Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo
Abstract
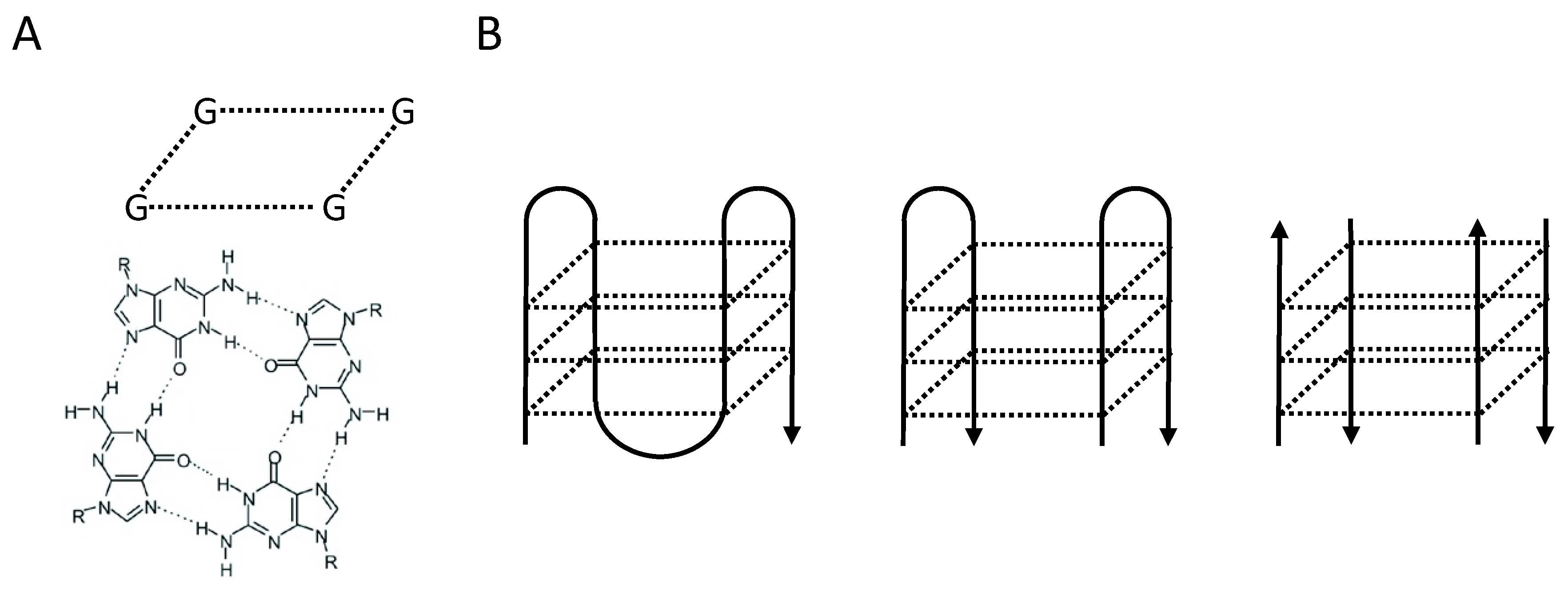
:1. G-Quadruplex Structures and G-Quadruplex Helicases
2. From Self-Renewal of Stem Cells to Telomeres
3. Hunting for Genes that Regulate Telomere Length
4. Discovery of dog-1
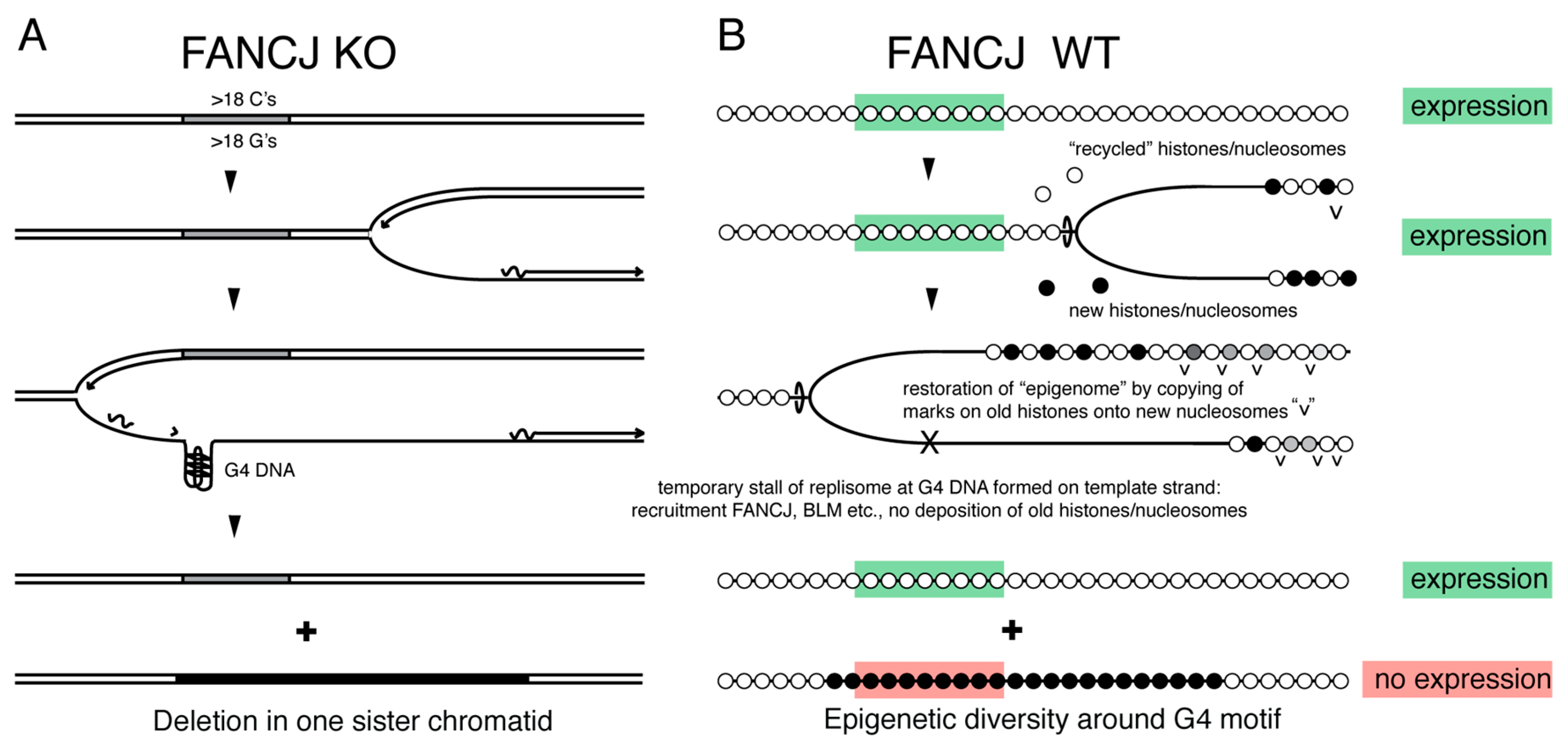
4.1. DOG-1/FANCJ Promotes Replication through G4 DNA
4.2. FANCJ Maintains Epigenetic Stability at G4 Motifs
5. Discovery of RTEL1
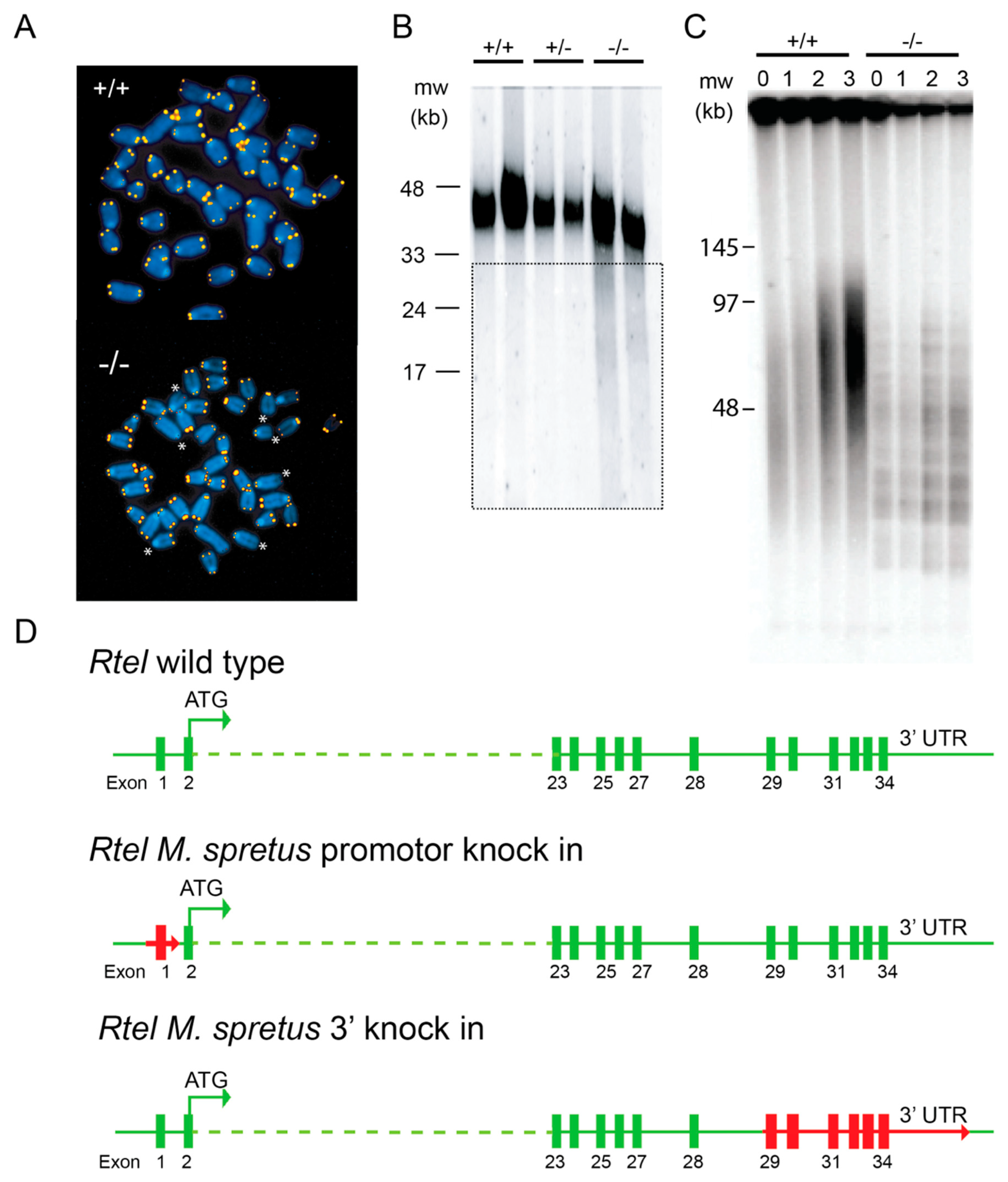
5.1. RTEL1 Maintains Genome Stability and Telomere Integrity
5.2. RTEL1 and Telomerase
5.3. What is Wrong with Rtel1 in M.spretus?
6. BLM is A Multifunctional Caretaker of Genome Stability
6.1. BLM Promotes Telomere Replication
6.2. BLM Prevents Replication fork Stalling and Recombination at G-Quadruplexes
6.3. BLM Deficiency Affects Transcription of Genes Containing G4 Motifs
7. Molecular Phenotypes That Inform in vivo Helicase Function
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lipps, H.J.; Rhodes, D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [PubMed]
- McManus, S.A.; Li, Y. Assessing the amount of quadruplex structures present within G(2)-tract synthetic random-sequence DNA libraries. PLoS ONE 2013, 8, e64131. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, S.; Curtis, E.A. Structure and Function of Multimeric G-Quadruplexes. Molecules 2019, 24, 3074. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.M.; Till, J.E.; Siminovitch, L.; McCulloch, E.A. Cytological evidence for a relationship between normal hemotopoietic colony-forming cells and cells of the lymphoid system. J. Exp. Med. 1968, 127, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Lemischka, I.R.; Raulet, D.H.; Mulligan, R.C. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell 1986, 45, 917–927. [Google Scholar] [CrossRef]
- Osawa, M.; Hanada, K.-I.; Hamada, H.; Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 1996, 273, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Lansdorp, P.M.; Dragowska, W.; Mayani, H. Ontogeny-related changes in proliferative potential of human hematopoietic cells. J. Exp. Med. 1993, 178, 787–791. [Google Scholar] [CrossRef]
- Vaziri, H.; Dragowska, W.; Allsopp, R.C.; Thomas, T.E.; Harley, C.B.; Lansdorp, P.M. Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc. Natl. Acad. Sci. USA 1994, 91, 9857–9860. [Google Scholar] [CrossRef]
- Lansdorp, P.M. Telomeres, stem cells, and hematology. Blood 2008, 111, 1759–1766. [Google Scholar] [CrossRef] [Green Version]
- Lansdorp, P.M.; Verwoerd, N.P.; Van De Rijke, F.M.; Dragowska, V.; Little, M.-T.; Dirks, R.W.; Raap, A.K.; Tanke, H.J. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996, 5, 685–691. [Google Scholar] [CrossRef]
- Rufer, N.; Dragowska, W.; Thornbury, G.; Roosnek, E.; Lansdorp, P.M. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat. Biotechnol. 1998, 16, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Aubert, G.; Baerlocher, G.M.; Vulto, I.; Poon, S.S.; Lansdorp, P.M. Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes. PLoS Genet. 2012, 8, e1002696. [Google Scholar] [CrossRef] [PubMed]
- Sekulovic, S.; Gylfadottir, V.; Vulto, I.; Gasparetto, M.; Even, Y.; Brookes, C.; Smith, C.; Eaves, C.J.; Lansdorp, P.M.; Rossi, F.M.; et al. Prolonged self-renewal activity unmasks telomerase control of telomere homeostasis and function of mouse hematopoietic stem cells. Blood 2011, 118, 1766–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, N.M.V.; Ryder, O.A.; Houck, M.L.; Charter, S.J.; Walker, W.; Forsyth, N.R.; Austad, S.N.; Venditti, C.; Pagel, M.; Shay, J.W.; et al. Comparative biology of mammalian telomeres: Hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell 2011, 10, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Baerlocher, G.M.; Vulto, I.; De Jong, G.; Lansdorp, P.M. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat. Protoc. 2006, 1, 2365–2376. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Hathcock, K.S.; Hande, M.P.; Lansdorp, P.M.; Seldin, M.F.; Hodes, R.J. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl. Acad. Sci. USA 1998, 95, 8648–8653. [Google Scholar] [CrossRef] [Green Version]
- Brenner, S. The Genetics of Caenorhabditis Elegans. Genetics 1974, 77, 71–94. [Google Scholar]
- Cheung, I.; Schertzer, M.; Rose, A.; Lansdorp, P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002, 31, 405–409. [Google Scholar] [CrossRef]
- Brenner, S. In the beginning was the worm. Genetics 2009, 182, 413–415. [Google Scholar] [CrossRef]
- Kruisselbrink, E.; Guryev, V.; Brouwer, K.; Pontier, D.B.; Cuppen, E.; Tijsterman, M. Mutagenic Capacity of Endogenous G4 DNA Underlies Genome Instability in FANCJ-Defective C. elegans. Curr. Boil. 2008, 18, 900–905. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.-Q.; Chen, Z.; Zheng, K.-W.; Chen, C.-Y.; Hao, Y.-H.; Tan, Z. G-quadruplex formation at the 3’ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011, 39, 6229–6237. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shin-Ya, K.; Brosh, R.M. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA To Defend Genomic Stability. Mol. Cell. Boil. 2008, 28, 4116–4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litman, R.; Peng, M.; Jin, Z.; Zhang, F.; Zhang, J.; Powell, S.; Andreassen, P.R.; Cantor, S.B. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 2005, 8, 255–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levran, O.; Attwooll, C.; Henry, R.T.; Milton, K.L.; Neveling, K.; Rio, P.; Batish, S.D.; Kalb, R.; Velleuer, E.; Barral, S.; et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 2005, 37, 931–933. [Google Scholar] [CrossRef]
- Levitus, M.; Waisfisz, Q.; Godthelp, B.C.; De Vries, Y.; Hussain, S.; Wiegant, W.W.; Elghalbzouri-Maghrani, E.; Steltenpool, J.; Rooimans, M.A.; Pals, G.; et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 2005, 37, 934–935. [Google Scholar] [CrossRef]
- London, T.B.C.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. FANCJ Is a Structure-specific DNA Helicase Associated with the Maintenance of Genomic G/C Tracts. J. Boil. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef] [Green Version]
- Schwab, R.A.; Nieminuszczy, J.; Shin-Ya, K.; Niedzwiedz, W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Boil. 2013, 201, 33–48. [Google Scholar] [CrossRef] [Green Version]
- Bosch, P.C.; Segura-Bayona, S.; Koole, W.; Heteren, J.T.; Dewar, J.M.; Tijsterman, M.; Knipscheer, P.; Van Heteren, J.T. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014, 33, 2521–2533. [Google Scholar] [CrossRef]
- Cantor, S.B.; Bell, D.W.; Ganesan, S.; Kass, E.M.; Drapkin, R.; Grossman, S.; Wahrer, D.C.; Sgroi, D.C.; Lane, W.S.; Haber, D.A.; et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 2001, 105, 149–160. [Google Scholar] [CrossRef]
- Brosh, R.M.; Cantor, S.B. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet. 2014, 5, 372. [Google Scholar] [CrossRef]
- Zhao, Y.; O’Neil, N.J.; Rose, A.M. Poly-G/poly-C tracts in the genomes of Caenorhabditis. BMC Genom. 2007, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkies, P.; Murat, P.; Phillips, L.G.; Patel, K.J.; Balasubramanian, S.; Sale, J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012, 40, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Suhasini, A.N.; Rawtani, N.A.; Wu, Y.; Sommers, J.A.; Sharma, S.; Mosedale, G.; North, P.S.; Cantor, S.B.; Hickson, I.D.; Brosh, R.M. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 2011, 30, 692–705. [Google Scholar] [CrossRef]
- Papadopoulou, C.; Guilbaud, G.; Schiavone, D.; Sale, J.E. Nucleotide Pool Depletion Induces G-Quadruplex-Dependent Perturbation of Gene Expression. Cell Rep. 2015, 13, 2491–2503. [Google Scholar] [CrossRef] [Green Version]
- Lansdorp, P.M. Immortal strands? Give me a break. Cell 2007, 129, 1244–1247. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Ding, H.; Schertzer, M.; Wu, X.; Gertsenstein, M.; Selig, S.; Kammori, M.; Tam, P.P. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell 2004, 117, 873–886. [Google Scholar] [CrossRef]
- Uringa, E.-J.; Lisaingo, K.; Pickett, H.A.; Brind’Amour, J.; Rohde, J.-H.; Zelensky, A.; Essers, J.; Lansdorp, P.M. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol. Boil. Cell 2012, 23, 2782–2792. [Google Scholar] [CrossRef]
- Barber, L.J.; Youds, J.L.; Ward, J.D.; McIlwraith, M.J.; O’Neil, N.J.; Petalcorin, M.I.; Martin, J.S.; Collis, S.J.; Cantor, S.B.; Auclair, M.; et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 2008, 135, 261–271. [Google Scholar] [CrossRef]
- Vannier, J.-B.; Pavicic-Kaltenbrunner, V.; Petalcorin, M.I.; Ding, H.; Boulton, S.J. RTEL1 Dismantles T Loops and Counteracts Telomeric G4-DNA to Maintain Telomere Integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannier, J.-B.; Sandhu, S.; Petalcorin, M.I.; Wu, X.; Nabi, Z.; Ding, H.; Boulton, S.J. RTEL1 Is a Replisome-Associated Helicase That Promotes Telomere and Genome-Wide Replication. Science 2013, 342, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Porreca, R.M.; Glousker, G.; Awad, A.; Fernandez, M.I.M.; Gibaud, A.; Naucke, C.; Cohen, S.B.; Bryan, T.M.; Tzfati, Y.; Draškovič, I.; et al. Human RTEL1 stabilizes long G-overhangs allowing telomerase-dependent over-extension. Nucleic Acids Res. 2018, 46, 4533–4545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Kan, Z.Y.; Yao, Y.; Wang, Q.; Hao, Y.H.; Tan, Z. G-quadruplex preferentially forms at the very 3’ end of vertebrate telomeric DNA. Nucleic Acids Res. 2008, 36, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Glousker, G.; Molczan, A.; Fox, A.J.; Lamm, N.; Dheekollu, J.; Weizman, O.-E.; Schertzer, M.; Wang, Z.; Vladimirova, O.; et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal–Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA 2013, 110, E3408–E3416. [Google Scholar] [CrossRef]
- Hemann, M.T.; Strong, M.A.; Hao, L.-Y.; Greider, C.W. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 2001, 107, 67–77. [Google Scholar] [CrossRef]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; De Lange, T. Mammalian telomeres end in a large duplex loo. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Margalef, P.; Kotsantis, P.; Borel, V.; Bellelli, R.; Panier, S.; Boulton, S.J. Stabilization of Reversed Replication Forks by Telomerase Drives Telomere Catastrophe. Cell 2018, 172, 439–453.e14. [Google Scholar] [CrossRef]
- Copeland, N.G.; Jenkins, N.A.; Court, D.L. Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001, 2, 769–779. [Google Scholar] [CrossRef]
- Cheng, K.; Wigley, D.B. DNA translocation mechanism of an XPD family helicase. eLife 2018, 7, 7. [Google Scholar] [CrossRef]
- Bloom, D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA. Am. J. Dis. Child. 1954, 88, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Ellis, N.A.; Groden, J.; Ye, T.-Z.; Straughen, J.; Lennon, D.J.; Ciocci, S.; Proytcheva, M.; German, J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 1995, 83, 655–666. [Google Scholar] [CrossRef]
- Chaganti, R.S.K.; Schönberg, S.; German, J. A Manyfold Increase in Sister Chromatid Exchanges in Bloom’s Syndrome Lymphocytes. Proc. Natl. Acad. Sci. USA 1974, 71, 4508–4512. [Google Scholar] [CrossRef] [PubMed]
- Karow, J.K.; Chakraverty, R.K.; Hickson, I.D. The Bloom’s Syndrome Gene Product Is a 3’-5’ DNA Helicase. J. Boil. Chem. 1997, 272, 30611–30614. [Google Scholar] [CrossRef]
- Karow, J.K.; Constantinou, A.; Li, J.-L.; West, S.C.; Hickson, I.D. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 2000, 97, 6504–6508. [Google Scholar] [CrossRef]
- Sun, H.; Karow, J.K.; Hickson, I.D.; Maizels, N. The Bloom’s Syndrome Helicase Unwinds G4 DNA. J. Boil. Chem. 1998, 273, 27587–27592. [Google Scholar] [CrossRef]
- Budhathoki, J.B.; Ray, S.; Urban, V.; Janscak, P.; Yodh, J.G.; Balci, H. RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 2014, 42, 11528–11545. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Hickson, I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 2003, 426, 870–874. [Google Scholar] [CrossRef]
- Cheok, C.F.; Wu, L.; Garcia, P.L.; Janscak, P.; Hickson, I.D. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005, 33, 3932–3941. [Google Scholar] [CrossRef]
- Ralf, C.; Hickson, I.D.; Wu, L. The Bloom’s Syndrome Helicase Can Promote the Regression of a Model Replication Fork. J. Boil. Chem. 2006, 281, 22839–22846. [Google Scholar] [CrossRef]
- Machwe, A.; Xiao, L.; Groden, J.; Orren, D.K. The Werner and Bloom Syndrome Proteins Catalyze Regression of a Model Replication Fork. Biochemistry 2006, 45, 13939–13946. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.-L.; North, P.S.; Hickson, I.D. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007, 26, 3397–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.L.; Hickson, I.D. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Boil. 2011, 22, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Barefield, C.; Karlseder, J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012, 40, 7358–7367. [Google Scholar] [CrossRef] [PubMed]
- Schawalder, J.; Paric, E.; Neff, N.F. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Boil. 2003, 4, 15. [Google Scholar]
- Lillard-Wetherell, K.; Machwe, A.; Langland, G.T.; Combs, K.A.; Behbehani, G.K.; Schonberg, S.A.; German, J.; Turchi, J.J.; Orren, D.K.; Groden, J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004, 13, 1919–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opresko, P.L.; von Kobbe, C.; Laine, J.P.; Harrigan, J.; Hickson, I.D.; Bohr, V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002, 277, 41110–41119. [Google Scholar] [CrossRef]
- Opresko, P.L.; Mason, P.A.; Podell, E.R.; Lei, M.; Hickson, I.D.; Cech, T.R.; Bohr, V.A.; Opresko, P. POT1 Stimulates RecQ Helicases WRN and BLM to Unwind Telomeric DNA Substrates. J. Boil. Chem. 2005, 280, 32069–32080. [Google Scholar] [CrossRef] [Green Version]
- Drosopoulos, W.C.; Kosiyatrakul, S.T.; Schildkraut, C.L. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Boil. 2015, 210, 191–208. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-Q.; Chen, C.-Y.; Xue, Y.; Hao, Y.-H.; Tan, Z. G-Quadruplex Hinders Translocation of BLM Helicase on DNA: A Real-Time Fluorescence Spectroscopic Unwinding Study and Comparison with Duplex Substrates. J. Am. Chem. Soc. 2010, 132, 10521–10527. [Google Scholar] [CrossRef]
- Falconer, E.; Hills, M.; Naumann, U.; Poon, S.S.S.; Chavez, E.A.; Sanders, A.D.; Zhao, Y.; Hirst, M.; Lansdorp, P.M. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods 2012, 9, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Sanders, A.D.; Falconer, E.; Hills, M.; Spierings, D.C.J.; Lansdorp, P.M. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat. Protoc. 2017, 12, 1151–1176. [Google Scholar] [CrossRef] [PubMed]
- Falconer, E.; Chavez, E.A.; Henderson, A.; Poon, S.S.; McKinney, S.; Brown, L.; Lansdorp, P.M. Identification of sister chromatids by DNA template strand sequences. Nature 2010, 463, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Goodwin, E.H. Strand-specific fluorescence in situ hybridization for determining orientation and direction of DNA sequences. Methods Mol. Biol. 1994, 33, 141–145. [Google Scholar] [PubMed]
- Latt, S.A. Microfluorometric Detection of Deoxyribonucleic Acid Replication in Human Metaphase Chromosomes. Proc. Natl. Acad. Sci. USA 1973, 70, 3395–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wietmarschen, N.; Lansdorp, P.M. Bromodeoxyuridine does not contribute to sister chromatid exchange events in normal or Bloom syndrome cells. Nucleic Acids Res. 2016, 44, 6787–6793. [Google Scholar] [CrossRef]
- van Wietmarschen, N.; Merzouk, S.; Halsema, N.; Spierings, D.C.; Guryev, V.; Lansdorp, P.M. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat. Commun. 2018, 9, 271. [Google Scholar] [CrossRef]
- Nguyen, G.H.; Tang, W.; Robles, A.I.; Beyer, R.P.; Gray, L.T.; Welsh, J.A.; Schetter, A.J.; Kumamoto, K.; Wang, X.W.; Hickson, I.D.; et al. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. USA 2014, 111, 9905–9910. [Google Scholar] [CrossRef] [Green Version]
- Smestad, J.A.; Maher, L.J., 3rd. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med. Genet. 2015, 16, 91. [Google Scholar] [CrossRef]
- Grierson, P.M.; Acharya, S.; Groden, J. Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription. Mutat. Res. 2013, 743, 89–96. [Google Scholar] [CrossRef]
- Grierson, P.M.; Lillard, K.; Behbehani, G.K.; Combs, K.A.; Bhattacharyya, S.; Acharya, S.; Groden, J. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum. Mol. Genet. 2012, 21, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Wanrooij, P.H.; Uhler, J.P.; Shi, Y.; Westerlund, F.; Falkenberg, M.; Gustafsson, C.M. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012, 40, 10334–10344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rout, M.; Sali, A. Principles for Integrative Structural Biology Studies. Cell 2019, 177, 1384–1403. [Google Scholar] [CrossRef] [PubMed]
- van Wietmarschen, N.; Moradian, A.; Morin, G.B.; Lansdorp, P.M.; Uringa, E.J. The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. J. Biol. Chem. 2012, 287, 43351–43358. [Google Scholar] [CrossRef] [PubMed]
- Gari, K.; Ortiz, A.M.L.; Borel, V.; Flynn, H.; Skehel, J.M.; Boulton, S.J. MMS19 Links Cytoplasmic Iron-Sulfur Cluster Assembly to DNA Metabolism. Science 2012, 337, 243–245. [Google Scholar] [CrossRef]
- Veatch, J.R.; McMurray, M.A.; Nelson, Z.W.; Gottschling, D.E. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 2009, 137, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, J.; Makrantoni, V.; Ingledew, W.J.; Stark, M.J.; White, M.F. The DNA Repair Helicases XPD and FancJ Have Essential Iron-Sulfur Domains. Mol. Cell 2006, 23, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.K.; Silva, R.M.B.; O’Brien, E. Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication. Annu. Rev. Biochem. 2019, 88, 163–190. [Google Scholar] [CrossRef] [Green Version]
- Einarson, O.J.; Sen, D. Self-biotinylation of DNA G-quadruplexes via intrinsic peroxidase activity. Nucleic Acids Res. 2017, 45, 9813–9822. [Google Scholar] [CrossRef]
- Robertson, M. Biology in the 1980s, plus or minus a decade. Nature 1980, 285, 358–359. [Google Scholar] [CrossRef]
- Sanders, C.M. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010, 430, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.-M.; Wu, W.-Q.; Duan, X.-L.; Liu, N.-N.; Li, H.-H.; Fu, J.; Dou, S.-X.; Li, M.; Xi, X.-G. Molecular mechanism of G-quadruplex unwinding helicase: Sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 2015, 466, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef] [Green Version]
- Ribeyre, C.; Lopes, J.; Boulé, J.-B.; Piazza, A.; Guédin, A.; Zakian, V.A.; Mergny, J.-L.; Nicolas, A. The Yeast Pif1 Helicase Prevents Genomic Instability Caused by G-Quadruplex-Forming CEB1 Sequences In Vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Voon, H.P.J.; Xella, B.; Scott, C.; Clynes, D.; Babbs, C.; Ayyub, H.; Kerry, J.; Sharpe, J.A.; Sloane-Stanley, J.A.; et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017, 18, 914–928. [Google Scholar] [CrossRef]
- Crabbe, L.; Verdun, R.E.; Haggblom, C.I.; Karlseder, J. Defective Telomere Lagging Strand Synthesis in Cells Lacking WRN Helicase Activity. Science 2004, 306, 1951–1953. [Google Scholar] [CrossRef]
- Johnson, J.E.; Cao, K.; Ryvkin, P.; Wang, L.S.; Johnson, F.B. Altered gene expression in the Werner and Bloom syndromes is associated with sequences with G-quadruplex forming potential. Nucleic Acids Res. 2010, 38, 1114–1122. [Google Scholar] [CrossRef]
- Huber, M.D.; Lee, D.C.; Maizels, N. G4 DNA unwinding by BLM and Sgs1p: Substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002, 30, 3954–3961. [Google Scholar] [CrossRef]
- Gray, L.T.; Vallur, A.C.; Eddy, J.; Maizels, N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Methods 2014, 10, 313–318. [Google Scholar] [CrossRef]
- Bharti, S.K.; Sommers, J.A.; George, F.; Kuper, J.; Hamon, F.; Shin-Ya, K.; Teulade-Fichou, M.-P.; Kisker, C.; Brosh, R.M. Specialization among Iron-Sulfur Cluster Helicases to Resolve G-quadruplex DNA Structures That Threaten Genomic Stability. J. Boil. Chem. 2013, 288, 28217–28229. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lansdorp, P.; van Wietmarschen, N. Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo. Genes 2019, 10, 870. https://doi.org/10.3390/genes10110870
Lansdorp P, van Wietmarschen N. Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo. Genes. 2019; 10(11):870. https://doi.org/10.3390/genes10110870
Chicago/Turabian StyleLansdorp, Peter, and Niek van Wietmarschen. 2019. "Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo" Genes 10, no. 11: 870. https://doi.org/10.3390/genes10110870



