Microorganisms 2020, 8(3), 356; https://doi.org/10.3390/microorganisms8030356 - 2 Mar 2020
Cited by 19 | Viewed by 4826
Abstract
Kinetoplastids are protozoa containing a range of ubiquitous free_living species–pathogens of invertebrates, vertebrates and even some plants. Some of them are causative agents of canine vector-borne diseases. Their diagnosis is often missing in a gold standard. Here, we proposed a molecular approach for
[...] Read more.
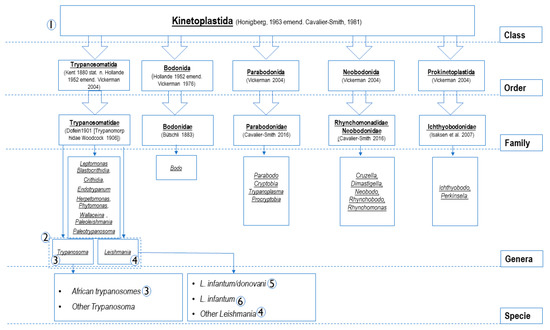
Kinetoplastids are protozoa containing a range of ubiquitous free_living species–pathogens of invertebrates, vertebrates and even some plants. Some of them are causative agents of canine vector-borne diseases. Their diagnosis is often missing in a gold standard. Here, we proposed a molecular approach for the diagnosis and study of Kinetoplastida. The TaqMan qPCR assays target the following genes: 24Sa LSU of Kinetoplastida, 28S LSU of Leishmania/ Trypanosoma spp., 5.8S rRNA of Trypanosoma spp., 18S SSU of Leishmania spp., kinetoplast minicircle DNA (kDNA) of L. donovani complex and kDNA of L. infantum, were designed, validated for their sensitivity (Se) and specificity (Sp) in silico and in vitro using a panel of known DNAs. They were then used to screen 369 blood samples (358 dogs, 2 equids, 9 monkeys). In addition, new 28S LSU primer sets are presented to use for Kinetoplastida’s identification by PCR/sequencing. All qPCRs showed consistently high analytical sensitivities and reproducibility. They detect approximately 0.01 parasite/ mL blood for the kDNA based- qPCRs and at least a single cell-equivalent of rDNA for the other systems. Based on the sequencing results, after screening, Se and Sp were: 0. 919 and 0.971, 0.853 and 0.979, 1.00 and 0.987, 0.826 and 0.995 for all of Kinetoplastida, Leishmania/ Trypanosoma, Trypanosoma, Leishmania spp. specific qPCRs, respectively. kDNA based qPCRs were more sensitive and specific (Se: 1.00; Sp: 0.997). PCR/sequencing allowed the detection of Kinetoplastids in animal blood samples such as L. infantum, L. guyanensis, T. congolense, T. evansi and Bodo spp. The molecular approach proposed here is useful for epidemiological studies, fundamental research such as screening for new Kinetoplastida species, diagnosis and therapeutic follow-up. In addition, researchers are free to choose the molecular tools adapted to their aims.
Full article
(This article belongs to the Special Issue Canine Vector Borne Diseases)
►
Show Figures