Changes in Expression of Genes Regulating Airway Inflammation Following a High-Fat Mixed Meal in Asthmatics
Abstract
:1. Introduction
2. Methods
2.1. Subject Recruitment
2.2. Study Design
2.3. Sample Collection and Processing
2.4. Whole Genome Gene Expression Microarrays
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Non-Esterified Plasma Fatty Acid Concentrations
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Asthmatics and Healthy Controls Group
| Asthma | Healthy | p-Value | |
|---|---|---|---|
| N | 11 | 8 | NA |
| Age | 48.5 (18.7) | 49.4 (18.8) | 0.925 |
| Gender (M|F) | 6|5 | 6|2 | 0.361 |
| BMI, mean (SD) | 30.6 (5.1) | 29.0 (5.5) | 0.514 |
| Smoking, ex|never | 1|10 | 5|3 | 0.013 |
| Atopy, n (%) | 9 (82) | 2 (25) | 0.013 |
| FEV1%, mean (SD) | 83.9 (25.3) | 101.8 (13.8) | 0.088 |
| FVC%, mean (SD) | 100.4 (21.1) | 104.8 (16.8) | 0.626 |
| FEV1/FVC%, mean (SD) | 67.3 (11.5) | 78.8 (6.6) | 0.021 |
| Total cell count (106/mL), median (Q1,Q3) | 4.8 (3.9–6.1) | 3.6 (1.2–8.2) | 0.457 |
| Neutrophils%, median (Q1,Q3) | 45.5 (23.8–68) | 33.8 (22.8–50.1) | 0.65 |
| Eosinophils%, median (Q1,Q3) | 2.8 (0.3–7.8) | 0.1 (0–2.8) | 0.141 |
| SNEFA (mg/L), mean (SD) | 156.7 (56.5) | 123.3 (54.9) | 0.215 |
| Total NEFA (mg/L), mean (SD) | 449.5 (170.3) | 376.5 (185.2) | 0.386 |
3.2. Effects of a High Fat Challenge on Sputum Gene Expression
| GO-Slim Category | Genes in the Reference List (n =) | Genes in the Experiment List (n =) | Genes Expected (n =) | Fold Enrichment | p-Value |
|---|---|---|---|---|---|
| immune system process | 1733 | 25 | 11.92 | 2.1 | 0.0003 |
| cellular defense response | 387 | 9 | 2.66 | 3.38 | 0.002 |
| cell adhesion | 890 | 14 | 6.12 | 2.29 | 0.004 |
| biological adhesion | 890 | 14 | 6.12 | 2.29 | 0.004 |
| metabolic process | 8613 | 75 | 59.25 | 1.27 | 0.006 |
3.3. Quantitative Real Time PCR (qPCR) Validation Of Sputum Results
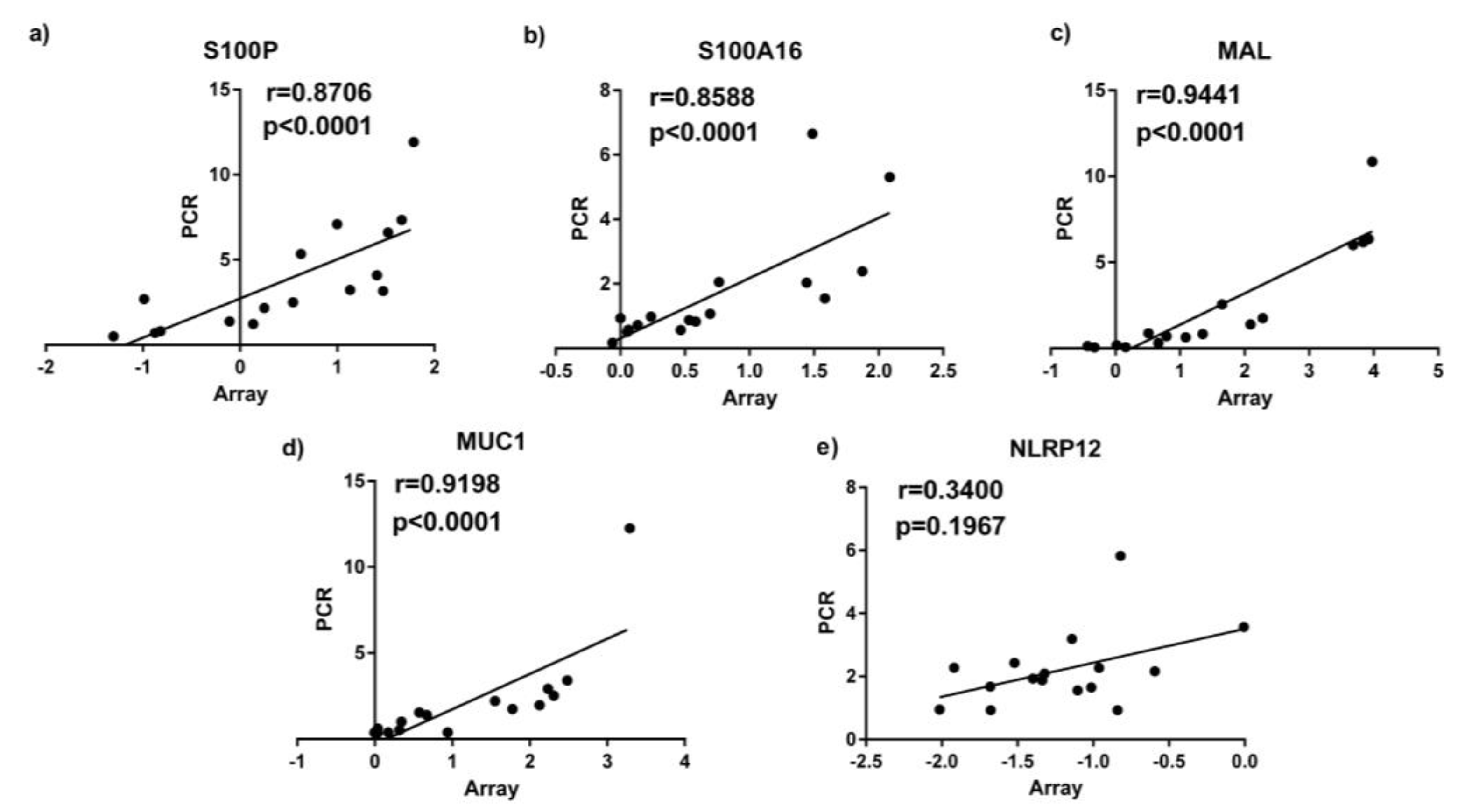
| Accession | Gene Symbol | Gene Name | Fold Change | ↑/↓ | Gene Function |
|---|---|---|---|---|---|
| NM_002371.2 | MAL | T-cell differentiation protein | 2.74 | ↑ | Lipid binding peptidase activator activity involved in apoptotic processes |
| NM_005656.2 | TMPRSS2 | Transmembrane protease, serine 2 | 2.56 | ↑ | Protein binding |
| NM_004433.3 | ELF3 | E74-like factor 3 | 2.09 | ↑ | RNA polymerase II core promoter proximal region sequence-specific DNA binding |
| NM_182502.2 | TMPRSS11B | Transmembrane protease, serine 11B | 1.95 | ↑ | Serine-type endopeptidase/peptidase activity |
| NM_144670.2 | A2ML1 | Alpha-2-macroglobulin-like 1 | 1.94 | ↑ | Peptidase activity;cytokine activity; serine-type endopeptidase inhibitor activity |
| NM_012431.1 | SEMA3E | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted | 1.93 | ↑ | Receptor binding |
| NM_080388.1 | S100A16 | S100 calcium binding protein A16 | 1.86 | ↑ | Novel calcium-binding signalling protein of dietary |
| NM_001093771.1 | TXNRD1 | Thioredoxin reductase 1 | 1.76 | ↑ | Thioredoxin-disulfide reductase activity |
| NM_005980.2 | S100P | S100 calcium binding protein P | 1.74 | ↑ | Calcium ion binding; receptor binding; signal transduction |
| NM_006238.2 | PPARD | Peroxisome proliferative activated receptor, delta | 1.52 | ↑ | NF-kappaB binding |
| NM_144687.1 | NLRP12 | NLR family, pyrin domain containing 12 | 1.51 | ↑ | Checkpoint of noncanonical NF-κb, inflammtion and tumorigenesis |
| NM_006613.3 | GRAP | GRB2-related adaptor protein | 1.55 | ↓ | SH3/SH2 adaptor activity |
| NM_002985.2 | CCL5 | Chemokine (C-C motif) ligand 5 | 2.28 | ↓ | Chemokine activity |
| NM_052931.3 | SLAMF6 | SLAM family member 6 | 1.90 | ↓ | Transmembrane receptor protein tyrosine kinase activity |
| NM_006120.2 | HLA-DMA | Major histocompatibility complex, class II, DM alpha | 1.86 | ↓ | MHC class II protein complex binding |
| NM_006725.2 | CD6 | CD6 molecule | 1.85 | ↓ | Oxidoreductase activity |
| NM_178562.2 | TSPAN33 | Tetraspanin 33 | 1.83 | ↓ | Receptor activity |
| NM_004454.1 | ETV5 | Ets variant gene 5 | 1.80 | ↓ | Sequence-specific DNA binding transcription factor activity |
| NM_016817.2 | OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71 kDa | 1.76 | ↓ | Nucleotidyltransferase activity; nucleic acid binding |
| NM_001040107.1 | HVCN1 | Hydrogen voltage-gated channel 1 | 1.7 | ↓ | Voltage-gated cation channel activity |
| NM_005202.1 | COL8A2 | Collagen, type VIII, alpha 2 | 1.63 | ↓ | Receptor activity |
| NM_001078.2 | VCAM1 | Vascular cell adhesion molecule 1 | 1.63 | ↓ | Phosphoprotein phosphatase activity |
| NM_175571.2 | GIMAP8 | GTPase, IMAP family member 8 | 1.58 | ↓ | Peptidase inhibitor activity |
| NM_000732.4 | CD3D | CD3d molecule, delta | 1.55 | ↓ | Protein heterodimerization activity |
| NM_012463.2 | ATP6V0A2 | ATPase, H+ transporting, lysosomal V0 subunit a2 | 1.51 | ↓ | Hydrolase activity; cation transmembrane transporter activity; hydrogen ion transmembrane transporter activity |
| Accession | Symbol | Fold Change | ↑/↓ | Gene Name |
|---|---|---|---|---|
| NM_002923.1 | RGS2 | 2.42 | ↑ | Regulator of G-protein signalling 2 |
| NM_002274.3 | KRT13 | 2.39 | ↑ | Keratin 13 |
| NM_006317.3 | BASP1 | 1.92 | ↑ | Brain abundant, membrane attached signal protein 1 |
| NM_002965.2 | S100A9 | 1.75 | ↑ | S100 calcium binding protein A9 (calgranulin B) |
| NM_058172.3 | ANTXR2 | 1.71 | ↑ | Anthrax toxin receptor 2 |
| NM_020226.3 | PRDM8 | 1.69 | ↑ | PR domain containing 8 |
| NM_004913.2 | C16orf7 | 1.62 | ↑ | Chromosome 16 open reading frame 7 |
| NM_003143.1 | SSBP1 | 1.55 | ↑ | Single-stranded DNA binding protein 1 |
| NM_024419.3 | PGS1 | 1.53 | ↑ | Phosphatidylglycerophosphate synthase 1 |
| NM_152361.1 | EID2B | 1.52 | ↑ | EP300 interacting inhibitor of differentiation 2B |
| NM_015104.1 | ATG2A | 1.51 | ↑ | ATG2 autophagy related 2 homolog A |
| NM_015352.1 | POFUT1 | 1.51 | ↑ | Protein O-fucosyltransferase 1 |
| NM_002443.2 | MSMB | 1.50 | ↑ | Microseminoprotein, beta |
| NM_017585.2 | SLC2A6 | 1.51 | ↓ | Solute carrier family 2, member 6 |
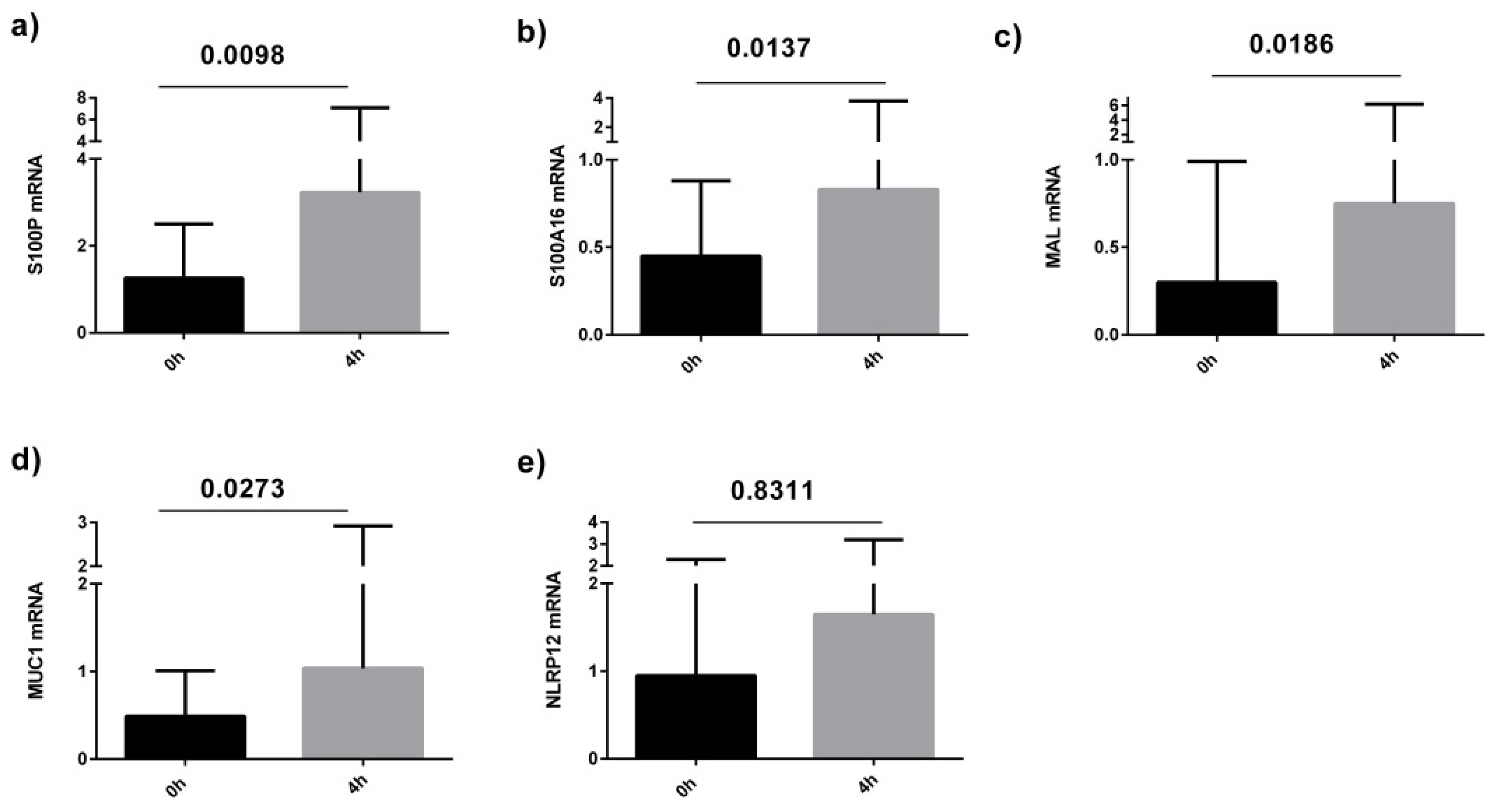
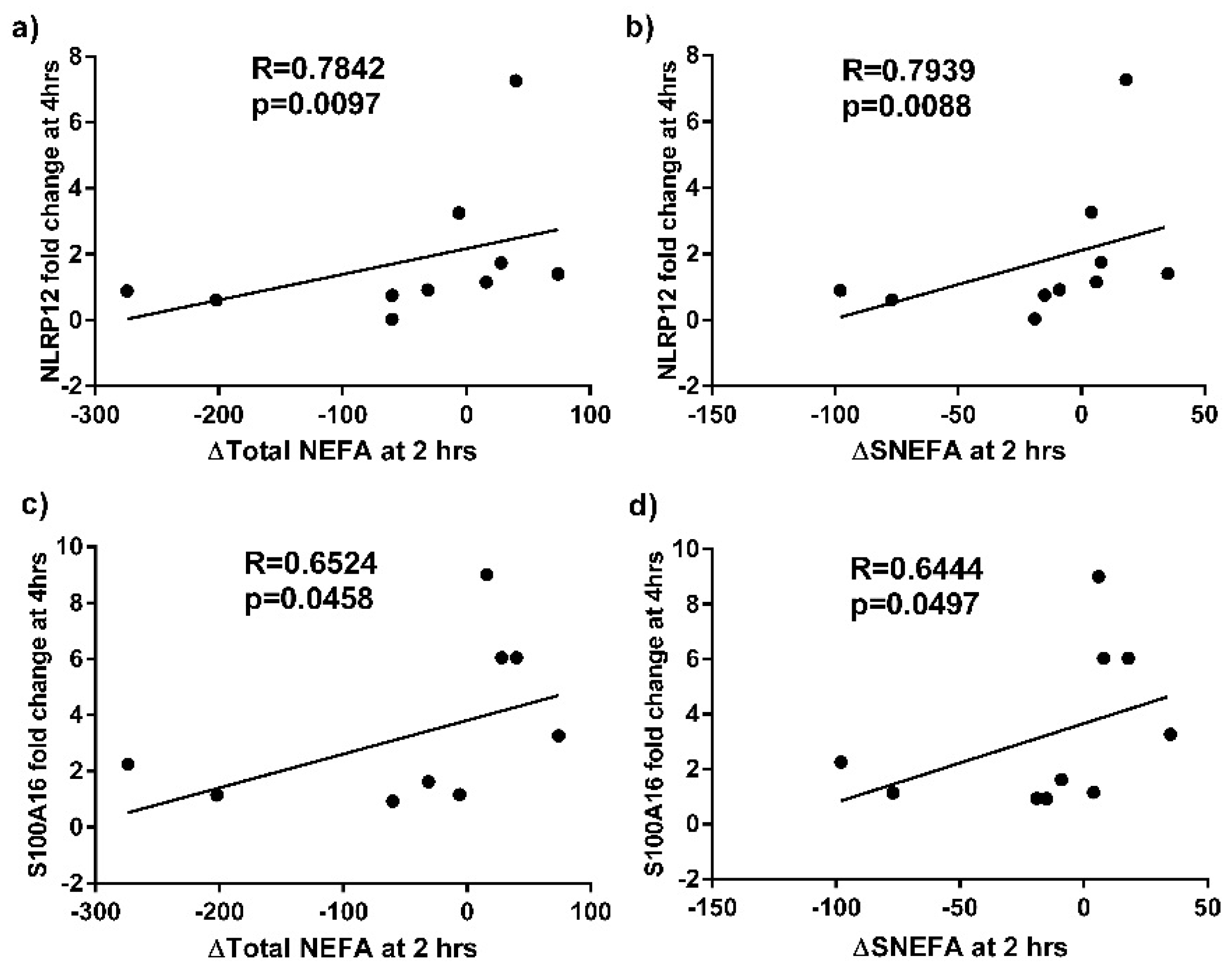
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Masoli, M.; Fabian, D.; Holt, S.; Beasley, R. The global burden of asthma: Executive summary of the GINA Dissemination Committee report. Allergy 2004, 59, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Grissell, T.V.; Douwes, J.; Scott, R.J.; Boyle, M.J.; Gibson, P.G. Innate immune activation in neutrophilic asthma and bronchiectasis. Thorax 2007, 62, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Jatakanon, A.; Uasuf, C.; Maziak, W.; Lim, S.; Chung, K.F.; Barnes, P.J. Neutrophilic inflammation in severe persistent asthma. Am. J. Respir. Crit. Care Med. 1999, 160, 1532–1539. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Scott, R.; Boyle, M.J.; Gibson, P.G. Inflammatory subtypes in asthma: Assessment and identification using induced sputum. Respirology 2006, 11, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, R.; Vinker, S.; Zakut, H.; Kizner, F.; Nakar, S.; Kitai, E. An Unusually High Prevalence of Asthma in Ethiopian Immigrants to Israel. Fam. Med. 1999, 31, 276–279. [Google Scholar] [PubMed]
- Gibson, P.G.; Henry, R.L.; Shah, S.; Powell, H.; Wang, H. Migration to a Western Country Increases Asthma Symptoms But Not Eosinophilic Airway Inflammation. Pediatr. Pulmonol. 2003, 36, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, A.; Barcella, M.; Bo, G.A.; Miadonna, A. Onset of allergy and asthma symptoms in extra-immigrants to Milan Itlay: Possilbe role of enviromental factors. Clin. Exp. Allergy 2003, 33, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Agostinis, F.; Foglia, C.; Landi, M.; Cottini, M.; Lombardi, C.; Canonica, G.W. GINA Report, Global Strategy for Asthma Management and Prevention. Allergy 2008, 63, 1637–1639. [Google Scholar] [CrossRef] [PubMed]
- Ajuwon, K.M.; Spurlock, M.E. Palmitate activates the NF-kappaB transcription factor and induces IL-6 and TNFalpha expression in 3T3-L1 adipocytes. J. Nutr. 2005, 135, 1841–1846. [Google Scholar] [PubMed]
- Soutar, A.; Seaton, A.; Brown, K. Bronchial reactivity and dietary antioxidants. Thorax 1997, 52, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Strom, K.; Janzon, L.; Mattisson, I.; Rosberg, H.E.; Arborelius, M. Asthma but not smoking-related airflow limitation is associated with a high fat diet in men: Results from the population study “Men born in 1914”, Malmo, Sweden. Monaldi Arch. Chest Dis. 1996, 51, 16–21. [Google Scholar] [PubMed]
- Bodner, C.; Godden, D.; Brown, K.; Little, J.; Ross, S.; Seaton, A. Antioxidant intake and adult-onset wheeze: A case–control study. Eur. Respir. J. 1999, 13, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.G.; Garg, M.L.; Gibson, P.G. A high-fat challenge increases airway inflammation and impairs bronchodilator recovery in asthma. J. Allergy Clin. Immunol. 2011, 127, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Kwon, M.J.; Huang, S.R.; Lee, J.Y.; Fukase, K.; Inohara, N.; Hwang, D.H. Differential modulation of Nods signaling pathways by fatty acids in human colonic epithelial HCT116 cells. J. Biol. Chem. 2007, 282, 11618–11628. [Google Scholar] [CrossRef] [PubMed]
- Gibson, P.G.; Simpson, J.L.; Saltos, N. Heterogeneity of airway inflammation in persistent asthma—Evidence of neutrophilic inflammation and increased sputum interleukin-8. Chest 2001, 119, 1329–1336. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Roth, J.; Oppenheim, J.J.; Tracey, K.J.; Vogl, T.; Feldmann, M.; Horwood, N. Alarmins: Awaiting a clinical response. J. Clin. Investig. 2012, 122, 2711–2719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.H.; Zhu, W.D.; Du, X.L.; Xin, J.; Xue, Y.; Zhang, Y.Y.; Li, D. S100A16 mediation of weight gain attenuation induced by dietary calcium. Metabolism 2012, 61, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, R.H.; Zhu, W.D.; Xue, Y.; Zhang, Y.Y.; Huang, Q.; Liu, M. S100A16 inhibits osteogenesis but stimulates adipogenesis. Mol. Biol. Rep. 2013, 40, 3465–3473. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, R.; Xin, J.; Sun, Y.; Li, J.; Wei, D.; Zhao, A.Z. Identification of S100A16 as a novel adipogenesis promoting factor in 3T3-L1 cells. Endocrinology 2011, 152, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Logsdon, C.D. S100P: A novel therapeutic target for cancer. Amino Acids 2011, 41, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.L.; Flier, J.S. TLR4 links innate immunity and fatty acid–induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef] [PubMed]
- Uysal, K.T.; Wiesbrock, S.M.; Marino, M.W.; Hotamisligil, G.S. Protection from obesity-induced insulin resistance in mice lacking TNF-α function. Nature 1997, 389, 610–614. [Google Scholar] [PubMed]
- Wellen, K.E.; Hotamisligil, G.S. Inflammation, stress, and diabetes. J. Clin. Investig. 2005, 115, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Lindén, S.K.; Sheng, Y.H.; Every, A.L.; Miles, K.M.; Skoog, E.C.; Sutton, P.; McGuckin, M.A. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009, 5, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Hollingsworth, M.A. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006, 16, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Hattrup, C.L.; Gendler, S.J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 2008, 70, 431–457. [Google Scholar] [CrossRef] [PubMed]
- Ueno, K.; Koga, T.; Kato, K.; Golenbock, D.T.; Gendler, S.J.; Kai, H.; Kim, K.C. MUC1 mucin is a negative regulator of toll-like receptor signaling. Am. J. Respir. Cell Mol. Biol. 2008, 38, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, Y.S.; Koga, T.; Treloar, A.; Kim, K.C. TNF-alpha is a key regulator of MUC1, an anti-inflammatory molecule, during airway Pseudomonas aeruginosa infection. Am. J. Respir. Cell Mol. Biol. 2011, 44, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Wang, L.; Manji, G.A.; Grenier, J.M.; Al-Garawi, A.; Merriam, S.; Lora, J.M.; Geddes, B.J. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J. Biol. Chem. 2002, 277, 29874–29880. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.L.; Taxman, D.J.; Linhoff, M.W.; Reed, W.; Ting, J.P. Cutting edge: Monarch-1: A pyrin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J. Immunol. 2003, 170, 5354–5358. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.L.; Lich, J.D.; Duncan, J.A.; Reed, W.; Rallabhandi, P.; Moore, C.; Kurtz, S. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J. Biol. Chem. 2005, 280, 39914–39924. [Google Scholar] [CrossRef] [PubMed]
- Ratsimandresy, R.A.; Dorfleutner, A.; Stehlik, C. An Update on PYRIN Domain-Containing Pattern Recognition Receptors: From Immunity to Pathology. Front. Immunol. 2013, 4, 440. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006, 440, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.B.; Tardivel, A.; Thorens, B.; Choi, I.; Tschopp, J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010, 11, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Gris, D.; Lei, Y.; Jha, S.; Zhang, L.; Huang, M.T.; Brickey, W.J. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 2011, 12, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Vandanmagsar, B.; Youm, Y.H.; Ravussin, A.; Galgani, J.E.; Stadler, K.; Mynatt, R.L.; Ravussin, E. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 2011, 17, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Frank, M. MAL, a proteolipid in glycosphingolipid enriched domains: Functional implications in myelin and beyond. Prog. Neurobiol. 2000, 60, 531–544. [Google Scholar] [CrossRef]
- Mori, Y.; Cai, K.; Cheng, Y.; Wang, S.; Paun, B.; Hamilton, J.P.; Jin, Z. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology 2006, 131, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C.; Bevan, S.J.; Newsholme, E.A. The inhibition of T-lymphocyte proliferation by fatty acids is via an eicosanoid-independent mechanism. Immunology 1992, 75, 108–115. [Google Scholar] [PubMed]
- Wood, L.G.; Scott, H.A.; Garg, M.L.; Gibson, P.G. Innate immune mechanisms linking non-esterified fatty acids and respiratory disease. Prog. Lipid Res. 2009, 48, 27–43. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Baines, K.J.; Gibson, P.G.; Wood, L.G. Changes in Expression of Genes Regulating Airway Inflammation Following a High-Fat Mixed Meal in Asthmatics. Nutrients 2016, 8, 30. https://doi.org/10.3390/nu8010030
Li Q, Baines KJ, Gibson PG, Wood LG. Changes in Expression of Genes Regulating Airway Inflammation Following a High-Fat Mixed Meal in Asthmatics. Nutrients. 2016; 8(1):30. https://doi.org/10.3390/nu8010030
Chicago/Turabian StyleLi, Qian, Katherine J. Baines, Peter G. Gibson, and Lisa G. Wood. 2016. "Changes in Expression of Genes Regulating Airway Inflammation Following a High-Fat Mixed Meal in Asthmatics" Nutrients 8, no. 1: 30. https://doi.org/10.3390/nu8010030





