The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies
Abstract
:1. Introduction
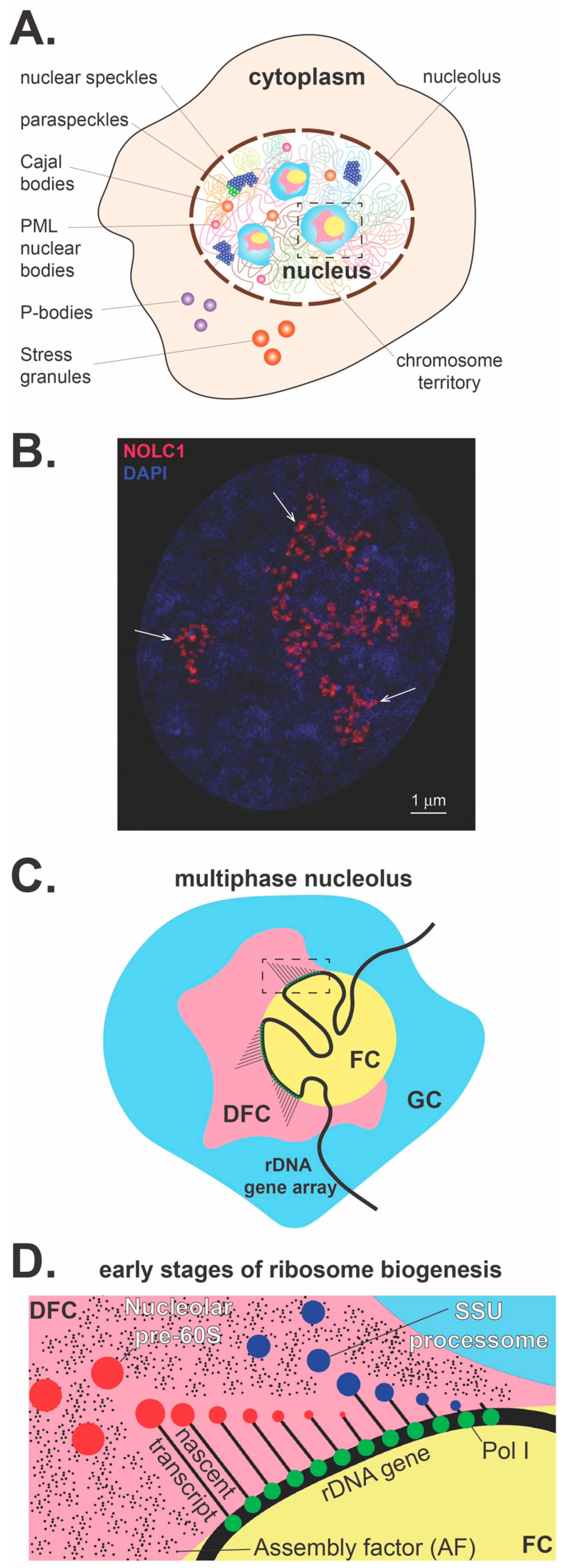
2. Phase Separation and the Dynamic Nature of the Nucleolus
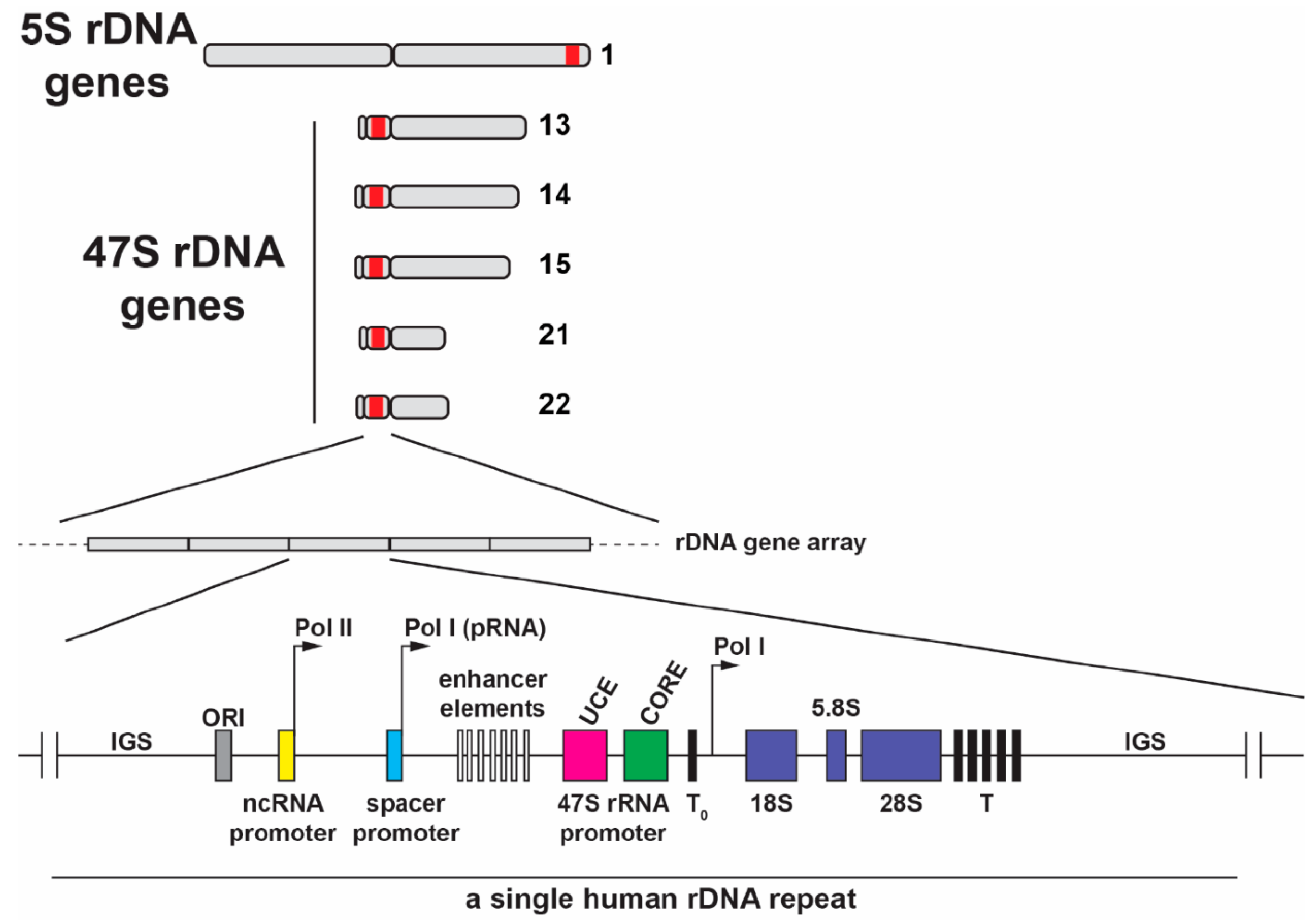
3. Organization of Ribosomal Genes
How Non-Coding RNA Maintains Heterochromatin by rDNA Silencing
4. Ribosomal Genes and Genomic Instability
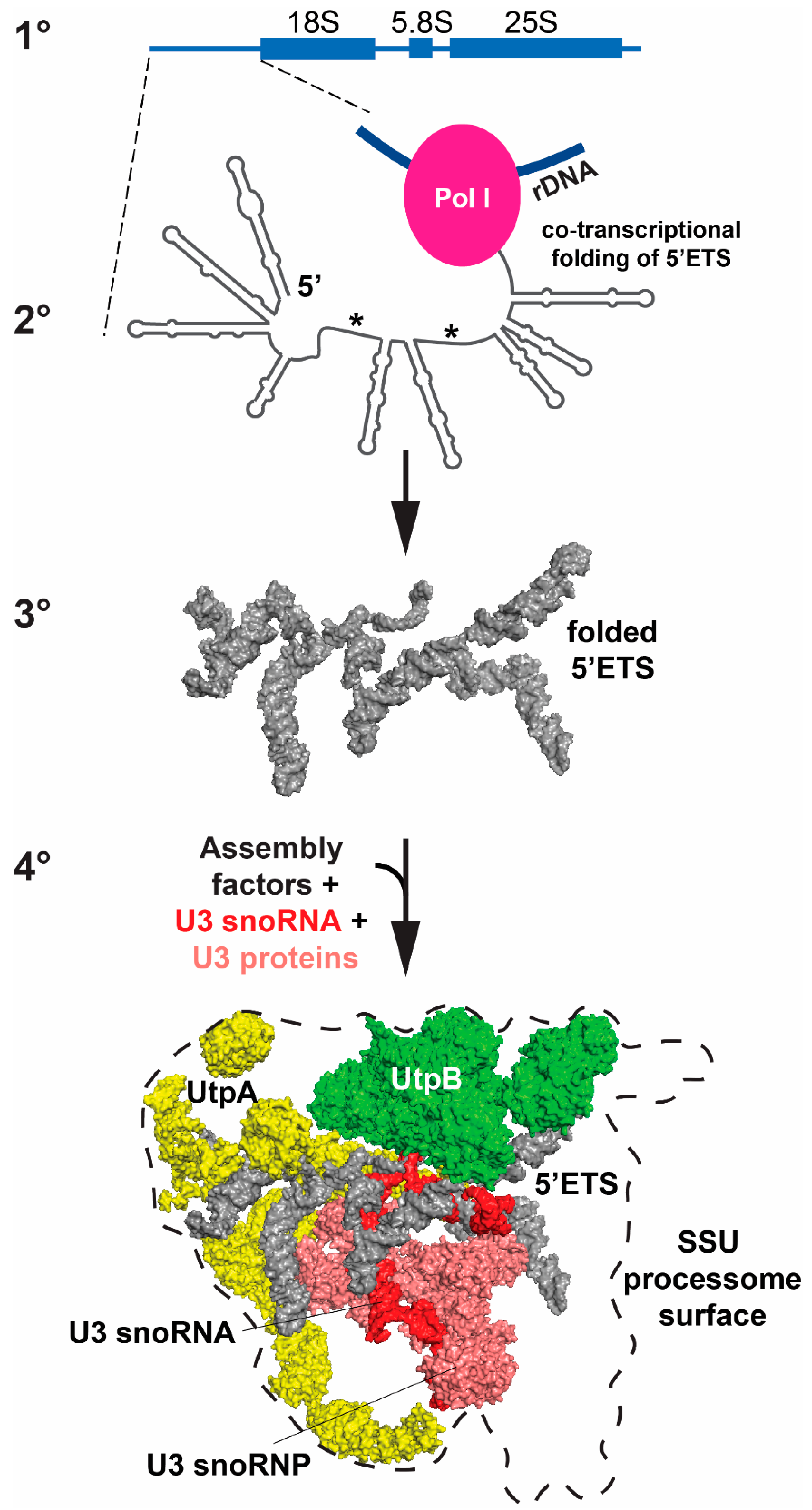
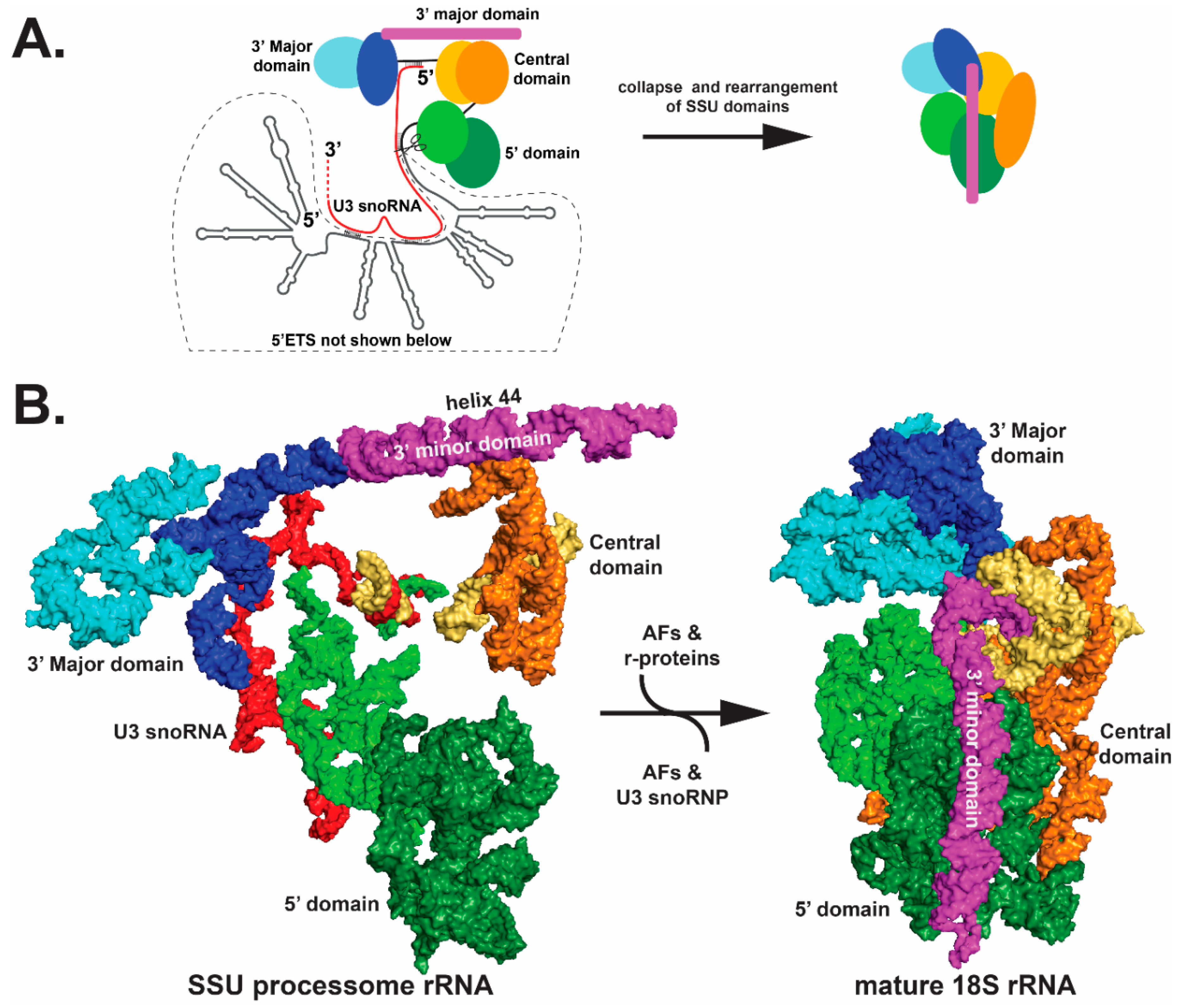
5. Structural Insights on Pre-rRNA Processing from Cryo-Electron Microscopy
6. Aging, Nucleolar Pathology and Ribosomopathies
7. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AF | assembly factor |
| BMC | biomolecular condensate |
| CPK | central pseudoknot |
| DFC | dense fibrillar component |
| ETS | external-transcribed spacer |
| FC | fibrillar left |
| GC | granular component |
| IDR | intrinsically disordered region |
| IGS | intergenic spacer |
| ITS | internal-transcribed spacer |
| LSU | Large subunit |
| NOR | nucleolus organizer region |
| NoRC | nucleolar repressive complex |
| pRNA | promoter-associated RNA |
| RNP | ribonucleoprotein |
| RNA pol | RNA polymerase |
| r-protein | ribosomal protein |
| rRNA | ribosomal RNA |
| snoRNP | small nucleolar ribonucleoprotein |
| SSU processome | Small Subunit processome |
References
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, I.A.; Bartek, J.; Dundr, M. Phase separated microenvironments inside the cell nucleus are linked to disease and regulate epigenetic state, transcription and RNA processing. Semin. Cell Dev. Biol. 2019, 90, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, H. Phase Separation, Transition, and Autophagic Degradation of Proteins in Development and Pathogenesis. Trends Cell Biol. 2019, 29, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, J.B.; Hyman, A.A.; Boke, E. Organization and Function of Non-dynamic Biomolecular Condensates. Trends Biochem. Sci. 2018, 43, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, I.A.; Dundr, M. Nuclear bodies: Built to boost. J. Cell Biol. 2016, 213, 509–511. [Google Scholar] [CrossRef]
- Stroberg, W.; Schnell, S. Do Cellular Condensates Accelerate Biochemical Reactions? Lessons from Microdroplet Chemistry. Biophys. J. 2018, 115, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [Green Version]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 2016, 5, e13571. [Google Scholar] [CrossRef]
- Fay, M.M.; Anderson, P.J. The Role of RNA in Biological Phase Separations. J. Mol. Biol. 2018, 430, 4685–4701. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Tollervey, D. Like attracts like: Getting RNA processing together in the nucleus. Science 2000, 288, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA 1972, 69, 3394–3398. [Google Scholar] [CrossRef] [PubMed]
- Kalmarova, M.; Smirnov, E.; Masata, M.; Koberna, K.; Ligasova, A.; Popov, A.; Raska, I. Positioning of NORs and NOR-bearing chromosomes in relation to nucleoli. J. Struct. Biol. 2007, 160, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parada, L.A.; McQueen, P.G.; Munson, P.J.; Misteli, T. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 2002, 12, 1692–1697. [Google Scholar] [CrossRef]
- Mao, Y.S.; Sunwoo, H.; Zhang, B.; Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011, 13, 95–101. [Google Scholar] [CrossRef]
- Shevtsov, S.P.; Dundr, M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011, 13, 167–173. [Google Scholar] [CrossRef]
- Chujo, T.; Hirose, T. Nuclear Bodies Built on Architectural Long Noncoding RNAs: Unifying Principles of Their Construction and Function. Mol. Cells 2017, 40, 889–896. [Google Scholar] [CrossRef]
- Chujo, T.; Yamazaki, T.; Kawaguchi, T.; Kurosaka, S.; Takumi, T.; Nakagawa, S.; Hirose, T. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017, 36, 1447–1462. [Google Scholar] [CrossRef]
- Michieletto, D.; Gilbert, N. Role of nuclear RNA in regulating chromatin structure and transcription. Curr. Opin. Cell Biol. 2019, 58, 120–125. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V.; Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem. Sci. 2019, 44, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Nguemaha, V.; Zhou, H.X. Liquid-Liquid Phase Separation of Patchy Particles Illuminates Diverse Effects of Regulatory Components on Protein Droplet Formation. Sci. Rep. 2018, 8, 6728. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.T.; Elbaum-Garfinkle, S.; Holehouse, A.S.; Chen, C.C.; Feric, M.; Arnold, C.B.; Priestley, R.D.; Pappu, R.V.; Brangwynne, C.P. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017, 9, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, D.; Glowatz, H.; Schlumpberger, M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr. Protoc. Mol. Biol. 2013, 103. [Google Scholar] [CrossRef]
- Albert, B.; Perez-Fernandez, J.; Leger-Silvestre, I.; Gadal, O. Regulation of ribosomal RNA production by RNA polymerase I: Does elongation come first? Genet. Res. Int. 2012, 2012, 276948. [Google Scholar] [CrossRef] [PubMed]
- Miller, O.L., Jr.; Beatty, B.R. Visualization of nucleolar genes. Science 1969, 164, 955–957. [Google Scholar] [CrossRef]
- Jackson, D.A.; Iborra, F.J.; Manders, E.M.; Cook, P.R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 1998, 9, 1523–1536. [Google Scholar] [CrossRef]
- Denissov, S.; Lessard, F.; Mayer, C.; Stefanovsky, V.; van Driel, M.; Grummt, I.; Moss, T.; Stunnenberg, H.G. A model for the topology of active ribosomal RNA genes. EMBO Rep. 2011, 12, 231–237. [Google Scholar] [CrossRef] [Green Version]
- Dundr, M. Nuclear bodies: Multifunctional companions of the genome. Curr. Opin. Cell Biol. 2012, 24, 415–422. [Google Scholar] [CrossRef]
- Koberna, K.; Malinsky, J.; Pliss, A.; Masata, M.; Vecerova, J.; Fialova, M.; Bednar, J.; Raska, I. Ribosomal genes in focus: New transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J. Cell Biol. 2002, 157, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Schofer, C.; Weipoltshammer, K. Nucleolus and chromatin. Histochem Cell Biol 2018, 150, 209–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelletier, J.; Thomas, G.; Volarevic, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Stults, D.M.; Killen, M.W.; Pierce, H.H.; Pierce, A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008, 18, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.G.; Branco, A.T.; Godinho, S.A.; Yu, S.; Lemos, B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 2485–2490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatica, A.; Tollervey, D. Making ribosomes. Curr. Opin. Cell Biol. 2002, 14, 313–318. [Google Scholar] [CrossRef]
- Bassler, J.; Hurt, E. Eukaryotic Ribosome Assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L., Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Melnikov, S.; Ben-Shem, A.; Garreau de Loubresse, N.; Jenner, L.; Yusupova, G.; Yusupov, M. One core, two shells: Bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012, 19, 560–567. [Google Scholar] [CrossRef]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Korner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Mangan, H.; Gailin, M.O.; McStay, B. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017, 284, 3977–3985. [Google Scholar] [CrossRef] [PubMed]
- Van Sluis, M.; McStay, B. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017, 46, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef]
- Nemeth, A.; Langst, G. Genome organization in and around the nucleolus. Trends Genet. 2011, 27, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Conti, C.; Schurra, C.; Lebofsky, R.; Edelstein, S.J.; Bensimon, A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005, 15, 1079–1085. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tao, X.; Jacob, M.D.; Bennett, C.A.; Ho, J.J.D.; Gonzalgo, M.L.; Audas, T.E.; Lee, S. Stress-Induced Low Complexity RNA Activates Physiological Amyloidogenesis. Cell Rep. 2018, 24, 1713–1721.e4. [Google Scholar] [CrossRef] [Green Version]
- Dillinger, S.; Straub, T.; Nemeth, A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS ONE 2017, 12, e0178821. [Google Scholar] [CrossRef]
- Iarovaia, O.V.; Minina, E.P.; Sheval, E.V.; Onichtchouk, D.; Dokudovskaya, S.; Razin, S.V.; Vassetzky, Y.S. Nucleolus: A Central Hub for Nuclear Functions. Trends Cell Biol. 2019, 29, 647–659. [Google Scholar] [CrossRef]
- McStay, B.; Grummt, I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008, 24, 131–157. [Google Scholar] [CrossRef]
- Conconi, A.; Widmer, R.M.; Koller, T.; Sogo, J.M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 1989, 57, 753–761. [Google Scholar] [CrossRef]
- Santoro, R.; Li, J.; Grummt, I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002, 32, 393–396. [Google Scholar] [CrossRef]
- Zentner, G.E.; Saiakhova, A.; Manaenkov, P.; Adams, M.D.; Scacheri, P.C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011, 39, 4949–4960. [Google Scholar] [CrossRef] [Green Version]
- Herdman, C.; Mars, J.C.; Stefanovsky, V.Y.; Tremblay, M.G.; Sabourin-Felix, M.; Lindsay, H.; Robinson, M.D.; Moss, T. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet. 2017, 13, e1006899. [Google Scholar] [CrossRef]
- Nguyen le, X.T.; Raval, A.; Garcia, J.S.; Mitchell, B.S. Regulation of ribosomal gene expression in cancer. J. Cell Physiol. 2015, 230, 1181–1188. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017, 13, e1006994. [Google Scholar] [CrossRef]
- Xu, B.; Li, H.; Perry, J.M.; Singh, V.P.; Unruh, J.; Yu, Z.; Zakari, M.; McDowell, W.; Li, L.; Gerton, J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017, 13, e1006771. [Google Scholar] [CrossRef]
- Sanij, E.; Poortinga, G.; Sharkey, K.; Hung, S.; Holloway, T.P.; Quin, J.; Robb, E.; Wong, L.H.; Thomas, W.G.; Stefanovsky, V.; et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008, 183, 1259–1274. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, R.J.; Earley, K.; Pontes, O.; Silva, M.; Chen, Z.J.; Neves, N.; Viegas, W.; Pikaard, C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 2004, 13, 599–609. [Google Scholar] [CrossRef]
- Penzo, M.; Casoli, L.; Pollutri, D.; Sicuro, L.; Ceccarelli, C.; Santini, D.; Taffurelli, M.; Govoni, M.; Brina, D.; Trere, D.; et al. JHDM1B expression regulates ribosome biogenesis and cancer cell growth in a p53 dependent manner. Int. J. Cancer 2015, 136, E272–E281. [Google Scholar] [CrossRef]
- Leone, S.; Bar, D.; Slabber, C.F.; Dalcher, D.; Santoro, R. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep. 2017, 18, 1248–1262. [Google Scholar] [CrossRef]
- Santoro, R.; Schmitz, K.M.; Sandoval, J.; Grummt, I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010, 11, 52–58. [Google Scholar] [CrossRef]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef]
- Guetg, C.; Scheifele, F.; Rosenthal, F.; Hottiger, M.O.; Santoro, R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell 2012, 45, 790–800. [Google Scholar] [CrossRef]
- Zhou, Y.; Grummt, I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 2005, 15, 1434–1438. [Google Scholar] [CrossRef]
- Gagnon-Kugler, T.; Langlois, F.; Stefanovsky, V.; Lessard, F.; Moss, T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell 2009, 35, 414–425. [Google Scholar] [CrossRef]
- Mayer, C.; Schmitz, K.M.; Li, J.; Grummt, I.; Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 2006, 22, 351–361. [Google Scholar] [CrossRef]
- Tsekrekou, M.; Stratigi, K.; Chatzinikolaou, G. The Nucleolus: In Genome Maintenance and Repair. Int. J. Mol. Sci 2017, 18, 1411. [Google Scholar] [CrossRef]
- Lindstrom, M.S.; Jurada, D.; Bursac, S.; Orsolic, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef]
- Ganley, A.R.; Kobayashi, T. Ribosomal DNA and cellular senescence: New evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 2014, 14, 49–59. [Google Scholar] [CrossRef]
- Killen, M.W.; Stults, D.M.; Adachi, N.; Hanakahi, L.; Pierce, A.J. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009, 18, 3417–3428. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, R.; Srivastava, R.; Ahn, S.H. The Epigenetic Pathways to Ribosomal DNA Silencing. Microbiol. Mol. Biol. Rev. 2016, 80, 545–563. [Google Scholar] [CrossRef] [Green Version]
- Harding, S.M.; Boiarsky, J.A.; Greenberg, R.A. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep. 2015, 13, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Kruhlak, M.; Crouch, E.E.; Orlov, M.; Montano, C.; Gorski, S.A.; Nussenzweig, A.; Misteli, T.; Phair, R.D.; Casellas, R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 2007, 447, 730–734. [Google Scholar] [CrossRef]
- Larsen, D.H.; Hari, F.; Clapperton, J.A.; Gwerder, M.; Gutsche, K.; Altmeyer, M.; Jungmichel, S.; Toledo, L.I.; Fink, D.; Rask, M.B.; et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell Biol. 2014, 16, 792–803. [Google Scholar] [CrossRef]
- Van Sluis, M.; McStay, B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015, 29, 1151–1163. [Google Scholar] [CrossRef] [Green Version]
- Warmerdam, D.O.; van den Berg, J.; Medema, R.H. Breaks in the 45S rDNA Lead to Recombination-Mediated Loss of Repeats. Cell Rep. 2016, 14, 2519–2527. [Google Scholar] [CrossRef] [Green Version]
- Korsholm, L.M.; Gal, Z.; Lin, L.; Quevedo, O.; Ahmad, D.A.; Dulina, E.; Luo, Y.; Bartek, J.; Larsen, D.H. Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Ben-Shem, A.; Garreau de Loubresse, N.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef]
- Barandun, J.; Chaker-Margot, M.; Hunziker, M.; Molloy, K.R.; Chait, B.T.; Klinge, S. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017, 24, 944–953. [Google Scholar] [CrossRef]
- Barrio-Garcia, C.; Thoms, M.; Flemming, D.; Kater, L.; Berninghausen, O.; Bassler, J.; Beckmann, R.; Hurt, E. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016, 23, 37–44. [Google Scholar] [CrossRef]
- Cheng, J.; Kellner, N.; Berninghausen, O.; Hurt, E.; Beckmann, R. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol 2017, 24, 954–964. [Google Scholar] [CrossRef]
- Greber, B.J.; Gerhardy, S.; Leitner, A.; Leibundgut, M.; Salem, M.; Boehringer, D.; Leulliot, N.; Aebersold, R.; Panse, V.G.; Ban, N. Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation. Cell 2016, 164, 91–102. [Google Scholar] [CrossRef]
- Kater, L.; Thoms, M.; Barrio-Garcia, C.; Cheng, J.; Ismail, S.; Ahmed, Y.L.; Bange, G.; Kressler, D.; Berninghausen, O.; Sinning, I.; et al. Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Cell 2017, 171, 1599–1610.e14. [Google Scholar] [CrossRef] [Green Version]
- Kornprobst, M.; Turk, M.; Kellner, N.; Cheng, J.; Flemming, D.; Kos-Braun, I.; Kos, M.; Thoms, M.; Berninghausen, O.; Beckmann, R.; et al. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell 2016, 166, 380–393. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Wu, S.; Li, N.; Chen, Y.; Yan, K.; Li, Z.; Zheng, L.; Lei, J.; Woolford, J.L., Jr.; Gao, N. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh. Nat. Struct. Mol. Biol. 2017, 24, 214–220. [Google Scholar] [CrossRef]
- Malyutin, A.G.; Musalgaonkar, S.; Patchett, S.; Frank, J.; Johnson, A.W. Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J. 2017, 36, 854–868. [Google Scholar] [CrossRef]
- Sanghai, Z.A.; Miller, L.; Molloy, K.R.; Barandun, J.; Hunziker, M.; Chaker-Margot, M.; Wang, J.; Chait, B.T.; Klinge, S. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 2018, 556, 126–129. [Google Scholar] [CrossRef]
- Sun, Q.; Zhu, X.; Qi, J.; An, W.; Lan, P.; Tan, D.; Chen, R.; Wang, B.; Zheng, S.; Zhang, C.; et al. Molecular architecture of the 90S small subunit pre-ribosome. eLife 2017, 6, e22086. [Google Scholar] [CrossRef]
- Wu, S.; Tutuncuoglu, B.; Yan, K.; Brown, H.; Zhang, Y.; Tan, D.; Gamalinda, M.; Yuan, Y.; Li, Z.; Jakovljevic, J.; et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 2016, 534, 133–137. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, X.; Zheng, S.; Tan, D.; Dong, M.Q.; Ye, K. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell 2019, 10, 120–130. [Google Scholar] [CrossRef]
- Sa-Moura, B.; Kornprobst, M.; Kharde, S.; Ahmed, Y.L.; Stier, G.; Kunze, R.; Sinning, I.; Hurt, E. Mpp10 represents a platform for the interaction of multiple factors within the 90S pre-ribosome. PLoS ONE 2017, 12, e0183272. [Google Scholar] [CrossRef]
- Sardana, R.; Liu, X.; Granneman, S.; Zhu, J.; Gill, M.; Papoulas, O.; Marcotte, E.M.; Tollervey, D.; Correll, C.C.; Johnson, A.W. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015, 13, e1002083. [Google Scholar] [CrossRef]
- Osheim, Y.N.; French, S.L.; Keck, K.M.; Champion, E.A.; Spasov, K.; Dragon, F.; Baserga, S.J.; Beyer, A.L. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 2004, 16, 943–954. [Google Scholar] [CrossRef]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef]
- Greber, B.J.; Boehringer, D.; Montellese, C.; Ban, N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2012, 19, 1228–1233. [Google Scholar] [CrossRef]
- Ghalei, H.; Trepreau, J.; Collins, J.C.; Bhaskaran, H.; Strunk, B.S.; Karbstein, K. The ATPase Fap7 Tests the Ability to Carry Out Translocation-like Conformational Changes and Releases Dim1 during 40S Ribosome Maturation. Mol. Cell 2017, 67, 990–1000.e3. [Google Scholar] [CrossRef] [Green Version]
- Lebaron, S.; Schneider, C.; van Nues, R.W.; Swiatkowska, A.; Walsh, D.; Bottcher, B.; Granneman, S.; Watkins, N.J.; Tollervey, D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012, 19, 744–753. [Google Scholar] [CrossRef] [Green Version]
- Strunk, B.S.; Novak, M.N.; Young, C.L.; Karbstein, K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 2012, 150, 111–121. [Google Scholar] [CrossRef]
- Derenzini, M.; Montanaro, L.; Trere, D. Ribosome biogenesis and cancer. Acta Histochem. 2017, 119, 190–197. [Google Scholar] [CrossRef]
- Derenzini, M.; Trere, D.; Pession, A.; Montanaro, L.; Sirri, V.; Ochs, R.L. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998, 152, 1291–1297. [Google Scholar]
- Tiku, V.; Antebi, A. Nucleolar Function in Lifespan Regulation. Trends Cell Biol. 2018, 28, 662–672. [Google Scholar] [CrossRef]
- Uppaluri, S.; Weber, S.C.; Brangwynne, C.P. Hierarchical Size Scaling during Multicellular Growth and Development. Cell Rep. 2016, 17, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Tiku, V.; Jain, C.; Raz, Y.; Nakamura, S.; Heestand, B.; Liu, W.; Spath, M.; Suchiman, H.E.D.; Muller, R.U.; Slagboom, P.E.; et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2017, 8, 16083. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef]
- Buchwalter, A.; Hetzer, M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017, 8, 328. [Google Scholar] [CrossRef]
- Aspesi, A.; Ellis, S.R. Rare ribosomopathies: Insights into mechanisms of cancer. Nat. Rev. Cancer 2019, 19, 228–238. [Google Scholar] [CrossRef]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358, eaan2755. [Google Scholar] [CrossRef]
- Freed, E.F.; Bleichert, F.; Dutca, L.M.; Baserga, S.J. When ribosomes go bad: Diseases of ribosome biogenesis. Mol. Biosyst. 2010, 6, 481–493. [Google Scholar] [CrossRef]
- Kurylo, C.M.; Parks, M.M.; Juette, M.F.; Zinshteyn, B.; Altman, R.B.; Thibado, J.K.; Vincent, C.T.; Blanchard, S.C. Endogenous rRNA Sequence Variation Can Regulate Stress Response Gene Expression and Phenotype. Cell Rep. 2018, 25, 236–248.e6. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correll, C.C.; Bartek, J.; Dundr, M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells 2019, 8, 869. https://doi.org/10.3390/cells8080869
Correll CC, Bartek J, Dundr M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells. 2019; 8(8):869. https://doi.org/10.3390/cells8080869
Chicago/Turabian StyleCorrell, Carl C., Jiri Bartek, and Miroslav Dundr. 2019. "The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies" Cells 8, no. 8: 869. https://doi.org/10.3390/cells8080869



