Teratogenic Rubella Virus Alters the Endodermal Differentiation Capacity of Human Induced Pluripotent Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cultivation
2.2. Directed and Undirected Differentiation of iPSCs
2.3. Embryoid Body Formation
2.4. Virus Infection and Interferon Assays
2.5. Calcein Live Cell Staining
2.6. RNA Isolation
2.7. Microarray Gene Expression Analysis and SOM Portrayal
2.8. Quantitative Real-Time PCR Analysis of Viral and Cellular RNA
2.9. Immunofluorescence
2.10. Statistical Analysis
3. Results
3.1. In the Presence of RV, iPSCs Maintain Pluripotent Properties and Lineage Identity after Initiation of Differentiation
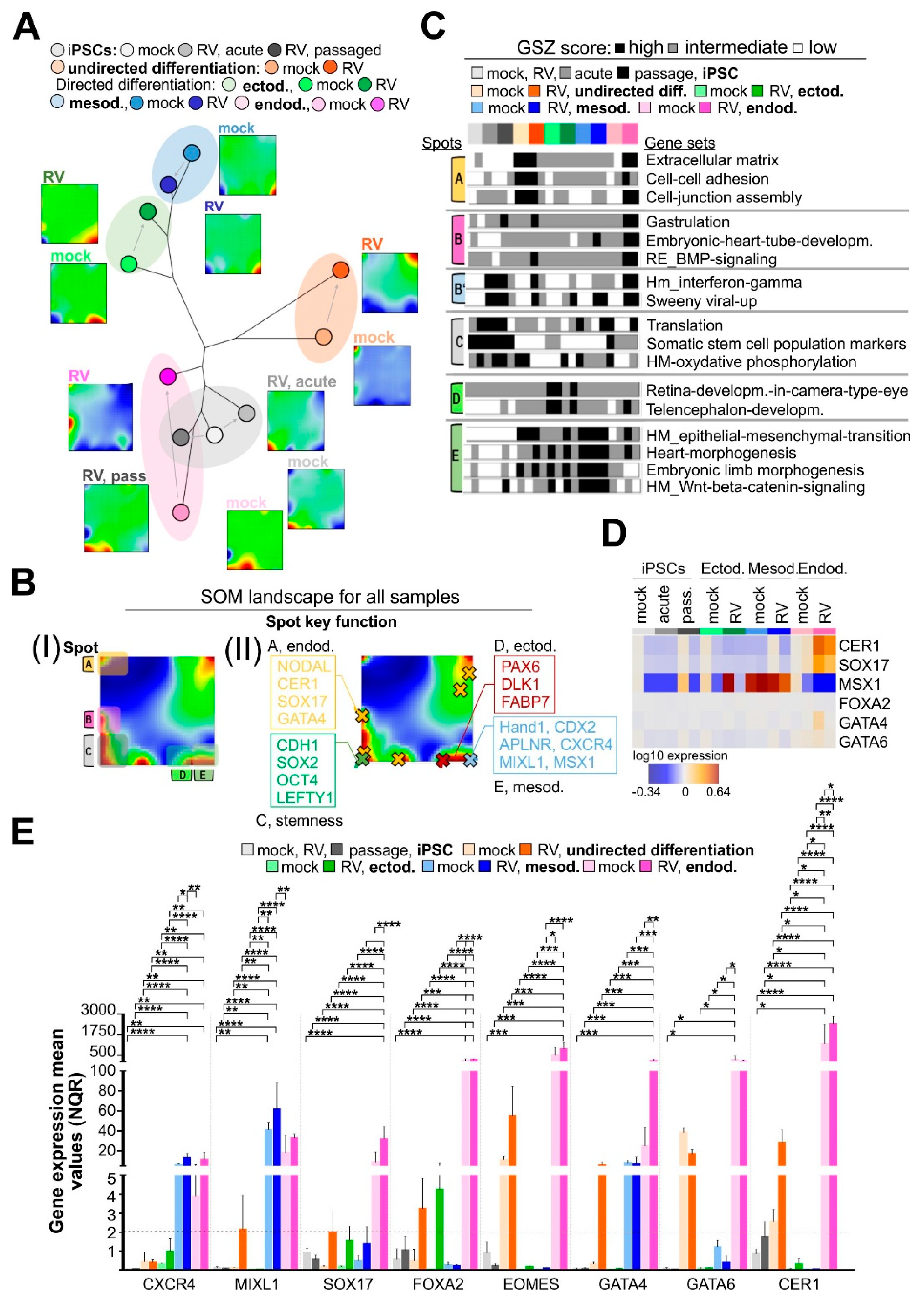
3.2. High-Resolution Transcriptomic Maps Reveal Modules of Coregulated Genes Promoted by RV Infection during Endodermal Differentiation
3.3. RV Infection Activates IFN Type III Response Pathways on iPSCs and Derived Lineages
3.4. RV Infection Is Associated with Chromatin Remodeling
3.5. RV Infection Impairs Aggregation of iPSCs into Embryoid Bodies
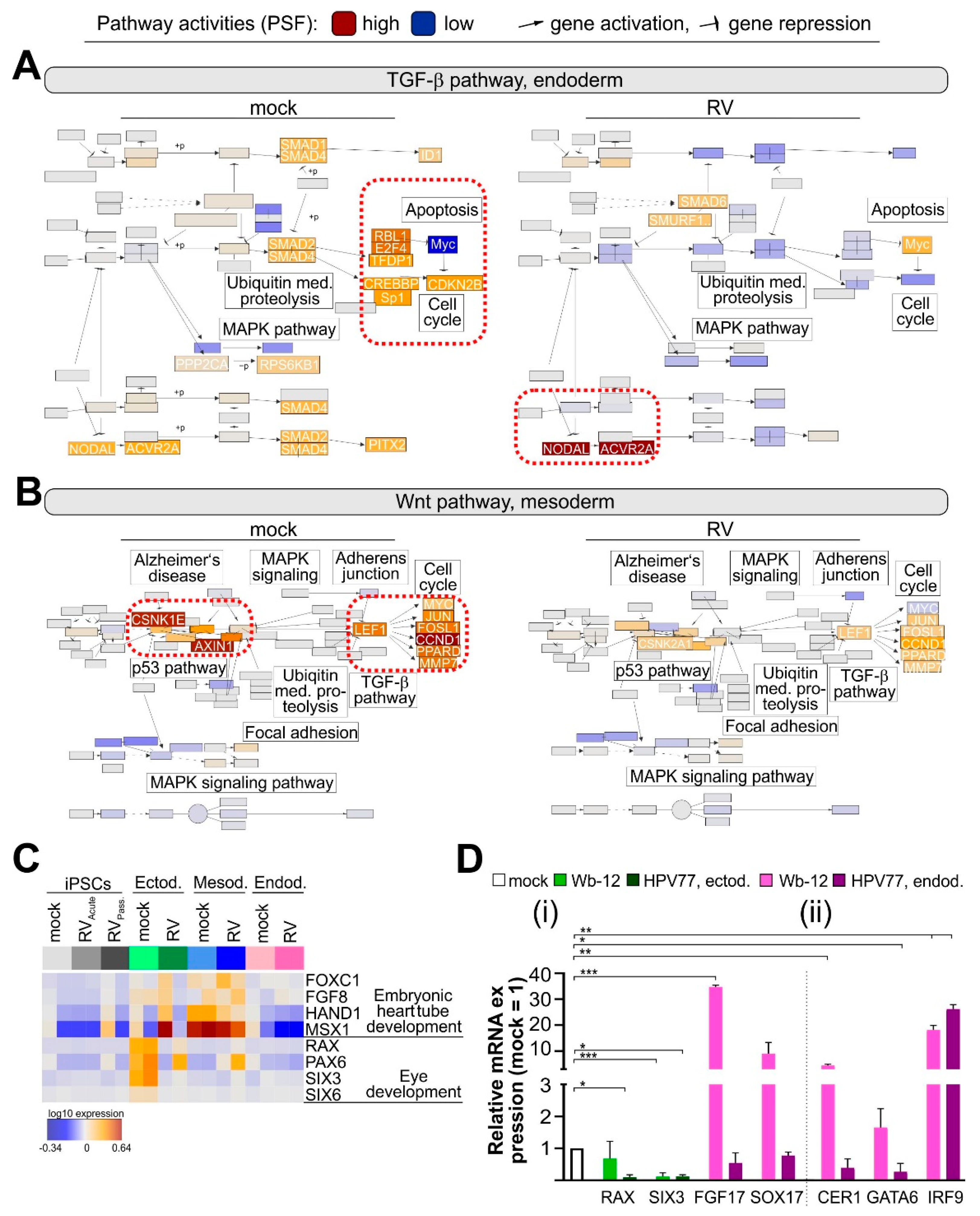
3.6. RV Infection Specifically Affects Developmental Pathways during Endodermal Differentiation
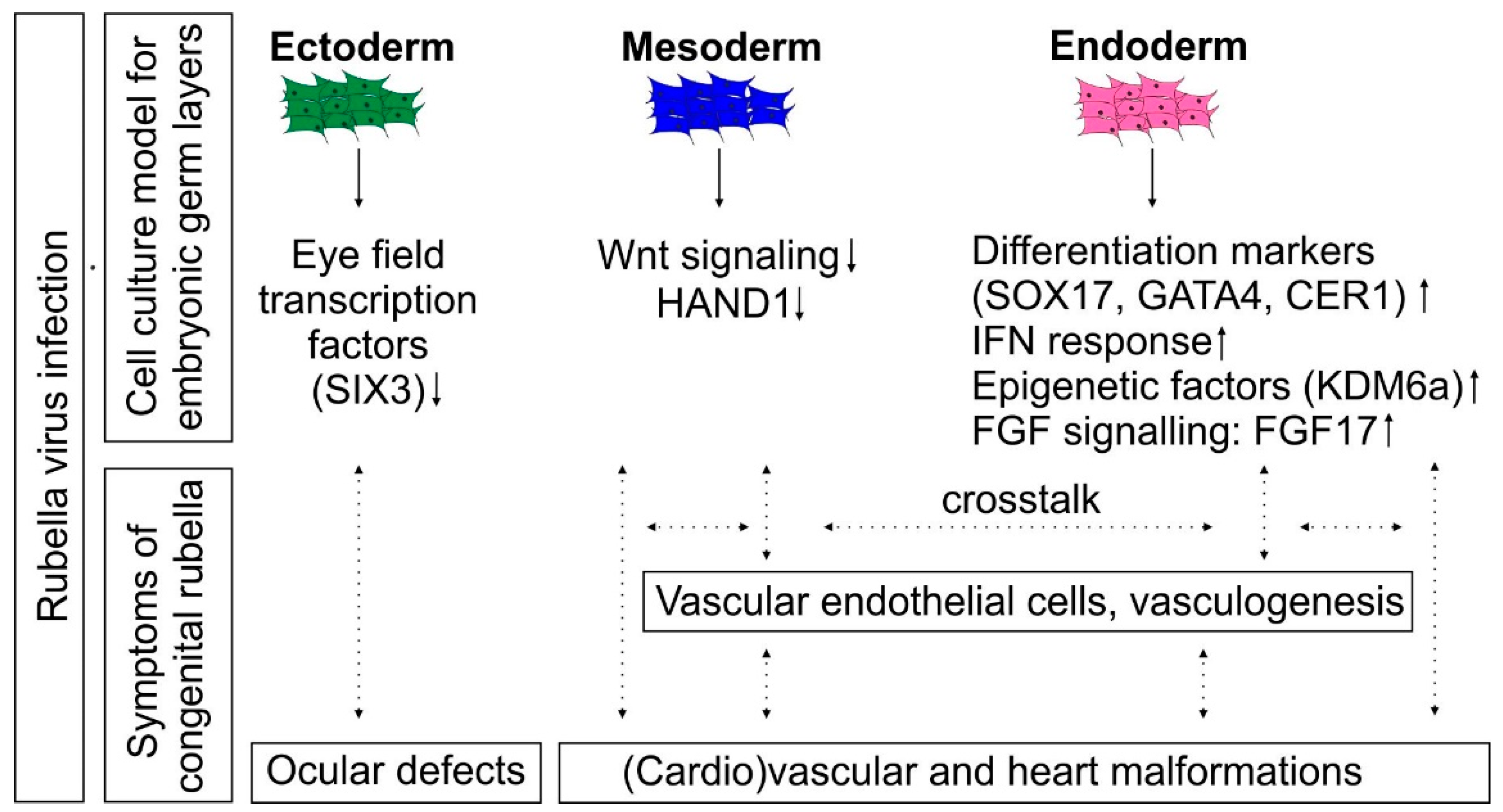
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dudgeon, J.A. Congenital rubella. J. Pediatr. 1975, 87, 1078–1086. [Google Scholar] [CrossRef]
- Duszak, R.S. Congenital rubella syndrome—Major review. Optometry 2009, 80, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Lazar, M.; Perelygina, L.; Martines, R.; Greer, P.; Paddock, C.D.; Peltecu, G.; Lupulescu, E.; Icenogle, J.; Zaki, S.R. Immunolocalization and distribution of rubella antigen in fatal congenital rubella syndrome. EBioMedicine 2016, 3, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Freij, B.J.; South, M.A.; Sever, J.L. Maternal rubella and the congenital rubella syndrome. Clin. Perinatol. 1988, 15, 247–257. [Google Scholar] [CrossRef]
- Bouthry, E.; Picone, O.; Hamdi, G.; Grangeot-Keros, L.; Ayoubi, J.M.; Vauloup-Fellous, C. Rubella and pregnancy: Diagnosis, management and outcomes. Prenat. Diagn. 2014, 34, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Enders, G.; Nickerl-Pacher, U.; Miller, E.; Cradock-Watson, J.E. Outcome of confirmed periconceptional maternal rubella. Lancet 1988, 1, 1445–1447. [Google Scholar] [CrossRef]
- Rossant, J.; Tam, P.P.L. Exploring early human embryo development. Science 2018, 360, 1075–1076. [Google Scholar] [CrossRef] [PubMed]
- Kugler, J.; Huhse, B.; Tralau, T.; Luch, A. Embryonic stem cells and the next generation of developmental toxicity testing. Expert Opin. Drug Metab. Toxicol. 2017, 13, 833–841. [Google Scholar] [CrossRef]
- Van Vliet, P.; Wu, S.M.; Zaffran, S.; Puceat, M. Early cardiac development: A view from stem cells to embryos. Cardiovasc. Res. 2012, 96, 352–362. [Google Scholar] [CrossRef]
- Simunovic, M.; Brivanlou, A.H. Embryoids, organoids and gastruloids: New approaches to understanding embryogenesis. Development 2017, 144, 976–985. [Google Scholar] [CrossRef]
- Meganathan, K.; Jagtap, S.; Wagh, V.; Winkler, J.; Gaspar, J.A.; Hildebrand, D.; Trusch, M.; Lehmann, K.; Hescheler, J.; Schluter, H.; et al. Identification of thalidomide-specific transcriptomics and proteomics signatures during differentiation of human embryonic stem cells. PLoS ONE 2012, 7, e44228. [Google Scholar] [CrossRef]
- Luz, A.L.; Tokar, E.J. Pluripotent stem cells in developmental toxicity testing: A review of methodological advances. Toxicol. Sci. 2018, 165, 31–39. [Google Scholar] [CrossRef]
- Hubner, D.; Jahn, K.; Pinkert, S.; Bohnke, J.; Jung, M.; Fechner, H.; Rujescu, D.; Liebert, U.G.; Claus, C. Infection of ipsc lines with miscarriage-associated coxsackievirus and measles virus and teratogenic rubella virus as a model for viral impairment of early human embryogenesis. ACS Infect. Dis. 2017, 3, 886–897. [Google Scholar] [CrossRef]
- Wu, X.; Dao Thi, V.L.; Huang, Y.; Billerbeck, E.; Saha, D.; Hoffmann, H.H.; Wang, Y.; Silva, L.A.V.; Sarbanes, S.; Sun, T.; et al. Intrinsic immunity shapes viral resistance of stem cells. Cell 2018, 172, 423–438. [Google Scholar] [CrossRef]
- Hong, X.X.; Carmichael, G.G. Innate immunity in pluripotent human cells: Attenuated response to interferon-beta. J. Biol. Chem. 2013, 288, 16196–16205. [Google Scholar] [CrossRef]
- Dziedzicka, D.; Markouli, C.; Barbe, L.; Spits, C.; Sermon, K.; Geens, M. A high proliferation rate is critical for reproducible and standardized embryoid body formation from laminin-521-based human pluripotent stem cell cultures. Stem Cell Rev. 2016, 12, 721–730. [Google Scholar] [CrossRef]
- Wirth, H.; Loffler, M.; von Bergen, M.; Binder, H. Expression cartography of human tissues using self organizing maps. BMC Bioinform. 2011, 12, 306. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Wirth, H.; von Bergen, M.; Murugaiyan, J.; Rosler, U.; Stokowy, T.; Binder, H. Maldi-typing of infectious algae of the genus prototheca using som portraits. J. Microbiol. Methods 2012, 88, 83–97. [Google Scholar] [CrossRef]
- Loffler-Wirth, H.; Kalcher, M.; Binder, H. Opossom: R-package for high-dimensional portraying of genome-wide expression landscapes on bioconductor. Bioinformatics 2015, 31, 3225–3227. [Google Scholar] [CrossRef]
- Nersisyan, L.; Löffler-Wirth, H.; Arakelyan, A.; Binder, H. Gene set- and pathway- centered knowledge discovery assigns transcriptional activation patterns in brain, blood, and colon cancer: A bioinformatics perspective. Int. J. Knowl. Discov. Bioinform. 2014, 4, 46–69. [Google Scholar] [CrossRef]
- Claus, C.; Bergs, S.; Emmrich, N.C.; Hubschen, J.M.; Mankertz, A.; Liebert, U.G. A sensitive one-step taqman amplification approach for detection of rubella virus clade i and ii genotypes in clinical samples. Arch. Virol. 2017, 162, 477–486. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. Qbase relative quantification framework and software for management and automated analysis of real-time quantitative pcr data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, K.; Liu, Y.; Fu, Y.; Gao, S.; Gong, P.; Wang, H.; Zhou, Z.; Zeng, M.; Wu, Z.; et al. Telomere heterogeneity linked to metabolism and pluripotency state revealed by simultaneous analysis of telomere length and rna-seq in the same human embryonic stem cell. BMC Biol. 2017, 15, 114. [Google Scholar] [CrossRef]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Chu, L.F.; Leng, N.; Zhang, J.; Hou, Z.; Mamott, D.; Vereide, D.T.; Choi, J.; Kendziorski, C.; Stewart, R.; Thomson, J.A. Single-cell rna-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol. 2016, 17, 173. [Google Scholar] [CrossRef]
- Hopp, L.; Loeffler-Wirth, H.; Nersisyan, L.; Arakelyan, A.; Binder, H. Footprints of sepsis framed within community acquired pneumonia in the blood transcriptome. Front Immunol 2018, 9, 1620. [Google Scholar] [CrossRef]
- Andres-Terre, M.; McGuire, H.M.; Pouliot, Y.; Bongen, E.; Sweeney, T.E.; Tato, C.M.; Khatri, P. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity 2015, 43, 1199–1211. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Wong, H.R.; Khatri, P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med. 2016, 8, 346ra91. [Google Scholar] [CrossRef]
- Schmidtke, L.; Schrick, K.; Saurin, S.; Kafer, R.; Gather, F.; Weinmann-Menke, J.; Kleinert, H.; Pautz, A. The kh-type splicing regulatory protein (ksrp) regulates type iii interferon expression post-transcriptionally. Biochem. J. 2019, 476, 333–352. [Google Scholar] [CrossRef]
- Winzen, R.; Thakur, B.K.; Dittrich-Breiholz, O.; Shah, M.; Redich, N.; Dhamija, S.; Kracht, M.; Holtmann, H. Functional analysis of ksrp interaction with the au-rich element of interleukin-8 and identification of inflammatory mrna targets. Mol. Cell. Biol. 2007, 27, 8388–8400. [Google Scholar] [CrossRef]
- Dambacher, S.; Hahn, M.; Schotta, G. Epigenetic regulation of development by histone lysine methylation. Heredity 2010, 105, 24–37. [Google Scholar] [CrossRef]
- Thalheim, T.; Hopp, L.; Binder, H.; Aust, G.; Galle, J. On the cooperation between epigenetics and transcription factor networks in the specification of tissue stem cells. Epigenomes 2018, 2, 20. [Google Scholar] [CrossRef]
- Grandy, R.A.; Whitfield, T.W.; Wu, H.; Fitzgerald, M.P.; VanOudenhove, J.J.; Zaidi, S.K.; Montecino, M.A.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; et al. Genome-wide studies reveal that h3k4me3 modification in bivalent genes is dynamically regulated during the pluripotent cell cycle and stabilized upon differentiation. Mol. Cell. Biol. 2016, 36, 615–627. [Google Scholar] [CrossRef]
- Bratt-Leal, A.M.; Carpenedo, R.L.; McDevitt, T.C. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnol. Prog. 2009, 25, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.T.; Che, S.Y.; Su, Z.; Zheng, S.Y.; Zhang, H.Y.; Yang, S.L.; Li, W.D.; Liu, J.P. Cd90 promotes cell migration, viability and sphere-forming ability of hepatocellular carcinoma cells. Int. J. Mol. Med. 2018, 41, 946–954. [Google Scholar] [CrossRef]
- Liu, C.; Peng, G.; Jing, N. Tgf-beta signaling pathway in early mouse development and embryonic stem cells. Acta Biochim. Biophys. Sin. 2018, 50, 68–73. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Gill, J.G.; Kyba, M.; Murphy, T.L.; Murphy, K.M. Canonical wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 2006, 133, 3787–3796. [Google Scholar] [CrossRef]
- Weed, L.S.; Mills, J.A. Strategies for retinal cell generation from human pluripotent stem cells. Stem Cell Investig. 2017, 4, 65. [Google Scholar] [CrossRef]
- Diacou, R.; Zhao, Y.; Zheng, D.; Cvekl, A.; Liu, W. Six3 and six6 are jointly required for the maintenance of multipotent retinal progenitors through both positive and negative regulation. Cell Rep. 2018, 25, 2510–2523. [Google Scholar] [CrossRef]
- Worley, K.E.; Rico-Varela, J.; Ho, D.; Wan, L.Q. Teratogen screening with human pluripotent stem cells. Integr. Biol. 2018, 10, 491–501. [Google Scholar] [CrossRef] [Green Version]
- Rathjen, J. The states of pluripotency: Pluripotent lineage development in the embryo and in the dish. ISRN Stem Cells 2014, 2014, 19. [Google Scholar] [CrossRef]
- Fu, Y.; Zhou, Z.; Wang, H.; Gong, P.; Guo, R.; Wang, J.; Lu, X.; Qi, F.; Liu, L. Ifitm1 suppresses expression of human endogenous retroviruses in human embryonic stem cells. FEBS Open Bio 2017, 7, 1102–1110. [Google Scholar] [CrossRef]
- Wolf, D.; Goff, S.P. Embryonic stem cells use zfp809 to silence retroviral dnas. Nature 2009, 458, 1201–1204. [Google Scholar] [CrossRef]
- Eggenberger, J.; Blanco-Melo, D.; Panis, M.; Brennand, K.J.; tenOever, B.R. Type i interferon response impairs differentiation potential of pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2019, 116, 1384–1393. [Google Scholar] [CrossRef]
- Pervolaraki, K.; Talemi, S.R.; Albrecht, D.; Bormann, F.; Bamford, C.; Mendoza, J.L.; Garcia, K.C.; McLauchlan, J.; Hofer, T.; Stanifer, M.L.; et al. Differential induction of interferon stimulated genes between type i and type iii interferons is independent of interferon receptor abundance. PLoS Pathog. 2018, 14, e1007420. [Google Scholar] [CrossRef]
- Zahedi-Amiri, A.; Sequiera, G.L.; Dhingra, S.; Coombs, K.M. Influenza a virus-triggered autophagy decreases the pluripotency of human-induced pluripotent stem cells. Cell Death Dis. 2019, 10, 337. [Google Scholar] [CrossRef]
- Heavner, W.; Pevny, L. Eye development and retinogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008391. [Google Scholar] [CrossRef]
- Geyer, H.; Bauer, M.; Neumann, J.; Ludde, A.; Rennert, P.; Friedrich, N.; Claus, C.; Perelygina, L.; Mankertz, A. Gene expression profiling of rubella virus infected primary endothelial cells of fetal and adult origin. Virol. J. 2016, 13, 21. [Google Scholar] [CrossRef]
- Lough, J.; Sugi, Y. Endoderm and heart development. Dev. Dyn. 2000, 217, 327–342. [Google Scholar] [CrossRef]
- Mulloy, B.; Rider, C.C. The bone morphogenetic proteins and their antagonists. Vitam. Horm. 2015, 99, 63–90. [Google Scholar]
- Foley, A.C.; Korol, O.; Timmer, A.M.; Mercola, M. Multiple functions of cerberus cooperate to induce heart downstream of nodal. Dev. Biol. 2007, 303, 57–65. [Google Scholar] [CrossRef]
- Vokes, S.A.; Krieg, P.A. Endoderm is required for vascular endothelial tube formation, but not for angioblast specification. Development 2002, 129, 775–785. [Google Scholar]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef]
- Hofstetter, C.; Kampka, J.M.; Huppertz, S.; Weber, H.; Schlosser, A.; Muller, A.M.; Becker, M. Inhibition of kdm6 activity during murine esc differentiation induces DNA damage. J. Cell Sci. 2016, 129, 788–803. [Google Scholar] [CrossRef]
- Fisher, J.B.; Pulakanti, K.; Rao, S.; Duncan, S.A. Gata6 is essential for endoderm formation from human pluripotent stem cells. Biol Open 2017, 6, 1084–1095. [Google Scholar] [CrossRef]
- Pfeiffer, M.J.; Quaranta, R.; Piccini, I.; Fell, J.; Rao, J.; Ropke, A.; Seebohm, G.; Greber, B. Cardiogenic programming of human pluripotent stem cells by dose-controlled activation of eomes. Nat. Commun. 2018, 9, 440. [Google Scholar] [CrossRef]
- Pfister, S.; Jones, V.J.; Power, M.; Truisi, G.L.; Khoo, P.L.; Steiner, K.A.; Kanai-Azuma, M.; Kanai, Y.; Tam, P.P.; Loebel, D.A. Sox17-dependent gene expression and early heart and gut development in sox17-deficient mouse embryos. Int. J. Dev. Biol. 2011, 55, 45–58. [Google Scholar] [CrossRef]
- Serrano, A.G.; Gandillet, A.; Pearson, S.; Lacaud, G.; Kouskoff, V. Contrasting effects of sox17- and sox18-sustained expression at the onset of blood specification. Blood 2010, 115, 3895–3898. [Google Scholar] [CrossRef]
- George, R.M.; Firulli, A.B. Hand factors in cardiac development. Anat. Rec. 2019, 302, 101–107. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bilz, N.C.; Willscher, E.; Binder, H.; Böhnke, J.; Stanifer, M.L.; Hübner, D.; Boulant, S.; Liebert, U.G.; Claus, C. Teratogenic Rubella Virus Alters the Endodermal Differentiation Capacity of Human Induced Pluripotent Stem Cells. Cells 2019, 8, 870. https://doi.org/10.3390/cells8080870
Bilz NC, Willscher E, Binder H, Böhnke J, Stanifer ML, Hübner D, Boulant S, Liebert UG, Claus C. Teratogenic Rubella Virus Alters the Endodermal Differentiation Capacity of Human Induced Pluripotent Stem Cells. Cells. 2019; 8(8):870. https://doi.org/10.3390/cells8080870
Chicago/Turabian StyleBilz, Nicole C., Edith Willscher, Hans Binder, Janik Böhnke, Megan L. Stanifer, Denise Hübner, Steeve Boulant, Uwe G. Liebert, and Claudia Claus. 2019. "Teratogenic Rubella Virus Alters the Endodermal Differentiation Capacity of Human Induced Pluripotent Stem Cells" Cells 8, no. 8: 870. https://doi.org/10.3390/cells8080870





