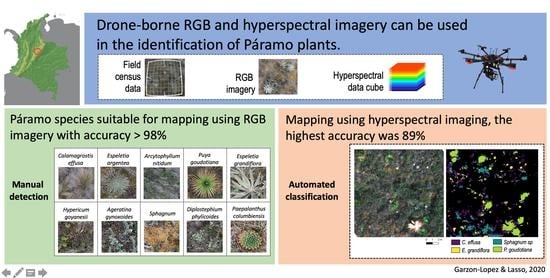
Species Classification in a Tropical Alpine Ecosystem Using UAV-Borne RGB and Hyperspectral Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Spectral and Field Data
2.3. RGB Imagery Analysis—Manual Detection
2.4. Hyperspectral Data Analysis—Automated Classification
3. Results
3.1. Manual Species Identification Using RGB Imagery
3.2. Automated Species Classification Using Hyperspectral Data
4. Discussion
4.1. Manual Species Identification Using RGB Imagery
4.2. Automated Species Identification Using Hyperspectral Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Madriñán, S.; Cortés, A.J.; Richardson, J.E. Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front. Genet. 2013, 4, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farley, K.A.; Bremer, L.L.; Harden, C.P.; Hartsig, J. Changes in carbon storage under alternative land uses in biodiverse Andean grasslands: Implications for payment for ecosystem services. Conserv. Lett. 2013, 6, 21–27. [Google Scholar] [CrossRef]
- Bueno, J.; Ritoré, S. Bioprospecting Model for a New Colombia Drug Discovery Initiative in the Pharmaceutical Industry. In Analysis of Science, Technology, and Innovation in Emerging Economies; Martínez, C.I.P., Poveda, A.C., Moreno, S.P.F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 37–63. [Google Scholar]
- Balthazar, V.; Vanacker, V.; Molina, A.; Lambin, E.F. Impacts of forest cover change on ecosystem services in high Andean mountains. Ecol. Indic. 2015, 48, 63–75. [Google Scholar] [CrossRef]
- Buytaert, W.; Celleri, R.; de Bievre, B.; Cisneros, F.; Wyseure, G.; Deckers, J.; Hofstede, R. Human impact on the hydrology of the Andean páramos. Earth-Sci. Rev. 2006, 79, 53–72. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E.; Kellner, J.R.; Wright, S.J. Operational Tree Species Mapping in a Diverse Tropical Forest with Airborne Imaging Spectroscopy. PLoS ONE 2015, 10, e0118403. [Google Scholar] [CrossRef]
- He, K.S.; Rocchini, D.; Neteler, M.; Nagendra, H. Benefits of hyperspectral remote sensing for tracking plant invasions: Plant invasion and hyperspectral remote sensing. Divers. Distrib. 2011, 17, 381–392. [Google Scholar] [CrossRef]
- Garzon-Lopez, C.X.; Bohlman, S.A.; Olff, H.; Jansen, P.A. Mapping Tropical Forest Trees Using High-Resolution Aerial Digital Photographs. Biotropica 2013, 45, 308–316. [Google Scholar] [CrossRef] [Green Version]
- Colomina, I.; Molina, P. Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef] [Green Version]
- Thenkabail, P.S.; Lyon, J.G.; Lyon, J.G. Hyperspectral Remote Sensing of Vegetation; CRC Press: Boca Raton, FA, USA, 2016. [Google Scholar]
- Yao, H.; Qin, R.; Chen, X. Unmanned Aerial Vehicle for Remote Sensing Applications—A Review. Remote Sens. 2019, 11, 1443. [Google Scholar] [CrossRef] [Green Version]
- Warner, T.A.; Nellis, M.D.; Foody, G.M. Remote Sensing Scale and Data Selection Issues. In The SAGE Handbook of Remote Sensing; SAGE Publications, Inc.: Oliver’s Yard, London, UK, 2009; pp. 2–17. [Google Scholar]
- Ewald, M.; Skowronek, S.; Aerts, R.; Lenoir, J.; Feilhauer, H.; van de Kerchove, R.; Honnay, O.; Somers, B.; Garzon-Lopez, C.X.; Rocchini, D.; et al. Assessing the impact of an invasive bryophyte on plant species richness using high resolution imaging spectroscopy. Ecol. Indic. 2020, 110, 105882. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Raczko, E.; Ochtyra, A.; Jarocińska, A. Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery. Remote Sens. 2018, 10, 570. [Google Scholar] [CrossRef] [Green Version]
- Martín, L.D.; Medina, J.; Upegui, E. Assessment of Image-Texture Improvement Applied to Unmanned Aerial Vehicle Imagery for the Identification of Biotic Stress in Espeletia. Case Study: Moorlands of Chingaza (Colombia). Cienc. E Ing. Neogranadina 2020, 30, 27–44. [Google Scholar] [CrossRef]
- Martínez, E. Análisis de la Respuesta Espectral de las Coberturas Vegetales de los Ecosistemas de Páramo Y Humedales a Partir de los Sensores Aerotransportados Ultracam D, Dji Phanton 3 Pro Y Mapir Nir. Casos de estudio humedal “El ocho”, Villamaria – Caldas. Master’s Thesis, Universidad Católica de Manizales, Caldas, Colombia, 2017. [Google Scholar]
- Abe, B.T.; Olugbara, O.O.; Marwala, T. Experimental comparison of support vector machines with random forests for hyperspectral image land cover classification. J. Earth Syst. Sci. 2014, 123, 779–790. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Gobakken, T.; Gianelle, D.; Næsset, E. Tree Species Classification in Boreal Forests with Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2632–2645. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Sluiter, R.; Pebesma, E.J. Comparing techniques for vegetation classification using multi- and hyperspectral images and ancillary environmental data. Int. J. Remote Sens. 2010, 31, 6143–6161. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Petropoulos, G.P.; Kontoes, C.C.; Keramitsoglou, I. Land cover mapping with emphasis to burnt area delineation using co-orbital ALI and Landsat TM imagery. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 344–355. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Pal, M.; Mather, P.M. Assessment of the effectiveness of support vector machines for hyperspectral data. Future Gener. Comput. Syst. 2004, 20, 1215–1225. [Google Scholar] [CrossRef]
- Shadman Roodposhti, M.; Aryal, J.; Lucieer, A.; Bryan, B.A. Uncertainty Assessment of Hyperspectral Image Classification: Deep Learning vs. Random Forest. Entropy 2019, 21, 78. [Google Scholar] [CrossRef] [Green Version]
- Leon-Garcia, I.V.; Lasso, E. High heat tolerance in plants from the Andean highlands: Implications for paramos in a warmer world. PLoS ONE 2019, 14, e0224218. [Google Scholar] [CrossRef] [Green Version]
- Madriñán, S.; Navas, A.; Garcia, M.W. Páramo Plants Online: A Web Resource to Study Páramo Plant Distributions, Dec. 2016. Available online: http://paramo.uniandes.edu.co/V3/ (accessed on 20 August 2020).
- Ariza Cortes, W. Caracterización Biótica del Complejo de Páramos Cruz Verde-Sumapaz en Jurisdicción de la CAM, CAR, CORMACARENA, CORPOORINOQUIA y la SDA; Instituto Alexander von Humboldt, Universidad Distrital Francisco Jose de Caldas: Columbia, Colombia, 2013. [Google Scholar]
- Rodriguez, C.R.R.; Duarte, C.; Ardila, J.O.G. Estudio de suelos y su relación con las plantas en el páramo el verjón ubicado en el municipio de choachí Cundinamarca. TECCIENCIA 2012, 6, 56–72. [Google Scholar]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geosp. Found. Project 2020. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Amini, S.; Homayouni, S.; Safari, A.; Darvishsefat, A.A. Object-based classification of hyperspectral data using Random Forest algorithm. Geo-Spat. Inf. Sci. 2018, 21, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (randomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Salas, E.A.L.; Subburayalu, S.K. Modified shape index for object-based random forest image classification of agricultural systems using airborne hyperspectral datasets. PLoS ONE 2019, 14, e0213356. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and Regression Training, R Package Ver. 6.0; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar]
- Meyer, H.; Reudenbach, C.; Wöllauer, S.; Nauss, T. Importance of spatial predictor variable selection in machine learning applications—Moving from data reproduction to spatial prediction. Ecol. Model. 2019, 411, 108815. [Google Scholar] [CrossRef] [Green Version]
- Chambers, J. Software for Data Analysis. Programming with R; Springer: New York, NY, USA, 2008. [Google Scholar]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling, R Package Ver. 3.3; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zelleis, A. Kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Dalponte, M.; Oerka, H.O. VarSel: Sequential Forward Floating Selection Using Jeffries-Matusita Distance, R Package Ver. 0.1; R Core Team: Vienna, Austria, 2016. [Google Scholar]
- Edzer, P. Simple Features for R: Standardized Support for Spatial Vector Data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef] [Green Version]
- Park, J.Y.; Muller-Landau, H.C.; Lichstein, J.W.; Rifai, S.W.; Dandois, J.P.; Bohlman, S.A. Quantifying Leaf Phenology of Individual Trees and Species in a Tropical Forest Using Unmanned Aerial Vehicle (UAV) Images. Remote Sens. 2019, 11, 1534. [Google Scholar] [CrossRef] [Green Version]
- Waite, C.E.; van der Heijden, G.M.F.; Field, R.; Boyd, D.S. A view from above: Unmanned aerial vehicles (UAVs) provide a new tool for assessing liana infestation in tropical forest canopies. J. Appl. Ecol. 2019, 56, 902–912. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, J.; Lian, J.; Fan, Z.; Ouyang, X.; Ye, W. Seeing the forest from drones: Testing the potential of lightweight drones as a tool for long-term forest monitoring. Biol. Conserv. 2016, 198, 60–69. [Google Scholar] [CrossRef]
- Sun, Y.; Yi, S.; Hou, F. Unmanned aerial vehicle methods makes species composition monitoring easier in grasslands. Ecol. Indic. 2018, 95, 825–830. [Google Scholar] [CrossRef]
- Woellner, R.; Wagner, T.C. Saving species, time and money: Application of unmanned aerial vehicles (UAVs) for monitoring of an endangered alpine river specialist in a small nature reserve. Biol. Conserv. 2019, 233, 162–175. [Google Scholar] [CrossRef]
- Cruzan, M.B.; Weinstein, B.G.; Grasty, M.R.; Kohrn, B.F.; Hendrickson, E.C.; Arredondo, T.M.; Thompson, P.G. Small unmanned aerial vehicles (micro-UAVs, drones) in plant ecology. Appl. Plant. Sci. 2016, 4, 1600041. [Google Scholar] [CrossRef]
- Tay, J.Y.L.; Erfmeier, A.; Kalwij, J.M. Reaching new heights: Can drones replace current methods to study plant population dynamics? Plant. Ecol. 2018, 219, 1139–1150. [Google Scholar] [CrossRef]
- Cortés, A.J.; Garzón, L.N.; Valencia, J.B.; Madriñán, S. On the Causes of Rapid Diversification in the Páramos: Isolation by Ecology and Genomic Divergence in Espeletia. Front. Plant. Sci. 2018, 9, 1700. [Google Scholar] [CrossRef]
- Jabaily, R.S.; Sytsma, K.J. Historical biogeography and life-history evolution of Andean Puya (Bromeliaceae). Bot. J. Linn. Soc. 2013, 171, 201–224. [Google Scholar] [CrossRef] [Green Version]
- Merchán-Gaitán, J.B.; Álvarez-Herrera, J.G.; Delgado-Merchán, M.V. Retención de agua en musgos de páramo de los municipios de Siachoque, Toca y Pesca (Boyacá). Rev. Colomb. Cienc. Hortícolas 2011, 5, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of Herbaceous Vegetation Using Airborne Hyperspectral Imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef] [Green Version]
- Hennessy, A.; Clarke, K.; Lewis, M. Hyperspectral Classification of Plants: A Review of Waveband Selection Generalisability. Remote Sens. 2020, 12, 113. [Google Scholar] [CrossRef] [Green Version]







| Species | Image | Individuals | Pixels |
|---|---|---|---|
| Calamagrostis effusa | A | 11 | 1365 |
| B | 8 | 1576 | |
| C | 13 | 940 | |
| D | 16 | 2791 | |
| Espeletia argentea | A | 0 | 0 |
| B | 11 | 1369 | |
| C | 24 | 2351 | |
| D | 19 | 2067 | |
| Espeletia grandiflora | A | 29 | 7605 |
| B | 0 | 0 | |
| C | 23 | 2960 | |
| D | 13 | 3286 | |
| Sphagnum sp. | A | 17 | 6888 |
| B | 11 | 1877 | |
| C | 23 | 2679 | |
| D | 11 | 2068 | |
| Puya gouditiana | A | 14 | 4226 |
| B | 12 | 2017 | |
| C | 23 | 4426 | |
| D | 15 | 5055 | |
| TOTAL | 4 | 293 | 55,546 |
| Species | Growth Form | Precision | Accuracy | Specificity | Sensitivity | Commission | Omission |
|---|---|---|---|---|---|---|---|
| Calamagrostis effusa | grass | 95.65 | 98.81 | 98.89 | 98.51 | 1.11 | 1.49 |
| Espeletia argentea | rosette | 98.63 | 99.02 | 99.57 | 97.30 | 0.43 | 2.70 |
| Hypericum juniperinum | shrub | 78.95 | 98.37 | 98.63 | 93.75 | 1.37 | 6.25 |
| Puya goudotiana | rosette | 97.50 | 98.69 | 99.62 | 92.86 | 0.38 | 7.14 |
| Arcytophyllum nitidum | shrub | 81.25 | 98.69 | 98.97 | 92.86 | 1.03 | 7.14 |
| Hypericum goyanessi | shrub | 90.00 | 97.81 | 98.57 | 92.31 | 1.43 | 7.69 |
| Espeletia grandiflora | rosette | 92.31 | 99.35 | 99.66 | 92.31 | 0.34 | 7.69 |
| Ageratina gynoxoides | shrub | 83.33 | 99.02 | 99.32 | 90.91 | 0.68 | 9.09 |
| Blechnum sp. | fern | 82.76 | 97.42 | 98.23 | 88.89 | 1.77 | 11.11 |
| Sphagnum sp. | moss | 95.00 | 98.70 | 99.65 | 86.36 | 0.35 | 13.64 |
| Diplostephium phylicoides | shrub | 75.00 | 99.02 | 99.33 | 85.71 | 0.67 | 14.29 |
| Paepalanthus columbiensis | rosette | 94.44 | 98.05 | 99.65 | 77.27 | 0.35 | 22.73 |
| Rhynchospora ruiziana | sedges | 66.67 | 98.37 | 99.33 | 57.14 | 0.67 | 42.86 |
| Monnina salicifolia | shrub | 83.33 | 98.37 | 99.66 | 55.56 | 0.34 | 44.44 |
| Acaena cylindristachya | forb | 80.00 | 98.05 | 99.67 | 44.44 | 0.33 | 55.56 |
| Aragoa abietina | shrub | 50.00 | 97.42 | 99.34 | 25.00 | 0.66 | 75.00 |
| Orthrosanthus chimboracensis | forb | 50.00 | 98.37 | 99.67 | 20.00 | 0.33 | 80.00 |
| Pentacalia vaccinioides | shrub | 50.00 | 97.73 | 99.67 | 14.29 | 0.33 | 85.71 |
| Bucquetia glutinosa | shrub | 25.00 | 97.11 | 99.01 | 14.29 | 0.99 | 85.71 |
| Valeriana pilosa | forb | 50.00 | 97.42 | 99.67 | 12.50 | 0.33 | 87.50 |
| Cross-Validation | Number of Classes | Classifier | Feature Selection | C.effusa | E. argentea | E.grandiflora | Sphagnum sp. | P. goudotiana | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | ||||
| Random | binary | RF | ALL | 35.10 | 98.73 | 27.93 | 98.93 | 44.20 | 96.43 | 50.48 | 95.80 | 94.34 | 51.95 |
| SFFS | 33.77 | 98.19 | 27.64 | 98.28 | 43.13 | 95.02 | 47.65 | 94.73 | 92.86 | 48.46 | |||
| RF | 26.37 | 97.77 | 22.30 | 98.03 | 34.85 | 93.97 | 38.46 | 92.87 | 91.49 | 38.73 | |||
| SVM | ALL | 34.99 | 99.33 | 32.96 | 99.57 | 44.04 | 97.79 | 54.91 | 97.17 | 96.47 | 50.09 | ||
| SFFS | 24.63 | 99.09 | 21.19 | 99.18 | 32.47 | 96.56 | 43.69 | 95.61 | 94.94 | 39.22 | |||
| RF | 14.72 | 99.36 | 11.51 | 99.50 | 23.85 | 97.82 | 32.22 | 95.55 | 96.42 | 21.88 | |||
| multiple | RF | ALL | 50.49 | 96.46 | 57.77 | 94.29 | 66.30 | 88.09 | 67.55 | 90.38 | 74.24 | 82.41 | |
| SFFS | 46.76 | 95.83 | 54.30 | 93.60 | 63.88 | 87.40 | 64.39 | 89.56 | 70.72 | 80.52 | |||
| RF | 39.47 | 95.01 | 47.26 | 91.79 | 54.47 | 83.81 | 55.64 | 85.73 | 61.03 | 76.86 | |||
| SVM | ALL | 52.52 | 97.01 | 61.71 | 94.58 | 65.78 | 89.45 | 69.65 | 91.21 | 75.94 | 82.08 | ||
| SFFS | 42.33 | 96.06 | 53.33 | 92.99 | 59.51 | 86.04 | 60.97 | 89.51 | 68.55 | 78.02 | |||
| RF | 29.96 | 96.43 | 45.48 | 92.07 | 52.84 | 85.10 | 54.95 | 84.25 | 62.32 | 73.54 | |||
| Spatial | binary | RF | ALL | 3.54 | 94.62 | 2.64 | 95.61 | 15.01 | 78.80 | 13.23 | 87.93 | 76.72 | 10.70 |
| SFFS | 5.08 | 93.70 | 3.70 | 95.36 | 17.06 | 77.21 | 15.04 | 86.94 | 75.46 | 12.17 | |||
| RF | 3.42 | 93.71 | 4.66 | 95.43 | 16.81 | 82.10 | 14.89 | 87.39 | 75.14 | 11.96 | |||
| SVM | ALL | 3.58 | 94.30 | 3.05 | 94.14 | 16.69 | 77.09 | 14.89 | 87.92 | 75.90 | 11.88 | ||
| SFFS | 4.48 | 94.23 | 3.98 | 94.35 | 17.14 | 78.10 | 15.13 | 87.49 | 75.54 | 14.44 | |||
| RF | 3.20 | 95.69 | 3.06 | 95.59 | 13.92 | 84.54 | 11.48 | 89.22 | 77.53 | 8.86 | |||
| multiple | RF | ALL | 8.75 | 91.32 | 9.84 | 84.27 | 25.86 | 64.54 | 20.21 | 80.44 | 25.96 | 59.85 | |
| SFFS | 9.78 | 91.15 | 9.13 | 83.68 | 26.89 | 65.49 | 21.68 | 80.02 | 25.46 | 61.02 | |||
| RF | 9.06 | 91.15 | 8.86 | 83.59 | 25.82 | 66.89 | 21.62 | 79.21 | 25.37 | 60.13 | |||
| SVM | ALL | 8.06 | 89.79 | 8.88 | 83.68 | 26.20 | 63.36 | 20.92 | 81.35 | 25.31 | 61.69 | ||
| SFFS | 9.83 | 89.55 | 9.74 | 82.82 | 27.79 | 66.49 | 22.60 | 81.33 | 26.85 | 63.01 | |||
| RF | 7.77 | 91.02 | 8.56 | 83.00 | 27.11 | 66.14 | 18.09 | 81.20 | 29.05 | 59.19 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garzon-Lopez, C.X.; Lasso, E. Species Classification in a Tropical Alpine Ecosystem Using UAV-Borne RGB and Hyperspectral Imagery. Drones 2020, 4, 69. https://doi.org/10.3390/drones4040069
Garzon-Lopez CX, Lasso E. Species Classification in a Tropical Alpine Ecosystem Using UAV-Borne RGB and Hyperspectral Imagery. Drones. 2020; 4(4):69. https://doi.org/10.3390/drones4040069
Chicago/Turabian StyleGarzon-Lopez, Carol X., and Eloisa Lasso. 2020. "Species Classification in a Tropical Alpine Ecosystem Using UAV-Borne RGB and Hyperspectral Imagery" Drones 4, no. 4: 69. https://doi.org/10.3390/drones4040069