations that satisfy constraints. Edges are feasible short paths that connect neighbor configurations. When adapted for protein conformations, the probabilistic exploration in PRM offers an advantage over combinatorial methods [99,105], as it allows efficiently sampling vast and complex conformational spaces of arbitrarily long protein chains [27,28,30].
Inspired by the success of RRT in exploring complex and vast conformational spaces, work in [32] adapts RRT to explore the closure space of a loop and sample an ensemble of closed loop conformations. Figure 7a illustrates the main idea in RRT. A tree is grown in conformational space, rooted at an initial conformation . The tree grows in iterations. At each iteration, the tree is expanded or pulled towards a randomly sampled conformation . The nearest node in the tree to is then determined. The actual conformation added to the tree, is the feasible conformation in the subpath connecting and . In [32], the subpath is a linear interpolation over the dihedral angles of the loop.
The RRT in [32] conducts its search over the closure space of the loop. The IK-based technique proposed in [132], the RLG algorithm, is employed to obtain the conformation. Briefly, RLG designates two active and one passive fragment in the loop chain. The passive fragment is limited to 6 consecutive dihedral angles so that exact IK techniques can be employed to obtain closed conformations for it. Forward kinematics is employed to sample conformations for the active fragments. Biased conformational sampling for the active fragments increases the probability of finding a feasible conformation for the passive fragment that closes the loop. The conformation of the active fragments are sampled one angle at a time to ensure that the endpoints are always reachable by the passive fragment, as illustrated in Figure 7b. Other details in [32] concern expediting the detection of steric clashes and conducting short energy minimizations on sampled conformations with few steric clashes. The actual growth of the RRT tree from to is achieved through a simple interpolation between the two conformations. The conformation is the closest to that is also energetically feasible.
Figure 7c shows a few distinct conformations obtained by the method in a few minutes for the “thumb”-loop in the glycoside hydrolase family 11 xylanase from Thermobacillus xylanilyticus (Tx-xyl) [150]. These conformations showcase the mobility of this loop and compare very well with known characterizations of the loop’s flexibility by detailed biophysical studies [151]. Specifically, the tip of the loop is able to move more than 10 Å in backbone RMSD in the direction towards the catalytic cleft from its conformation in the crystallographic structure of the enzyme.
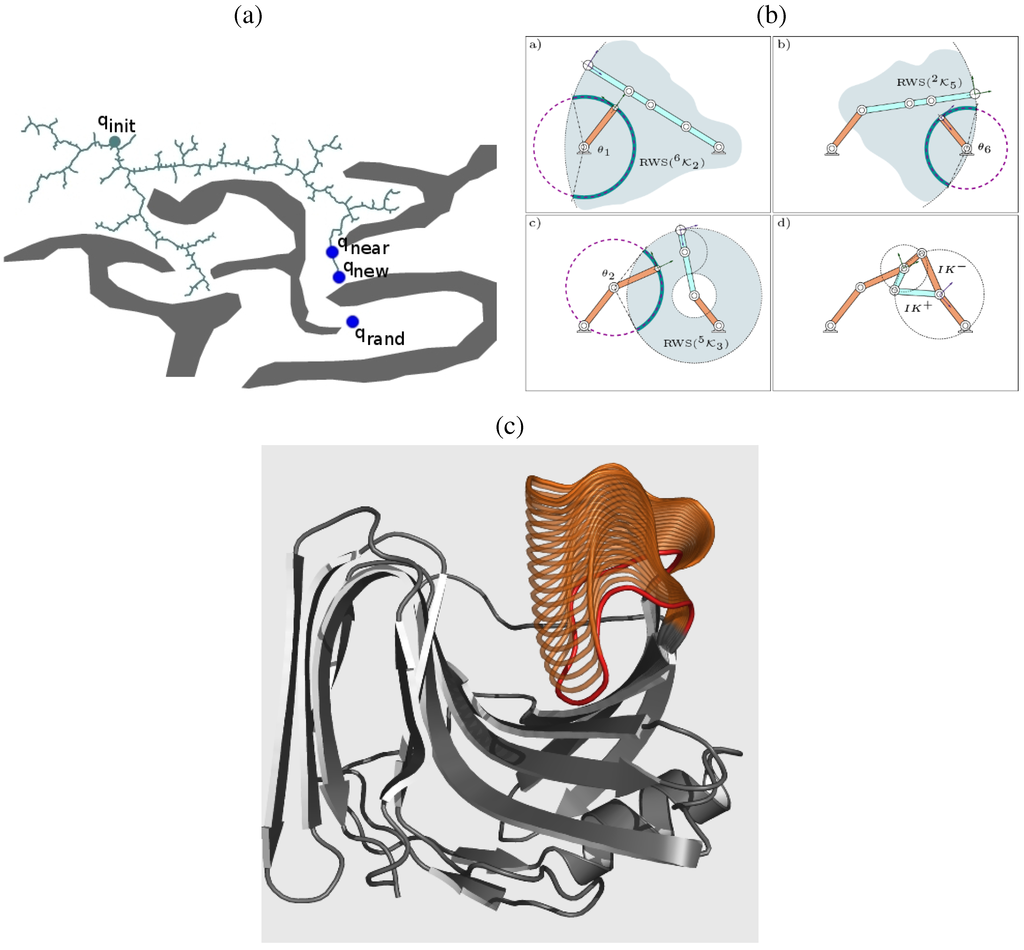
Figure 7.
(a) Figure illustrates the general RRT search algorithm for a point robot in a 2D workspace. Contributed by Erion Plaku. (b) Figure illustrates the RLG algorithm. The algorithm is based on the decomposition of the loop into several fragments. The algorithm performs a biased sampling of the conformation of the left and right fragments, which increases the probability of finding a feasible closed conformation for the middle fragment through exact IK techniques. (c) Figure shows of the “thumb”-loop in the glycoside hydrolase family 11 xylanase from Thermobacillus xylanilyticus computed by embedding RLG in the RRT algorithm. Only a few minutes on a Sun Blade 100 Workstation with a 500-MHz UltraSPARC-IIe processor are required to compute the loop conformations shown here. Contributed by Juan Cortes.
6.4. The Fragment Ensemble Method
The Fragment Ensemble Method (FEM) proposed in [70] addresses equilibrium mobility in the loop modeling problem. FEM generates an ensemble of low-energy loop conformations that complete the given protein structure. The method combines a statistical mechanics formulation with an efficient exploration of conformational space, exploiting analogies between protein chains and robot kinematic chains [26,27]. In particular, a loop fragment is modeled as an open kinematic chain.
FEM proceeds in stages, first sampling open loop conformations, then closing these conformations, and finally structurally and energetically refining them in the context of the rest of the protein structure. The process is illustrated in Figure 8. The open conformations are sampled uniformly at random in the space for a chain n rotatable dihedral bonds. Each open conformation is closed with CCD. The loop is modeled at a backbone level of detail until many closed conformations are obtained. All-atom detail is added onto each closed conformation by adding missing side chains. The resulting conformations are energetically refined in order to improve atomic interactions both within the loop and between the loop and the rest of the protein structure. In order to place most of the burden on the loop, a pseudo-energy function combines physics-based terms with a dampening term that limits the extent of atomic motions outside the loop. The optimization protocol combines conjugate gradient minimization in Cartesian space with small angular movements in the self motion manifold in order to deform closed loop conformations for the purpose of improving energetic interactions without breaking closure [70].
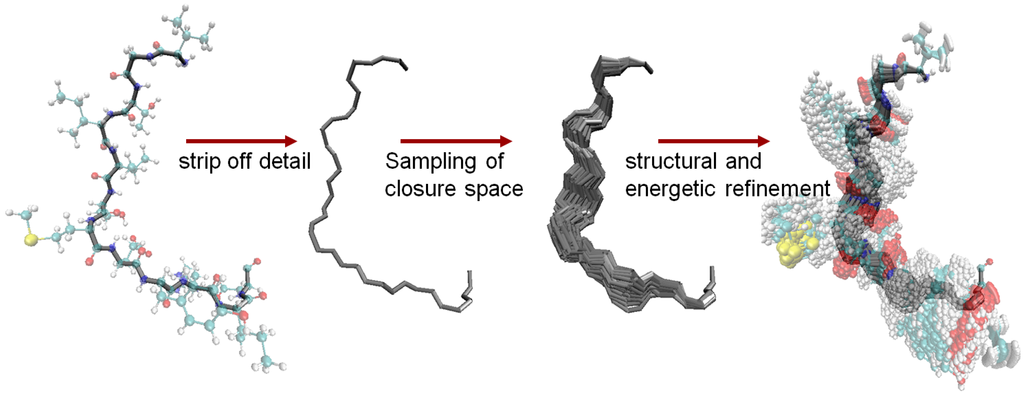
Figure 8.
Top: Schematic illustration of FEM. Sampled open loop conformations are subjected to CCD. The resulting closed conformations are only at a backbone level of detail. Structural (missing side chains) and energetic detail (in the form of a short energy optimization protocol) is then added to these conformations to obtain an ensemble of low-energy all-atom closed loop conformations. Bottom: Conformational ensembles are shown for three loops of 12 (in cytochrome inhibitor 2), 30 (in α-Lactalbumin), and 20 amino acids (in the variable surface antigen) in (a), (b), and (c), respectively. The known native structures of the loop are drawn in opaque, whereas computed conformations are drawn in transparent. The structure drawn for the loop in (c) is the one of lowest energy among computed conformations, as this loop is missing in the crystal structure of the protein. Figures on comparisons with experimental and simulation data, originally appearing in [70], are reprinted with permission of John Wiley & Sons, Inc. Copyright 2011 John Wiley & Sons, Inc.
The employment of CCD in the context of probabilistic sampling of open loop conformations allows FEM to explore the space of arbitrarily long loops. Loops modeled in [70] range from 12 to 30 amino acids. In particular, FEM is applied to characterize loop structure and mobility both in strongly stable and completely disordered loops (selected applications are shown in Figure 8a,b). A statistical mechanics formulation of the obtained loop conformational ensemble in [70] is employed as a natural way to associate a weight (Boltzmann factor) to each computed loop conformation and so obtain an equilibrium conformational ensemble. This is an obvious advantage over other geometry-based ab-initio methods applied to proteins [68,69,134]. In particular, the weighted ensemble facilitates measuring thermodynamic data as weighted averages over fluctuations in the ensemble and so directly comparing with published experimental and simulation data. Fluctuations measured over generated loop conformational ensembles agree well with published experimental and simulation data [70,71]. Figure 8 compares computed fluctuations to experimental ones derived from B-factors for the loop in cytochrome inhibitor 2, fluctuations obtained by simulation studies [152] for the loop in α-Lactalbumin, and fluctuations predicted from sequence data [153] for the missing loop in the variable surface antigen.
The agreement with experimental data that measure equilibrium fluctuations in [70] and the simulation-based analysis in [71] provide empirical evidence that FEM is not severely limited in its sampling capability. The employment of optimization-based IK techniques, however, complicates theoretical analysis into coverage of the closure space. Further application of FEM to model equilibrium fluctuations of consecutive overlapping fragments (not just loops) in protein chains shows that the approach is capable of reproducing equilibrium local fluctuations of protein chains [154]. This is perhaps further evidence that the basic framework in FEM may be promising to generally model equilibrium mobility in loops.
7. Conclusions
The loop modeling literature is rich and growing steadily. In light of this, it is not possible to have a complete review of work in loop modeling. Instead, this paper focuses on methods that implement geometry- and energy-based approaches to model loops and their equilibrium flexibilities. Other methods that employ different approaches are also being applied to loop modeling. For instance, the method in [155], which employs HMMs, is a recent example of machine learning methods. Additionally, a whole suite of methods exist that employ rigidity analysis of a protein’s native structure to identify flexible regions and employ them for modeling local fluctuations around the given structure [16,17,18,156,157,158]. While these methods have not been employed to model the equilibrium flexibility of loops in protein structures, they can be promising in narrowing down the relevant degrees of freedom to specific flexible regions in the given loop.
The various examples listed in this review illustrate that modeling equilibrium loop mobility in proteins is important for understanding biological function. The methods described in [32,70,136,140], also referred to as sampling-based methods, present promising first steps in this direction. Moreover, the employment of IK techniques to address kinematic constraints in sampling-based methods is allowing new applications on modeling the equilibrium flexibility not only of specific protein fragments, such as loops, but of entire protein chains [71,154,159]. IK techniques are efficient and allow sampling-based methods to spend computational resources on sampling a large number of closed conformations. Further work is needed, however, to understand the ability of sampling-based methods in obtaining a representative view of the closure space populated by the loop at equilibrium.
Currently, sampling-based methods cannot guarantee that they model all relevant regions of the closure space. Often, the validation relies on comparing various aspects of the obtained conformational ensembles with other experimental or simulation data on the loops at hand. An accurate characterization of loop equilibrium mobility may rely on obtaining a broad view of the constrained conformational space of the loop. This requires methods that can enhance sampling of the loop’s closure space. Further progress is needed in expediting the process of obtaining closed physically-reasonable loop conformations without relying on expensive optimization protocols. This will allow devoting available computational resources to the exploration component of search-based methods rather than the optimization of specific conformations to render them physically reasonable.
Another open problem in loop modeling is how to directly incorporate constraints other than those posed by the stationary anchor in loop modeling methods. The rigidity analysis methods listed above promise to allow incorporation of additional constraints, but they have not been employed in loop modeling so far. Incorporating additional constraints is a great area of interest, as it may not only to expedite, for instance, the generation of closed conformations that are also free of steric clashes, but also possibly directly realize observations made in the wet laboratory on closed loop conformations. The recent method in [143] illustrates that the incorporation of additional distance or angle constraints is possibly less challenging in the close-and-relax approach to loop modeling.
Most of the current work in loop modeling limits the conformational search to the loop at hand, ignoring potential loop-induced motions in the rest of the protein structure to accommodate a loop conformation. Characterization of dihydrofolate reductase actually shows that small fluctuations are observed in the rest of the protein structure in concert with the motions in the M20 loop [15]. Additionally, when loops are in active sites of proteins and interact with ligands or other proteins, their motions may often be induced by the presence of the partner molecule. Differences in loop length and conformation in a family of related proteins often correlate with the specificity of ligand binding. Ligands may induce conformational changes in the loops with which they interact. Such scenarios are not uncommon, but it remains unclear how to address them in an efficient manner.
With the exception of some work in loop modeling that considers environments, such as a crystal or side chains surrounding the loop, most methods do not model motions outside the loop that may change the probability of occurrence and so the physiological relevance of a particular computed loop conformation. In cases when loops interact with ligands, ideally, modeling of the loop and the ligand should be conducted simultaneously. This, however, increases the dimensionality of the search space. One way to address this limitation without the actual presence of the ligand is to obtain a broader, ensemble view of the different conformations that the chosen loop can assume under native conditions. The presence of the ligand can then be employed to discriminate against loop conformations that do not interact favorably with the ligand. This approach follows the view that conformations are not truly induced. Instead, they are populated by a system even in isolation, albeit with lower probabilities. The presence of different conformations for the ligand or the rest of the protein may change the occurrence probabilities of certain conformations sampled in isolation.
Early work in [47] concluded that accuracy in loop modeling was limited primarily by the energy function rather than by the thoroughness of the optimization protocol. While further research is needed in improving energy functions, the review here has shown that search plays a central role both in improving accuracy and in modeling equilibrium flexibility. The main focus of recent loop modeling methods remains the development of efficient and accurate techniques for either closing open loop conformations or relaxing closed distorted conformations. As in the general structure prediction framework, the investigation of powerful search frameworks for loop modeling is fertile territory for further progress.
From a user’s perspective, it is currently hard to determine which loop modeling method to employ. Various considerations of accuracy versus efficiency and one loop conformation of high accuracy versus a large ensemble of relevant loop conformations come into play. As this review has shown, comparable accuracies can be obtained from different methods. Moreover, the positive development in the loop modeling community is the pace with which new methods are pursued to further improve the state of the art. Despite the open problems listed in this review, the current interest, as demonstrated by contributions of different communities, promises great advances in loop modeling.
Acknowledgements
This work was supported in part by NSF grant No. 1016995 (AS), by the Texas Higher Education Board NHARP01907 (LK), and by the John and Ann Doerr Fund for Computational Biomedicine at Rice University (LK). We are indebted to the authors of the highlighted methods for contributing images in this review.
References
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; Kuriyan, J. Molecular Dynamics and protein function. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 6679–6685. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Koga, N.; Takada, S.; Onuchic, J.N.; Wolynes, P.G. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 11844–11849. [Google Scholar] [CrossRef] [PubMed]
- Hub, J.S.; de Groot, B.L. Detection of functional modes in protein dynamics. PLoS Comp. Biol. 2009, 5, e1000480. [Google Scholar] [CrossRef] [PubMed]
- Jenzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual Molecular Dynamics. J. Mol. Graph. Model. 1996, 14, 33–38. Available online: http://www.ks.uiuc.edu/Research/vmd/ (accessed on 6 February 2012). [Google Scholar] [CrossRef]
- Decanniere, K.; Muyldermans, S.; Wyns, L. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes? J. Mol. Biol. 2000, 300, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Likitvivatanavong, S.; Aimanova, K.G.; Gill, S.S. Loop residues of the receptor binding domain of Bacillus thuringiensis Cry11Ba toxin are important for mosquitocidal activity. FEBS Lett. 2007, 583, 2021–2030. [Google Scholar] [CrossRef] [PubMed]
- Lepsik, M.; Field, M.J. Binding of calcium and other metal ions to the EF-hand loops of calmodulin studied by quantum chemical calculations and molecular dynamics simulations. J. Phys. Chem. 2007, 111, 10012–10022. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.M.; Marintcheva, B.; Cook, T.; Lee, S.J.; Tabor, S.; Richardson, C.C. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 5096–5101. [Google Scholar] [CrossRef] [PubMed]
- Swedberg, J.E.; de Veer, S.J.; Sit, K.C.; Reboul, C.F.; Buckle, A.M.; Harris, J.M. Mastering the canonical loop of serine protease inhibitors: Enhancing potency by optimising the internal hydrogen bond network. PLoS One 2011, 6, e19302. [Google Scholar] [CrossRef] [PubMed]
- Thanki, N.; Zeelen, J.P.; Mathieu, M.; Laenicke, R.; Abagyan, R.A.; Wierenga, R.K.; Schliebs, W. Protein engineering with monomeric triosephosphate isomerase (monoTIM): The modelling and structure verification of a seven residue loop. Protein Eng. 1997, 10, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliand, G.; Bhat, T.N.; Weissig, H.; N., S.I.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Eicken, C.; Sharma, V.; Klabunde, T.; Lawrenz, M.B.; Hardham, J.M.; Norris, S.J.; Sacchettini, J.C. Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia Burgdorferi. J. Biol. Chem. 2002, 277, 21691–21696. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.R.; Dyson, H.J.; Wright, P.E. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomolec. Struct. 2004, 33, 119–140. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.J.; Rader, A.J.; Kuhn, L.A.; Thorpe, M.F. Protein flexibility predictions using graph theory. Protein. Struct. Funct. Bioinf. 2001, 44, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Mamonova, T.; Hespenheide, B.; Straub, R.; Thorpe, M.F.; Kurnikova, M. Protein flexibility using constraints from molecular dynamics simulations. J. Phys. Biol. 2005, 2, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Fox, N.; Jagodzinski, F.; Li, Y.; Streinu, I. KINARI-Web: A server for protein rigidity analysis. Nucleic Acids Res. 2011, 39, W177–W183. [Google Scholar] [CrossRef] [PubMed]
- Dunbrack Jr., R.L. Comparative modeling of CASP3 targets using PSI-BLAST and SCWRL. Protein. Struct. Funct. Bioinf. 1999, 37, 81–87. [Google Scholar] [CrossRef]
- Bradley, P.; Misura, K.M.S.; Baker, D. Toward high-resolution de novo structure prediction for dmall proteins. Science 2005, 309, 1868–1871. [Google Scholar] [CrossRef] [PubMed]
- Craig, J. Introduction to Robotics: Mechanics and Control, 2nd ed.; Addison-Wesley: Boston, MA, USA, 1989; p. 450. [Google Scholar]
- Zhang, M.; Kavraki, L.E. A new method for fast and accurate derivation of molecular conformations. Chem. Inf. Comput. Sci. 2002, 42, 64–70. [Google Scholar] [CrossRef]
- Zhang, M.; Kavraki, L.E. Finding solutions of the inverse kinematics problem in computer-aided drug design. In Currents in Computational Molecular Biology; Florea, L., Walenz, B., Hannenhalli, S., Eds.; ACM Press: Washington, DC, USA, 2002; Number TR02-385; pp. 214–215. [Google Scholar]
- Engh, R.A.; Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. 1991, A47, 392–400. [Google Scholar] [CrossRef]
- Abayagan, R.; Totrov, M.; Kuznetsov, D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994, 15, 488–506. [Google Scholar] [CrossRef]
- Manocha, D.; Zhu, Y. Kinematic manipulation of molecular chains subject to rigid constraints. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 285–293. [Google Scholar] [PubMed]
- Singh, A.P.; Latombe, J.C.; Brutlag, D.L. A motion planning approach to flexible ligand binding. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 7, 252–261. [Google Scholar]
- Apaydin, M.S.; Singh, A.P.; Brutlag, D.L.; Latombe, J.C. Capturing molecular energy landscapes with probabilistic conformational roadmaps. Proc. IEEE Int. Conf. Robot. Autom. 2001, 1, 932–939. [Google Scholar]
- Amato, N.M.; Dill, K.A.; Song, G. Using motion planning to map protein folding landscapes and analyze folding kinetics of known native structures. J. Comp. Biol. 2002, 10, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Apaydin, M.S.; Brutlag, D.L.; Guestrin, C.; Hsu, D.; Latombe, J.C. Stochastic roadmap simulation: An efficient representation and algorithm for analyzing molecular motion. J. Comp. Biol. 2003, 10, 257–281. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Amato, N.M. A Motion planning approach to folding: From paper craft to protein folding. IEEE Trans. Robot. Autom. 2004, 20, 60–71. [Google Scholar] [CrossRef]
- Cortes, J.; Simeon, T.; de Angulo, R.; Guieysse, D.; Remaud-Simeon, M.; Tran, V. A path planning approach for computing large-amplitude motions of flexible molecules. Bioinformatics 2005, 21, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Streinu, I.; Brock, O. A methodology for efficiently sampling the conformation space of molecular structures. J. Phys. Biol. 2005, 2, S108–S115. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Jernigan, R.L.; Chirikjian, G.S. Efficient generation of feasible pathways for protein conformationa transitions. Biophys. J. 2002, 83, 1620–1630. [Google Scholar] [CrossRef]
- Georgiev, I.; Donald, B.R. Dead-end elimintation with backbone flexibility. Bioinformatics 2007, 23, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.H.; Apaydin, M.S.; Brutlag, D.L.; Hsu, D.; Latombe, J.C. Using stochastic roadmap simulation to predict experimental quantities in protein folding kinetics: Folding rates and phi-values. J. Comp. Biol. 2007, 14, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Kirillova, S.; Cortes, J.; Stefaniu, A.; Simeon, T. An NMA-guided path planning approach for computing large-amplitude conformational changes in proteins. Protein. Struct. Funct. Bioinf. 2008, 70, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Dunbrack, R.L., Jr.; Cohen, F.E. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997, 6, 1661–1681. [Google Scholar] [CrossRef] [PubMed]
- Clementi, C. Coarse-grained models of protein folding: Toy-models or predictive tools? Curr. Opinion Struct. Biol. 2008, 18, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Chan, H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997, 4, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Socci, N.D.; Onuchic, J.N.; Wolynes, P.G. Protein folding mechanisms and the multidimensional folding funnel. Protein. Struct. Funct. Bioinf. 1998, 32, 136–158. [Google Scholar] [CrossRef]
- Onuchic, J.N.; Wolynes, P.G. Theory of protein folding. Curr. Opinion Struct. Biol. 1997, 14, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Onuchic, J.N.; Luthey-Schulten, Z.; Wolynes, P.G. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 1997, 48, 545–600. [Google Scholar] [CrossRef] [PubMed]
- Ejtehadi, M.R.; Avall, S.P.; Plotkin, S.S. Three-body interactions improve the prediction of rate and mechanism in protein folding models. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15088–15093. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, P.J.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Protein. Struct. Funct. Bioinf. 2004, 55, 351–367. Available online: http://www.francisco.compchem.ucsf.edu/~jacobson/Software:PLOP (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Do, R.K.; Sali, A. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. Available online: http://modbase.compbio.ucsf.edu/modloop/ (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Soto, C.S.; Honig, B. Evaluating conformational free energies: The colony energy and its application to the problem of loop prediction. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 7432–7437. Available online: http://wiki.c2b2.columbia.edu/honiglab public/index.php/Software:Loopy (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Smith, K.; Honig, B. Evaluation of the conformational free energies of loops in proteins. Proteins 1994, 18, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Kolodny, R.; Guibas, L.; Levitt, M.; Koehl, P. Inverse kinematics in biology: The protein loop closure problem. Int. J. Robot. Res. 2005, 24, 151–163. [Google Scholar] [CrossRef]
- Primrose, E.J.F. On the input-output equation of the general 7R- mechanism. Mech. Mach. Theory 1986, 21, 509–510. [Google Scholar] [CrossRef]
- Manocha, D.; Canny, J. Efficient inverse kinematics for general 6R manipulator. IEEE Trans. Robot. Autom. 1994, 10, 648–657. [Google Scholar] [CrossRef]
- Manocha, D.; Zhu, Y.; Wright, W. Conformational analysis of molecular chains using nano-kinematics. Comput. Appl. Biosci. 1995, 11, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Go, N.; Scheraga, H.J. Ring closure and local conformational deformations of chain molecules. Macromolecules 1970, 3, 178–187. [Google Scholar] [CrossRef]
- Bruccoleri, R.E.; Karplus, M. Chain closure with bond angle variations. Macromolecules 1985, 18, 2676–2773. [Google Scholar] [CrossRef]
- Palmer, K.A.; Scheraga, H.A. Standard-geometry chains fitted to x-ray derived structures: Validation of the rigid-geometry approximation. I. chain closure through a limited search of “loop” conformations. J. Comput. Chem. 1991, 12, 505–526. [Google Scholar] [CrossRef]
- Wedemeyer, W.J.; Scheraga, H.J. Exact analytical loop closure in proteins using polynomial equations. J. Comput. Chem. 1999, 20, 819–844. [Google Scholar] [CrossRef]
- Coutsias, E.A.; Seok, C.; Jacobson, M.; Dill, K. A kinematic view of loop closure. J. Comput. Chem. 2004, 25, 510–528. Available online: http://dillgroup. ucsf.edu/loop closure/ (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; White, R.A.; Wang, L.; Goldman, R.; Kavraki, L.E.; Hasset, B. Improving conformational searches by geometric screening. Bioinformatics 2005, 21, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Chirikjian, G.S. General methods for computing hyper-redundant manipulator inverse kinematics. In Proceedings of the 1993 IEEE/RSJ International Conference on Intelligent Robots and Systems ’93, IROS ’93, Yokohama, Japan, 26–30 July 1993; Volume 2, pp. 1067–1073.
- Zhang, M.; Kavraki, L.E. Solving molecular inverse kinematics problems for protein folding and drug design. In Currents in Computational Molecular Biology; ACM Press: New York, NY, USA, 2002; pp. 214–215. [Google Scholar]
- Fine, R.M.; Wang, H.J.; Shenkin, P.; Yarmush, D.; Levinthal, C. Predicting antibody hypervariable loop conformations. II: Minimization and molecular dynamics studies of MCPC603 from many randomly generated loop conformations. Proteins 1986, 1, 342–362. [Google Scholar] [CrossRef] [PubMed]
- Shenkin, P.S.; Yarmush, D.L.; Fine, R.; Wang, H.J.; Levinthal, C. Predicting antibody hypervariable loop conformations. I: Ensembles of random conformations for ring-like structures. Biopolymers 1987, 26, 2053–2085. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.T.; Chen, C.C. A combined optimization method for solving the inverse kinematics problem of mechanical manipulators. IEEE Trans. Robot. Autom. 1991, 7, 489–499. [Google Scholar] [CrossRef]
- Ring, C.S.; Kneller, D.G.; Langridge, R.; Cohen, F.E. Taxonomy and conformational analysis of loops in proteins. J. Mol. Biol. 1992, 224, 685–699. [Google Scholar] [CrossRef]
- Canutescu, A.A.; Dunbrack, R.L. Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 2003, 12, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Lotan, I. Algorithms exploiting the chain structure of proteins. Ph.D. Thesis, Stanford University, Stanford, CA, USA, 2004. [Google Scholar]
- Lotan, I.; van den Bedem, H.; Deacon, A.M.; Latombe, J.C. Computing protein structures from electron density maps: The mising loop problem. In Algorithmic Foundations of Robotics VI; Erdman, M., Hsu, D., Overmars, M., van der Stappen, F., Eds.; Springer: Berlin, Germany, 2004; pp. 153–168. [Google Scholar]
- van den Bedem, H.; Lotan, I.; Latombe, J.C.; Deacon, A.M. Real-space protein-model completion: An inverse-kinematics approach. Acta Crystallogr. 2005, D61, 2–13. Avaiable online: https://simtk.org/home/looptk/Software:LoopTK (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Shehu, A.; Clementi, C.; Kavraki, L.E. Modeling protein conformational ensembles: From missing loops to equilibrium fluctuations. Protein. Struct. Funct. Bioinf. 2006, 65, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Shehu, A.; Clementi, C.; Kavraki, L.E. Sampling conformation space to model equilibrium fluctuations in proteins. Algorithmica 2007, 48, 303–327. [Google Scholar] [CrossRef]
- Zhu, K.; Pincus, D.L.; Zhao, S.; Friesner, R.A. Long loop prediction using the protein local optimization program. Protein. Struct. Funct. Bioinf. 2006, 65, 438–452. [Google Scholar] [CrossRef] [PubMed]
- Sellers, B.D.; Zhu, K.; Zhao, S.; Friesner, R.A.; Jacobson, M.P. Toward better refinement of comparative models: Predicting loops in inexact environments. Protein. Struct. Funct. Bioinf. 2008, 72, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C.; Lesk, A.M.; Prilusky, J.; Manning, N.O. Canonical structures for the hypervariable loops of immunoglobulins. J. Mol. Biol. 1987, 196, 901–917. [Google Scholar] [CrossRef]
- Claessens, M.; Cutsem, E.; Lasters, I.; Wodak, S. Modeling the polypeptide backbone with ‘spare parts’ from known protein structures. Protein Eng. 1989, 4, 335–345. [Google Scholar] [CrossRef]
- Summers, N.L.; Karplus, M. Modeling of globular proteins. A distance-based data search procedure for the construction of insertion/deletion regions and Pro- non-Pro mutations. J. Mol. Biol. 1990, 216, 991–1016. [Google Scholar] [CrossRef]
- Tramontano, A.; Lesk, A. Common features of the conformations of antigen-binding loops in immunoglobulins and application to modeling loop conformations. Proteins 1992, 13, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M. Accurate modeling of protein conformation by automatic segment matching. J. Mol. Biol. 1992, 226, 507–533. [Google Scholar] [CrossRef]
- Topham, C.M.; McLeod, A.; Eisenmenger, F.; Overington, J.P.; Johnson, M.; Blundell, T. Fragment ranking in modeling of protein structure: Conformationally constrained environmental amino acid substitution tables. J. Mol. Biol. 1993, 229, 194–220. [Google Scholar] [CrossRef] [PubMed]
- Lessel, U.; Schomburg, D. Similarities between protein structures. Protein Eng. 1994, 7, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.C.R.; Thornton, J.M. Structural families in loops of homologous proteins—Automatic classification, modeling and application to antibodies. J. Mol. Biol. 1996, 263, 800–815. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Z.; Lai, L. Protein loops on structurally similar scaffolds: Database and conformational analysis. Biopolymers 1999, 49, 481–495. [Google Scholar] [CrossRef]
- Jones, T.A.; Thirup, S. Using known substructures in protein model building and crystallography. EMBO J. 1986, 5, 819–822. [Google Scholar] [PubMed]
- Chothia, C.; Lesk, A.; Tramontano, A.; Levitt, M.S.; Gill, S.J.; Air, G.; Sheriff, S.; Padla, E.; Davies, D.; Tulip, W.R.; et al. Conformation of immunoglobulin hypervariable regions. Nature 1989, 342, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Morea, V.; Tramontano, A.; Rustici, M.; Chothia, C.; Lesk, A.M. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 1998, 275, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Fidelis, K.; Stern, P.S.; Bacon, D.; Moult, J. Comparison of systematic search and database methods for constructing segments of protein structure. Protein Eng. 1994, 7, 953–960. [Google Scholar] [CrossRef] [PubMed]
- van Vlijmen, H.W.T.; Karplus, M. PDB-based protein loop prediction: Parameters for selection and methods for optimization. J. Mol. Biol. 1997, 267, 975–1001. [Google Scholar] [CrossRef] [PubMed]
- Moult, J.; James, M.N.G. An algorithm for determining the conformation of polypeptide segments in proteins by systematic search. Proteins 1986, 1, 146–163. [Google Scholar] [CrossRef] [PubMed]
- Du, P.C.; Andrec, M.; Levy, R.M. Have we seen all structures corresponding to short protein fragments in the Protein Data Bank? An update. Protein Eng. 2003, 16, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Tossato, C.E.; Bindewald, E.; Hesser, J.; Maenner, R. A divide and conquer approach to fast loop modeling. Protein Eng. 2002, 15, 279–286. [Google Scholar] [CrossRef]
- Lee, J.; Lee, D.; Park, H.; Coutsias, E.A.; Seok, C. Protein loop modeling by using fragment assembly and analytical loop closure. Protein. Struct. Funct. Bioinf. 2010, 78, 3428–3436. [Google Scholar] [CrossRef] [PubMed]
- Hansson, T.; Oostenbrink, C.; van Gunsteren, W.F. Molecular dynamics simulations. Curr. Opinion Struct. Biol. 2002, 12, 190–196. [Google Scholar] [CrossRef]
- Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N.; Teller, A.H.; Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953, 21, 1087–1092. [Google Scholar] [CrossRef]
- van Gunsteren, W.F.; Bakowies, D.; Baron, R.; Chandrasekhar, I.; Christen, M.; Daura, X.; Gee, P.; Geerke, D.P.; Glättli, A.; Hünenberger, P.H.; et al. Biomolecular modeling: Goals, problems, perspectives. Angew. Chem. Int. Ed. Engl. 2006, 45, 4064–4092. [Google Scholar] [CrossRef] [PubMed]
- Bruccoleri, R.E.; Haber, E.; Novotny, J. Structure of antibody hypervariable loops reproduced by a conformational search algorithm. Nature 1988, 335, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Bruccoleri, R.E.; Karplus, M. Prediction of the folding of short poly-peptide segments by uniform conformational sampling. Biopolymers 1987, 26, 137–168. [Google Scholar] [CrossRef] [PubMed]
- Bruccoleri, R.E. Application of systematic conformational search to protein modeling. Mol. Simulat. 1993, 10, 151–174. [Google Scholar] [CrossRef]
- Brower, R.C.; Vasmatzis, G.; Silverman, M.; DeLisi, C. Exhaustive conformational search and simulated annealing for models of lattice peptides. Biopolymers 1993, 33, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Deane, C.M.; Blundell, T.L. A novel exhaustive search algorithm for predicting the conformation of polypeptide segments in proteins. Proteins 2000, 40, 135–144. [Google Scholar] [CrossRef]
- DePristo, M.A.; de Bakker, P.I.; Lovell, S.C.; Blundell, T.L. Ab initio construction of polypeptide fragments: Efficient generation of accurate, representative ensembles. Protein. Struct. Funct. Bioinf. 2003, 51, 41–55. Available online: http://mordred.bioc.cam.ac.uk/~rapper/Software:RAPPER (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Bruccoleri, R.E.; Karplus, M. Conformational Sampling using high temperature molecular dynamics. Biopolymers 1990, 29, 1847–1862. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.H.; Scheraga, H.A. Pattern recognition in the prediction of protein structure. I. Tripeptide conformational probabilities calculated from the amino acid sequence. J. Comput. Chem. 1989, 10, 770–797. [Google Scholar] [CrossRef]
- Lambert, M.H.; Scheraga, H.A. Pattern recognition in the prediction of protein structure. II. Chain conformation from a probability-directed search procedure. J. Comput. Chem. 1989, 10, 798–816. [Google Scholar] [CrossRef]
- Lambert, M.H.; Scheraga, H.A. Pattern recognition in the prediction of protein structure. III. An importance-sampling minimization procedure. J. Comput. Chem. 1989, 10, 817–831. [Google Scholar] [CrossRef]
- Dudek, M.; Scheraga, H.J. Protein structure prediction using a combination of sequence homology and global energy minimization. I. Global energy minimization of surface loops. J. Comput. Chem. 1990, 11, 121–151. [Google Scholar] [CrossRef]
- Dudek, M.J.; Ramnarayan, K.; Ponder, J.W. Protein structure prediction using a combination of sequence homology and global energy minimization II. Energy functions. J. Comput. Chem. 1998, 19, 548–573. [Google Scholar] [CrossRef]
- Tanner, J.J.; Nell, L.J.; McCammon, J.A. Anti-insulin antibody structure and conformation. II. Molecular Dynamics with explicit solvent. Biopolymers 1992, 32, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rao, U.; Teeter, M.M. Improvement of turn prediction by molecular dynamics: A case study of a1-Purothionin. Protein Eng. 1993, 6, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, N.; Higo, J.; Kidera, A. Free energy landscapes of short peptides by enhanced conformational sampling. J. Mol. Biol. 2000, 296, 197–216. [Google Scholar] [CrossRef] [PubMed]
- McGarrah, D.B.; Judson, R.S. Analysis of the genetic algorithm method of molecular conformation determination. J. Comput. Chem. 1993, 14, 1385–1395. [Google Scholar] [CrossRef]
- Pedersen, J.T.; Moult, J. Ab initio structure prediction for small polypeptides and protein fragments using genetic algorithms. Proteins 1995, 23, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S.; DeLisi, C. Determining minimum energy conformations of polypeptides by dynamic programming. Biopolymers 1990, 29, 1755–1772. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, A.V.; Reva, B.A. Search for the stable state of a short chain in a molecular field. Protein Eng. 1992, 5, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Abagyan, R.; Totrov, M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994, 235, 983–1002. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.S.; Mathiowetz, A.M.; Chan, S.I.; Goddard, W.A. De novo prediction of polypeptide conformations using dihedral probability grid Monte Carlo methodology. Protein Sci. 1995, 4, 1203–1216. [Google Scholar] [CrossRef] [PubMed]
- Rapp, C.S.; Friesner, R.A. Prediction of loop geometries using a generalized born model of solvation effects. Proteins 1999, 35, 173–183. [Google Scholar] [CrossRef]
- Higo, J.; Collura, V.; Garnier, J. Development of an extended simulated annealing method: Application to the modeling of complementary determining regions of immunoglobulins. Biopolymers 1992, 32, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Collura, V.; Higo, J.; Garnier, J. Modelling of protein loops by simulated annealing. Protein Sci. 1993, 2, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Carlacci, L.; Englander, L. The loop problem in proteins: A Monte Carlo simulated annealing approach. Biopolymers 1993, 33, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- Carlacci, L.; Englander, S.W. Loop problem in proteins: Developments on the Monte Carlo simulated annealing approach. J. Comput. Chem. 1996, 17, 1002–1012. [Google Scholar] [CrossRef]
- Vasmatzis, G.; C.Brower, R.; DeLisi, C. Predicting immunoglobulin-like hypervariable loops. Biopolymers 1994, 34, 1669–1680. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, R.; Zheng, Q.; Vajda, S.; DeLisi, C. Computing the structure of bound peptides. Application to antigen recognition by class I major histocompatibility complex receptors. J. Mol. Biol. 1993, 234, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Rosenfeld, R.; DeLisi, C.; Kyle, D.J. Multiple copy sampling in protein loop modelling: Computational efficiency and sensitivity to dihedral perturbations. Protein Sci. 1994, 3, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, R.; Rosenfeld, R. Simultaneous modeling of multiple loops in proteins. Protein Sci. 1995, 4, 496–505. [Google Scholar]
- Kidera, A. Enhanced conformational sampling in Monte Carlo simulations of proteins: Application to a constrained peptide. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 9886–9889. [Google Scholar] [CrossRef] [PubMed]
- Koehl, P.; Delarue, M. New mean field self-consistent formalism providing simultaneously both gap closure and side chain positioning in protein homology modelling. Nat. Struct. Biol. 1995, 2, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Samudrala, R.; Moult, J. A graph-theoretic algorithm for comparative modeling of protein structure. J. Mol. Biol. 1998, 279, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.S.; Fasnacht, M.; Zhu, J.; Forrest, L.; Honig, B. Loop modeling: Sampling, filtering, and scoring. Protein. Struct. Funct. Bioinf. 2008, 70, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Yakey, J.; LaValle, S.M.; Kavraki, L.E. Randomized path planning for linkages with closed kinematic chains. IEEE Trans. Robot. Autom. 2001, 17, 951–959. [Google Scholar] [CrossRef]
- Han, L.; Amato, N.M. A kinematics-based probabilistic roadmap method for closed chain systems. In Algorithmic and Computational Robotics: New Directions; Donald, B.R., Lynch, K.M., Rus, D., Eds.; AK Peters: MA, USA, 2001; pp. 233–246. [Google Scholar]
- Xie, D.; Amato, N.M. A kinematics-based probabilistic roadmap method for high DOF closed chain systems. In Proceedings of the 2004 IEEE International Conference on Robotics and Automation, 26 April–1 May 2004; Volume 1, pp. 473–478.
- Cortes, J.; Simeon, T.; Laumond, J.P. A Random Loop Generator for planning the motions of closed kinematic chains using PRM methods. In Proceedings of the 2002 IEEE International Conference on Robotics and Automation, 2002; Volume 2, pp. 2141–2146.
- Cortes, J.; Simeon, T. Probabilistic motion planning for parallel mechanisms. In Proceedings of the 2003 IEEE International Conference on Robotics and Automation, 14–19 September 2003; Volume 3, pp. 4354–4359.
- Cortes, J.; Simeon, T.; Remauld-Simeon, M.; Tran, V. Geometric algorithms for the conformational analysis of long protein loops. J. Comput. Chem. 2004, 25, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Milgram, R.J.; Liu, G.; Latombe, J.C. On the structure of the inverse kinematics map of a fragment of protein backbone. J. Comput. Chem. 2008, 29, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Dhanik, A.; Marz, N.; Propper, R.; Kou, C.; Liu, G.; van den Bedem, H.; Latombe, J.C.; Halperin-Landsberg, I.; Altman, R.B. Efficient algorithms to explore conformation spaces of flexible protein loops. In Proceedings of the IEEE/ACM Transactions on Computational Biology and Bioinformatics, October–December 2008; pp. 534–545.
- Ko, J.; Lee, D.; Park, H.; Coutsias, E.A.; Lee, J.; Seok, C. The FALC-Loop web server for protein loop modeling. Nucleic Acids Res. 2011, 39, W210–W214. Available online: http://falc-loop.seoklab.org/ (accessed on 6 February 2012). [Google Scholar] [CrossRef] [PubMed]
- Mandell, D.J.; Coutsias, E.A.; Kortemme, T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 2009, 6, 510–528. [Google Scholar] [CrossRef] [PubMed]
- Coutsias, E.A.; Seok, C.; Wester, M.J.; Dill, K.A. Resultants and loop closure. Int. J. Quantum Chem. 2006, 106, 176–189. [Google Scholar] [CrossRef]
- Nilmeier, J.; Hua, L.; Coutsias, E.; Jacobson, M.P. Assessing loop flexibility by hierarchical Monte Carlo sampling. J. Chem. Theory Comput. 2011, 7, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Rosenfeld, R.; Vajda, S.; DeLisi, C. Loop closure via bond scaling and relaxation. J. Comput. Chem. 1992, 14, 556–565. [Google Scholar] [CrossRef]
- Zheng, Q.; Rosenfeld, R.; Vajda, S.; DeLisi, C. Determining protein loop conformation using scaling-relaxation techniques. Protein Sci. 1993, 2, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, F.; Rassokhin, D.N.; Agrafiotis, D.K. A Self-organizing algorithm for modeling protein loops. PLoS Comp. Biol. 2009, 5, e1000478. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Rata, I.; Jakobsson, E. Sampling multiple scoring functions can improve protein loop structure prediction accuracy. J. Chem. Inf. Model. 2011, 51, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, S.; Zhou, Y. Accurate and efficient loop selections by the DFIRE-based all-atom statistical potential. Protein Sci. 2004, 13, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Fitzkee, N.C.; Fleming, P.J.; Rose, G.D. The protein coil library: A structural database of nonhelix, nonstrand fragments derived from the PDB. Protein. Struct. Funct. Bioinf. 2005, 58, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Storn, R.; Price, K. Differential evolution—A simple and efficient heuristic for global optimization over continuous spaces. J. Global Optim. 2004, 11, 341–359. [Google Scholar] [CrossRef]
- LaValle, S.M.; Kuffner, J.J. Randomized kinodynamic planning. Int. J. Robot. Res. 2001, 20, 378–400. [Google Scholar] [CrossRef]
- Kavraki, L.E.; Svetska, P.; Latombe, J.C.; Overmars, M. Probabilistic roadmaps for path planning in high-dimensional configuration spaces. IEEE Trans. Robot. Autom. 1996, 12, 566–580. [Google Scholar] [CrossRef]
- Debeire-Gosselin, M.; Loonis, M.; Samain, E.; Debeire, P. Purification and properties of a 22kDa endoxylanase excreted by a new strain of thermophilic bacterium. Xylans and Xylanases; Elsevier Science Publishers: Amsterdam, The Netherlands, 1997; pp. 463–466. [Google Scholar]
- Harris, G.W.; Pickersgill, R.; Connerton, I.; Debeire, P.; Touzel, J.P.; Breton, C.; Pérez, S. Structural basis of the properties of an industrially relevant thermophilic xylanase. Protein. Struct. Funct. Bioinf. 1997, 29, 77–86. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Pacci, E.; Dobson, C.; Karplus, M. Rare fluctuations of native proteins sampled by equilibrium hydrogen exchange. J. Am. Chem. Soc. 2003, 125, 15686–15687. [Google Scholar] [CrossRef]
- Li, X.; Romero, P.; Rani, M.; Dunker, A.K.; Obradovic, Z. Sequence complexity of disordered protein. Protein. Struct. Funct. Bioinf. 2001, 42, 38–48. [Google Scholar]
- Shehu, A.; Kavraki, L.E.; Clementi, C. On the characterization of protein native state ensembles. Biophys. J. 2007, 92, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.; Chavan, A.G.; Day, R.; Lennox, K.P.; Sukhanov, P.; Dahl, D.B.; Vannucci, M.; Tsai, J. Near-native protein loop sampling using nonparametric density estimation accommodating sparcity. PLoS Comp. Biol. 2011, 7, e1002234. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, M.F.; Ming, L. Macromolecular flexibility. Phil. Mag. 2004, 84, 1323–31137. [Google Scholar] [CrossRef]
- Wells, S.; Menor, S.; Hespenheide, B.; Thorpe, M.F. Constrained geometric simulation of diffusive motion in proteins. J. Phys. Biol. 2005, 2, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Farrell, D.W.; Speranskiy, K.; Thorpe, M.F. Generating stereochemically acceptable protein pathways. Protein. Struct. Funct. Bioinf. 2010, 78, 2908–2921. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Zhang, L.; Latombe, J.C. Sampling-based exploration of folded state of a protein under kinematic and geometric constraints. Protein. Struct. Funct. Bioinf. 2011, 80, 25–43. [Google Scholar] [CrossRef] [PubMed]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/.)