Estimating the Threshold of Detection on Tree Crown Defoliation Using Vegetation Indices from UAS Multispectral Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. UAS Image Acquisition and Processing
2.3. Vegetation Indices
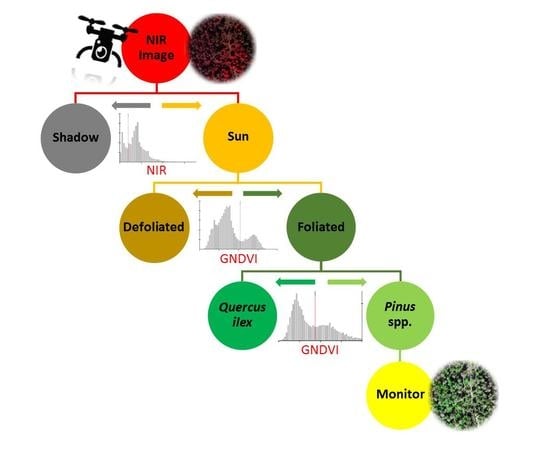
2.4. Pixel-Based Thresholding Analysis
2.4.1. Shadow Removal
2.4.2. Defoliation Detection
2.4.3. Foliated Species Discrimination
2.5. Object-Based Random Forest
2.6. Validation and Accuracy Assessment
3. Results
3.1. Pixel-Based Thresholding Analysis
3.1.1. Shadow Removal
3.1.2. Defoliation Detection
3.1.3. Foliated Species Discrimination
3.2. Object-Based Random Forest
3.3. Validation and Accuracy Assessment
4. Discussion
4.1. Shadow Removal
4.2. Defoliation Detection
4.3. Foliated Species Discrimination
4.4. Classification Techniques
4.5. Future Research Directions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A




References
- Collins, M.; Knutti, R.; Arblaster, J.; Dufresne, J.-L.; Fichefet, T.; Friedlingstein, P.; Gao, X.; Gutowski, W.J.; Johns, T.; Krinner, G.; et al. Long–term Climate Change: Projections, Commitments and Irreversibility. In Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013; pp. 1029–1136. [Google Scholar]
- Netherer, S.; Schopf, A. Potential Effects of Climate Change on Insect Herbivores in European Forests–General Aspects and the Pine Processionary Moth as Specific Example. For. Ecol. Manag. 2010, 259, 831–838. [Google Scholar] [CrossRef]
- Robinet, C.; Roques, A. Direct Impacts of Recent Climate Warming on Insect Populations. Integr. Zool. 2010, 5, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Battisti, A.; Larsson, S. Climate Change and Insect Pest Distribution Range. In Climate Change and Insect Pests; Björkman, C., Niemelä, P., Eds.; CAB International: Oxfordshire, UK; Boston, MA, USA, 2015; pp. 1–15. [Google Scholar]
- Battisti, A.; Larsson, S.; Roques, A. Processionary Moths and Associated Urtication Risk: Global Change–Driven Effects. Annu. Rev. Entomol. 2016, 62, 323–342. [Google Scholar] [CrossRef] [PubMed]
- Roques, A. Processionary Moths and Climate Change: An Update; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Battisti, A.; Stastny, M.; Netherer, S.; Robinet, C.; Schopf, A.; Roques, A.; Larsson, S. Expansion of Geographic Range in the Pine Processionary Moth Caused by Increased Winter Temperatures. Ecol. Appl. 2005, 15, 2084–2096. [Google Scholar] [CrossRef]
- Robinet, C.; Baier, P.; Pennerstorfer, J.; Schopf, A.; Roques, A. Modelling the Effects of Climate Change on the Potential Feeding Activity of Thaumetopoea Pityocampa (Den. & Schiff.) (Lep., Notodontidae) in France. Glob. Ecol. Biogeogr. 2007, 16, 460–471. [Google Scholar]
- FAO; Plan Bleu. State of Mediterranean Forests 2018; Food and Agriculture Organization of the United Nations: Rome, Italy; Plan Bleu: Valbonne, France, 2018; p. 308. [Google Scholar]
- Näsi, R.; Honkavaara, E.; Lyytikäinen-Saarenmaa, P.; Blomqvist, M.; Litkey, P.; Hakala, T.; Viljanen, N.; Kantola, T.; Tanhuanpää, T.; Holopainen, M. Using UAV–Based Photogrammetry and Hyperspectral Imaging for Mapping Bark Beetle Damage at Tree–Level. Remote Sens. 2015, 7, 15467–15493. [Google Scholar] [CrossRef]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing Very High Resolution UAV Imagery for Monitoring Forest Health during a Simulated Disease Outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Lehmann, J.R.K.; Nieberding, F.; Prinz, T.; Knoth, C. Analysis of Unmanned Aerial System–Based CIR Images in Forestry–a New Perspective to Monitor Pest Infestation Levels. Forests 2015, 6, 594–612. [Google Scholar] [CrossRef]
- Brovkina, O.; Cienciala, E.; Surový, P.; Janata, P. Unmanned Aerial Vehicles (UAV) for Assessment of Qualitative Classification of Norway Spruce in Temperate Forest Stands. Geo-Spat. Inf. Sci. 2018, 21, 12–20. [Google Scholar] [CrossRef]
- Cardil, A.; Vepakomma, U.; Brotons, L. Assessing Pine Processionary Moth Defoliation Using Unmanned Aerial Systems. Forests 2017, 8, 402. [Google Scholar] [CrossRef]
- Hentz, A.M.K.; Strager, M.P. Cicada Damage Detection Based on UAV Spectral and 3D Data. Silvilaser 2017, 10, 95–96. [Google Scholar]
- Cardil, A.; Otsu, K.; Pla, M.; Silva, C.A.; Brotons, L. Quantifying Pine Processionary Moth Defoliation in a Pine–Oak Mixed Forest Using Unmanned Aerial Systems and Multispectral Imagery. PLoS ONE 2019, 14, e0213027. [Google Scholar] [CrossRef] [PubMed]
- Rullan-Silva, C.D.; Olthoff, A.E.; Delgado de la Mata, J.A.; Pajares-Alonso, J.A. Remote Monitoring of Forest Insect Defoliation—A Review. For. Syst. 2013, 22, 377. [Google Scholar] [CrossRef]
- ESA. Spatial Resolutions Sentinel-2 MSI User Guides. Available online: https://sentinel.esa.int/web/sentinel/missions/sentinel-2/instrument-payload/resolution-and-swath (accessed on 16 March 2019).
- Manfreda, S.; McCabe, M.F.; Miller, P.E.; Lucas, R.; Madrigal, V.P.; Mallinis, G.; Dor, E.B.; Helman, D.; Estes, L.; Ciraolo, G.; et al. On the Use of Unmanned Aerial Systems for Environmental Monitoring. Remote Sens. 2018, 10, 641. [Google Scholar] [CrossRef]
- Roncat, A.; Morsdorf, F.; Briese, C.; Wagner, W.; Pfeifer, N. Laser Pulse Interaction with Forest Canopy: Geometric and Radiometric Issues. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 27, pp. 19–41. [Google Scholar]
- Kantola, T.; Vastaranta, M.; Lyytikäinen-Saarenmaa, P.; Holopainen, M.; Kankare, V.; Talvitie, M.; Hyyppä, J. Classification of Needle Loss of Individual Scots Pine Trees by Means of Airborne Laser Scanning. Forests 2013, 4, 386–403. [Google Scholar] [CrossRef] [Green Version]
- Pajares, G. Overview and Current Status of Remote Sensing Applications Based on Unmanned Aerial Vehicles (UAVs). Photogramm. Eng. Remote Sens. 2015, 81, 281–330. [Google Scholar] [CrossRef] [Green Version]
- Torresan, C.; Berton, A.; Carotenuto, F.; Filippo, S.; Gennaro, D.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; et al. International Journal of Remote Sensing Forestry Applications of UAVs in Europe: A Review Forestry Applications of UAVs in Europe: A Review. Int. J. Remote Sens. 2017, 38, 8–10. [Google Scholar] [CrossRef]
- Michez, A.; Piégay, H.; Lisein, J.; Claessens, H.; Lejeune, P. Classification of Riparian Forest Species and Health Condition Using Multi–Temporal and Hyperspatial Imagery from Unmanned Aerial System. Environ. Monit. Assess. 2016, 188, 1–19. [Google Scholar] [CrossRef]
- Smigaj, M.; Gaulton, R.; Barr, S.L.; Suárez, J.C. Uav–Borne Thermal Imaging for Forest Health Monitoring: Detection of Disease–Induced Canopy Temperature Increase. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, XL–3/W3, 349–354. [Google Scholar] [CrossRef]
- Dare, P.M. Shadow Analysis in High–Resolution Satellite Imagery of Urban Areas. Photogramm. Eng. Remote Sens. 2005, 71, 169–177. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray–Level Histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, D.; Jing, L.; Shi, P. Shadow Information Recovery in Urban Areas from Very High Resolution Satellite Imagery. Int. J. Remote Sens. 2007, 28, 3249–3254. [Google Scholar] [CrossRef]
- Adeline, K.R.M.; Chen, M.; Briottet, X.; Pang, S.K.; Paparoditis, N. Shadow Detection in Very High Spatial Resolution Aerial Images: A Comparative Study. ISPRS J. Photogramm. Remote Sens. 2013, 80, 21–38. [Google Scholar] [CrossRef]
- Chang, C.J. Evaluation of Automatic Shadow Detection Approaches Using ADS–40 High Radiometric Resolution Aerial Images at High Mountainous Region. J. Remote Sens. GIS 2016, 5, 1–5. [Google Scholar] [CrossRef]
- Aasen, H.; Honkavaara, E.; Lucieer, A.; Zarco-Tejada, P.J. Quantitative Remote Sensing at Ultra–High Resolution with UAV Spectroscopy: A Review of Sensor Technology, Measurement Procedures, and Data Correctionworkflows. Remote Sens. 2018, 10, 1091. [Google Scholar] [CrossRef]
- Tang, L.; Shao, G. Drone Remote Sensing for Forestry Research and Practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Blaschke, T. Object Based Image Analysis for Remote Sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Weih, R.C.; Riggan, N.D. Object–Based Classification vs. Pixel–Based Classification: Comparitive Importance of Multi–Resolution Imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. S41 2010, XXXVIII, 1–6. [Google Scholar]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A Comparison of Pixel-Based and Object-Based Image Analysis with Selected Machine Learning Algorithms for the Classification of Agricultural Landscapes Using SPOT-5 HRG Imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Qian, Y.; Zhou, W.; Yan, J.; Li, W.; Han, L. Comparing Machine Learning Classifiers for Object–Based Land Cover Classification Using Very High Resolution Imagery. Remote Sens. 2015, 7, 153–168. [Google Scholar] [CrossRef]
- Hossain, M.D.; Chen, D. Segmentation for Object–Based Image Analysis (OBIA): A Review of Algorithms and Challenges from Remote Sensing Perspective. ISPRS J. Photogramm. Remote Sens. 2019, 150, 115–134. [Google Scholar] [CrossRef]
- Agisoft LLC. Tutorial Intermediate Level: Radiometric Calibration Using Reflectance Panels in PhotoScan; Agisoft LLC: St. Petersburg, Russia, 2018. [Google Scholar]
- Pla, M.; Bota, G.; Duane, A.; Balagu, J.; Curc, A.; Guti, R. Calibrating Sentinel-2 Imagery with Multispectral UAV Derived Information to Quantify Damages in Mediterranean Rice Crops Caused by Western Swamphen ( Porphyrio Porphyrio). Drones 2019, 3, 45. [Google Scholar] [CrossRef]
- ICGC. Orthophoto in colour of Catalonia 25cm (OF–25C) v4.0. Available online: https://ide.cat/geonetwork/srv/eng/catalog.search#/metadata/ortofoto–25cm–v4r0–color–2017 (accessed on 16 January 2019).
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS–MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Rouse, J.W.; Hass, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. Third Earth Resour. Technol. Satell. Symp. 1973, 1, 309–317. [Google Scholar]
- Wang, F.; Huang, J.; Tang, Y.; Wang, X. New Vegetation Index and Its Application in Estimating Leaf Area Index of Rice. Rice Sci. 2007, 14, 195–203. [Google Scholar] [CrossRef]
- Chevrel, S.; Belocky, R.; Grösel, K. Monitoring and Assessing the Environmental Impact of Mining in Europe Using Advanced Earth Observation Techniques—MINEO. In Proceedings of the 16th Conference Environmental Communication in the Information Society, Vienna, Austria, 25–27 September 2002; pp. 519–526. [Google Scholar]
- Shahtahmassebi, A.; Yang, N.; Wang, K.; Moore, N.; Shen, Z. Review of Shadow Detection and De–Shadowing Methods in Remote Sensing. Chin. Geogr. Sci. 2013, 23, 403–420. [Google Scholar] [CrossRef]
- Riemann Hershey, R.; Befort, W.A. Aerial Photo Guide to New England Forest Cover Types; U.S. Department of Agriculture, Forest Service: Radnor, PA, USA, 1995; p. 70.
- Aronoff, S. Remote Sensing for GIS Managers. ESRI Press: Redlands, Calif, 2005. [Google Scholar]
- Mohan, M.; Silva, C.A.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, A.T.; Dia, M. Individual Tree Detection from Unmanned Aerial Vehicle (UAV) Derived Canopy Height Model in an Open Canopy Mixed Conifer Forest. Forests 2017, 8, 340. [Google Scholar] [CrossRef]
- Pla, M.; Duane, A.; Brotons, L. Potencial de Las Imágenes UAV Como Datos de Verdad Terreno Para La Clasificación de La Severidad de Quema de Imágenes Landsat: Aproximaciones a Un Producto Útil Para La Gestión Post Incendio. Rev. Teledetec. 2017, 2017, 91–102. [Google Scholar] [CrossRef]
- Otsu, K.; Pla, M.; Vayreda, J.; Brotons, L. Calibrating the Severity of Forest Defoliation by Pine Processionary Moth with Landsat and UAV Imagery. Sensors 2018, 18, 3278. [Google Scholar] [CrossRef]
- Shahtahmassebi, A.R.; Wang, K.; Shen, Z.; Deng, J.; Zhu, W.; Han, N.; Lin, F.; Moore, N. Evaluation on the Two Filling Functions for the Recovery of Forest Information in Mountainous Shadows on Landsat ETM + Image. J. Mt. Sci. 2011, 8, 414–426. [Google Scholar] [CrossRef]
- Miura, H.; Midorikawa, S. Detection of Slope Failure Areas Due to the 2004 Niigata–Ken Chuetsu Earthquake Using High–Resolution Satellite Images and Digital Elevation Model. J. JAEEJournal Japan Assoc. Earthq. Eng. 2013, 7, 1–14. [Google Scholar]
- Lu, D. Detection and Substitution of Clouds/Hazes and Their Cast Shadows on IKONOS Images. Int. J. Remote Sens. 2007, 28, 4027–4035. [Google Scholar] [CrossRef]
- Martinuzzi, S.; Gould, W.A.; Ramos González, O.M. Creating Cloud–Free Landsat ETM+ Data Sets in Tropical Landscapes: Cloud and Cloud–Shadow Removal; U.S. Department of Agriculture, Forest Service: Rio Piedras, PR, USA, 2006; p. 12.
- Richter, R.; Müller, A. De-Shadowing of Satellite/Airborne Imagery. Int. J. Remote Sens. 2005, 26, 3137–3148. [Google Scholar] [CrossRef]
- Schläpfer, D.; Richter, R.; Kellenberger, T. Atmospheric and Topographic Correction of Photogrammetric Airborne Digital Scanner Data (Atcor–Ads). In Proceedings of the EuroSDR—EUROCOW, Barcelona, Spain, 8–10 February 2012; p. 5. [Google Scholar]
- Xiao, Q.; McPherson, E.G. Tree Health Mapping with Multispectral Remote Sensing Data at UC Davis, California. Urban Ecosyst. 2005, 8, 349–361. [Google Scholar] [CrossRef]
- Masaitis, G.; Mozgeris, G.; Augustaitis, A. Spectral Reflectance Properties of Healthy and Stressed Coniferous Trees. iForest Biogeosciences For. 2013, 6, 30–36. [Google Scholar] [CrossRef]
- Harwin, S.; Lucieer, A. Assessing the Accuracy of Georeferenced Point Clouds Produced via Multi–View Stereopsis from Unmanned Aerial Vehicle (UAV) Imagery. Remote Sens. 2012, 4, 1573–1599. [Google Scholar] [CrossRef]
- Dandois, J.P.; Ellis, E.C. Remote Sensing of Environment High Spatial Resolution Three–Dimensional Mapping of Vegetation Spectral Dynamics Using Computer Vision. Remote Sens. Environ. 2013, 136, 259–276. [Google Scholar] [CrossRef]
- Mathews, A.J.; Jensen, J.L.R. Visualizing and Quantifying Vineyard Canopy LAI Using an Unmanned Aerial Vehicle (UAV) Collected High Density Structure from Motion Point Cloud. Remote Sens. 2013, 5, 2164–2183. [Google Scholar] [CrossRef] [Green Version]
- Wallace, L.; Lucieer, A.; Malenovskỳ, Z.; Turner, D.; Vopěnka, P. Assessment of Forest Structure Using Two UAV Techniques: A Comparison of Airborne Laser Scanning and Structure from Motion (SfM) Point Clouds. Forests 2016, 7, 62. [Google Scholar] [CrossRef]
- Gini, R.; Passoni, D.; Pinto, L.; Sona, G. Use of Unmanned Aerial Systems for Multispectral Survey and Tree Classification: A Test in a Park Area of Northern Italy. Eur. J. Remote Sens. 2014, 47, 251–269. [Google Scholar] [CrossRef]
- Lisein, J.; Michez, A.; Claessens, H.; Lejeune, P. Discrimination of Deciduous Tree Species from Time Series of Unmanned Aerial System Imagery. PLoS ONE 2015, 10, e0141006. [Google Scholar] [CrossRef] [PubMed]
- Baena, S.; Moat, J.; Whaley, O.; Boyd, D.S. Identifying Species from the Air: UAVs and the Very High Resolution Challenge for Plant Conservation. PLoS ONE 2017, 12, e0188714. [Google Scholar] [CrossRef] [PubMed]
- Yoder, B.J.; Waring, R.H. The Normalized Difference Vegetation Index of Small Douglas–Fir Canopies with Varying Chlorophyll Concentrations. Remote Sens. Environ. 1994, 49, 81–91. [Google Scholar] [CrossRef]
- Baldridge, A.M.; Hook, S.J.; Grove, C.I.; Rivera, G. Remote Sensing of Environment The ASTER Spectral Library Version 2.0. Remote Sens. Environ. 2009, 113, 711–715. [Google Scholar] [CrossRef]
- Motohka, T.; Nasahara, K.N.; Oguma, H.; Tsuchida, S. Applicability of Green–Red Vegetation Index for Remote Sensing of Vegetation Phenology. Remote Sens. 2010, 2, 2369–2387. [Google Scholar] [CrossRef]
- NASA. Landsat 8 Bands Landsat Science. Available online: https://landsat.gsfc.nasa.gov/landsat-8/landsat-8-bands (accessed on 16 March 2019).
- Fraser, R.H.; van der Sluijs, J.; Hall, R.J. Calibrating Satellite–Based Indices of Burn Severity from UAV–Derived Metrics of a Burned Boreal Forest in NWT, Canada. Remote Sens. 2017, 9, 279. [Google Scholar] [CrossRef]










| Site | Codo | Hostal | Bosquet | Olius | ||||
|---|---|---|---|---|---|---|---|---|
| RGB | NIR | RGB | NIR | RGB | NIR | RGB | NIR | |
| Date (dd/mm/yy) | 26/11/2017 | 19/01/2018 | 23/01/2018 | 30/01/2018 | ||||
| Time (duration) | 12:43–12:50 | 12:05–12:14 | 12:16–12:22 | 11:55–12:03 | ||||
| Elevation (m) | 1300 | 820 | 620 | 720 | ||||
| Flight height (m) | 95 | 78 | 76 | 85 | ||||
| Area (ha) | 14.1 | 16.2 | 7.4 | 26.3 | ||||
| Number of images | 210 | 333 | 155 | 344 | ||||
| Data size (GB) | 0.93 | 0.49 | 1.65 | 0.78 | 0.39 | 0.36 | 1.05 | 0.80 |
| Processing time (h) | 4.8 | 3.3 | 7.6 | 5.0 | 3.9 | 2.4 | 5.7 | 3.8 |
| Software platform | Microsoft Windows 7 (64 bits) | |||||||
| Ground resolution (cm/pix) | 2.32 | 8.64 | 1.90 | 6.82 | 1.80 | 6.58 | 2.12 | 7.49 |
| RMS re-projection error (pix) | 2.45 | 0.66 | 2.51 | 0.70 | 2.34 | 0.64 | 2.19 | 0.62 |
| Index | Acronym | Formula | |
|---|---|---|---|
| Normalized Difference Vegetation Index | NDVI | [42] | |
| Green Normalized Difference Vegetation Index | GNDVI | [41] | |
| Green–Red Normalized Difference Vegetation Index | GRNDVI | [43] | |
| Normalized Difference Vegetation Index Red Edge | NDVIRE | [44] |
| Classification | Index | Codo | Hostal | Bosquet | Olius | Total Average |
|---|---|---|---|---|---|---|
| Shadow | NIR | 17 | 23 | 27 | 28 | 24 |
| Defoliated | NDVI | 0.584 | 0.529 | 0.481 | 0.490 | 0.52 |
| GNDVI | 0.561 | - | - | 0.393 | - | |
| GRNDVI | 0.295 | 0.254 | 0.171 | 0.175 | 0.22 | |
| NDVIRE | 0.515 | 0.475 | 0.416 | 0.431 | 0.46 | |
| Species | NDVI | - | - | - | - | - |
| GNDVI | 0.681 | - | - | 0.631 | - | |
| GRNDVI | 0.539 | - | - | - | - | |
| NDVIRE | - | - | - | - | - |
| Class | Predicted | ||||
|---|---|---|---|---|---|
| Shadow | Sun | Total | Producer’s Accuracy | ||
| Observed | Shadow | 167 | 13 | 180 | 93% |
| Sun | 8 | 212 | 220 | 96% | |
| Total | 175 | 225 | 400 | ||
| User’s Accuracy | 95% | 94% | 95% | ||
| Index | Codo | Hostal | Bosquet | Olius | Total |
|---|---|---|---|---|---|
| NIR | 96% | 93% | 96% | 94% | 95% |
| Index | Codo | Hostal | Bosquet | Olius | Total |
|---|---|---|---|---|---|
| NDVI | 93% | 91% | 97% | 98% | 95% |
| GNDVI | 91% | - | - | 86% | - |
| GRNDVI | 93% | 84% | 95% | 97% | 92% |
| NDVIRE | 94% | 90% | 97% | 97% | 95% |
| Index | Codo | Hostal | Bosquet | Olius | Total |
|---|---|---|---|---|---|
| GNDVI | 96% | - | - | 93% | - |
| Class | Predicted | ||||||
|---|---|---|---|---|---|---|---|
| Shadow | Defoliated | Pine | Oak | Total | Producer’s Accuracy | ||
| Observed | Shadow | 22 | 1 | 3 | 0 | 26 | 85% |
| Defoliated | 0 | 31 | 0 | 0 | 31 | 100% | |
| Pine | 0 | 1 | 21 | 2 | 24 | 88% | |
| Oak | 0 | 0 | 0 | 19 | 19 | 100% | |
| Total | 22 | 33 | 24 | 21 | 100 | - | |
| User’s Accuracy | 100% | 94% | 88% | 90% | - | 93% | |
| Class | Predicted | ||||||
|---|---|---|---|---|---|---|---|
| Shadow | Defoliated | Pine | Oak | Total | Producer’s Accuracy | ||
| Observed | Shadow | 22 | 1 | 1 | 0 | 24 | 92% |
| Defoliated | 0 | 26 | 0 | 0 | 26 | 100% | |
| Pine | 0 | 2 | 26 | 0 | 28 | 93% | |
| Oak | 4 | 0 | 1 | 17 | 22 | 77% | |
| Total | 26 | 29 | 28 | 17 | 100 | - | |
| User’s Accuracy | 85% | 90% | 93% | 100% | - | 91% | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otsu, K.; Pla, M.; Duane, A.; Cardil, A.; Brotons, L. Estimating the Threshold of Detection on Tree Crown Defoliation Using Vegetation Indices from UAS Multispectral Imagery. Drones 2019, 3, 80. https://doi.org/10.3390/drones3040080
Otsu K, Pla M, Duane A, Cardil A, Brotons L. Estimating the Threshold of Detection on Tree Crown Defoliation Using Vegetation Indices from UAS Multispectral Imagery. Drones. 2019; 3(4):80. https://doi.org/10.3390/drones3040080
Chicago/Turabian StyleOtsu, Kaori, Magda Pla, Andrea Duane, Adrián Cardil, and Lluís Brotons. 2019. "Estimating the Threshold of Detection on Tree Crown Defoliation Using Vegetation Indices from UAS Multispectral Imagery" Drones 3, no. 4: 80. https://doi.org/10.3390/drones3040080