1. Introduction
The widespread use of cross-sectional imaging, such as computed tomography (CT) and magnetic resonance imaging (MRI), has resulted in a high incidence of incidentally detected pancreatic cystic lesions (PCLs). There is currently an “epidemic” of such lesions, with 15–45% of asymptomatic patients having a pancreatic cyst identified in cross-sectional abdominal imaging studies [
1]. PCLs encompass a broad range of lesions, ranging from benign cysts to mucinous pre-malignant lesions that carry a risk of progressing to high-grade dysplasia or adenocarcinoma (HGD-Ca). Surgical interventions, such as pancreaticoduodenectomy (Whipple’s procedure), total pancreatectomy, and distal pancreatectomy, aim to resect malignant lesions with HGD-Ca. Conversely, patients with only low-grade dysplasia (LGD) can be managed through serial imaging monitoring [
2,
3,
4].
PCLs are classified into mucinous and non-mucinous lesions. Non-mucinous lesions include cystic neuroendocrine tumors, solid pseudopapillary neoplasms, and serous cystadenomas. Mucinous cysts include intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs), both of which are pancreatic cancer precursors. IPMNs are classified as main-duct (MD) IPMNs, which represent cystic dilation of the main pancreatic duct, and branch-duct (BD) IPMNs, which are cysts that lie in communication with the main duct. BD-IPMNs are the most common PCLs, with the reported risk of malignancy ranging between 6 and 46% [
5,
6,
7]. The current standard of care for risk stratifying PCLs involves the use of endoscopic ultrasound (EUS)-guided fine needle aspiration (FNA) and analysis of cyst fluid, including measurements of carcinoembryonic antigen (CEA), cytology, amylase, and glucose levels. However, these modalities provide only 65–75% accuracy in identifying high-grade dysplasia or adenocarcinoma (HGD-Ca) [
8].
It is estimated that two-thirds of surgically resected PCLs demonstrate only LGD or benign neoplasms, indicating a significant rate of overtreatment and unnecessary surgeries [
9]. Considering the high morbidity and mortality of Whipple procedures and pancreatectomies, these data point to an unacceptably high false-positive rate with current diagnostic modalities [
10,
11]. Conversely, several series report that up to 37% of invasive cancers are discovered during routine follow-ups of suspected BD-IPMNs, suggesting possible delays in diagnosis [
12]. Application of the Fukuoka International Consensus Guidelines specifically intended for IPMNs also continues to contribute to missed cancers at one end and surgical overtreatment and unnecessary pancreatic resections at the other [
4,
8,
13,
14,
15,
16,
17].
Unlike solid tumors, where tissue biopsy often guides diagnosis, there are currently no standard-of-care, consistently reliable options for obtaining tissue from the PCL epithelium prior to resection. Additionally, IPMNs can demonstrate a wide range of histologic features within a single cyst that vary from low-grade dysplasia (LGD) to invasive cancer, suggesting that intracystic micro-biopsies may not precisely sample the area with the highest degree of malignant progression [
5,
6]. This creates a need for more precise diagnostic techniques that use existing information, such as cross-sectional imaging, cyst fluid (CEA, glucose, cytology) analysis, next-generation sequencing (NGS) of cyst fluid, and novel imaging biomarkers (confocal endomicroscopy). EUS-guided needle-based confocal laser endomicroscopy (EUS-nCLE) features of IPMNs are highlighted in
Figure 1.
1.1. Artificial Intelligence
Artificial intelligence (AI) is a technology that aims to mimic human intelligence to perform tasks, such as object recognition and decision making. AI encompasses several branches, among which machine learning (ML) approaches have attracted the most attention in recent years.
ML constructs models that could perform tasks by learning from data. For instance, to differentiate dog images from cat images, ML requires a dataset of cat and dog images, each of which is manually labeled with the ground-truth category so that it can train the model (i.e., a neural network classifier) to correctly classify images.
Many kinds of ML models and algorithms have been developed. Logistic regression linearly combines the features in a data sample [
18]. Decision trees build tree-structured decision rules to classify a data sample; random forests build multiple trees and output their joint decisions (e.g., majority voting) to make the classification more stable [
19,
20]. Support vector machines (SVMs) could construct complicated decision rules by applying the kernel tricks, which are a set of mathematical techniques used to transform the input data into a higher-dimensional feature space. Kernel tricks enable SVMs to classify non-linearly separable data by implicitly computing dot products between transformed feature vectors without explicitly calculating the higher-dimensional feature space [
21,
22]. Most of these algorithms require informative features (often hand-crafted) to be extracted from data samples.
Recently, deep learning approaches that aim to learn neural network (NN)-based models have shown unprecedented results in many application domains such as image recognition [
23,
24]. Compared to the aforementioned methods, NN-based models can often learn to perform feature extraction and decision (e.g., classification) in an end-to-end fashion. In image recognition and computer vision, convolutional neural networks (CNNs), which consist of layers of convolution filters, are widely applied to capture image patterns [
25]. Several representative CNN models include AlexNet, VGGNet, and ResNet for image classification [
26,
27,
28]; Faster R-CNN and YOLO for object detection (i.e., localize objects in an image with bounding boxes and classify them) [
29,
30]; U-Nets and DeepLab for semantic segmentation (i.e., classify each image pixel into a semantic category) [
31,
32]; and Mask R-CNN for instance segmentation, which localizes and segments objects using object masks [
33].
1.2. Challenges in AI Application to Pancreas Imaging
There are multiple considerations when designing an AI algorithm to risk stratify PCLs, which include steps from identifying the pancreas in a CT or MRI image, to selecting imaging features that can predict the malignant progression of a cyst. The interpretation of pancreatic lesions presents a unique challenge for AI. First, the pancreas occupies a relatively small area (approximately 1.3%) in cross-sectional images [
34]. It is also both irregularly shaped and highly variable in its location relative to other organs [
35]. Pancreatic lesions can also often have similar radiographic features to the surrounding tissue.
Previous models have required significant pre-processing of images prior to AI interpretation. Wei et al. developed an SVM system to diagnose serous cystadenomas. This system utilized CT radiomics features combined with regions of interest that were marked by a radiologist [
36]. Similarly, Chakraborty et al. employed manually segmented pancreas images, with manual outlining of the pancreas head, body, and tail, to train AI models for predicting high-risk IPMNs [
37]. While such models may achieve high accuracies, the need for human pre-processing of images ultimately detracts from the goal of leveraging AI to reduce specialist workload (
Table 1).
There has been a recent shift toward more end-to-end, independent, or self-supervised predictive algorithms for the interpretation of CT images. Javed et al. presented the first framework for automated 3D pancreas segmentation in CT images, enabling the identification and delineation of the pancreas without manual intervention [
41]. In a similar vein, Lim et al. validated a CNN model for automating pancreas segmentation, achieving a mean precision of 0.87 in an internal cohort and 0.78 for external validation [
42]. Si et al. developed a deep learning model for diagnosing pancreatic tumors, including IPMNs and pancreatic ductal adenocarcinoma (PDAC), based on unedited, non-annotated abdominal CT images [
34]. They used images from 347 patients and reported an AUC of 0.871 and accuracy of 82.7% for all tumor types. Remarkably, the model achieved 100% accuracy for IPMN diagnosis, with surgical histopathology as the reference standard. A manual review of the images required approximately 8 minutes per patient, whereas the model only needed an average of 18.6 seconds, demonstrating that a fully end-to-end predictive algorithm can significantly reduce specialist workload and healthcare utilization.
2. Materials and Methods
In this review, we highlight existing applications for the AI-powered diagnosis of PCLs and describe an ongoing study performed by our group to improve their accuracy and generalizability.
A literature search was performed in February 2023 via PubMed and Embase for publications within the last 20 years (2003–2023). Articles were included if they constituted primary literature describing original AI algorithms for diagnosing or predicting malignant potential of pancreatic cystic lesions. The search was conducted by three authors (J.J., T.C., S.J.K.). Search terms included synonyms of “IPMNs”, “PCLs”, “artificial intelligence”, and “machine learning”. Review articles and clinical case reports were excluded.
The primary objective of this review is to outline an ongoing multicenter, prospective initiative that details the study methodology for enhancing and prospectively evaluating a CNN-AI algorithm based on nCLE to improve presurgical risk stratification for BD-IPMNs. Furthermore, we suggest and assess an integrated diagnostic approach, which incorporates nCLE, cyst fluid analysis, and standard-of-care variables, with the aim of enhancing the accuracy of IPMN risk stratification.
3. Results
3.1. Utility and Accuracy of EUS-nCLE
EUS-nCLE provides real-time microscopic analysis of PCL epithelium without the need for high-risk endoscopic biopsy or surgical excision [
43,
44]. The PCL is visualized using EUS, and intravenous fluorescein is injected 2–3 minutes prior to imaging. A 19-gauge FNA needle preloaded with an nCLE miniprobe is advanced into the cyst until it opposes the cyst epithelium. The miniprobe is moved throughout the cyst cavity to assess different areas of its internal lining for approximately 6 minutes. After the video sequence is acquired, the miniprobe is removed, and the cyst fluid is aspirated for further analysis [
45].
In our previous studies, we demonstrated that quantitative analysis of PCL epithelium using EUS-nCLE outperformed the current standard of care in diagnosing HGD-Ca and LGD in IPMNs [
46,
47,
48]. These findings highlight the potential of EUS-nCLE to enhance diagnostic capabilities in this context.
We discovered several features that can be visualized on EUS-nCLE, which correspond to a higher histologic grade. For example, papillary epithelial width, as measured on nCLE images, suggests cellular atypia, while increased epithelial darkness on images is associated with nuclear stratification. These characteristics demonstrated high sensitivity in predicting HGD-Ca in an analysis of 26 BD-IPMNs. Papillary epithelial “width” and “darkness” exhibited sensitivities of 90% and 91%, respectively [
48]. Surgical histopathology served as the reference standard in this analysis. Additionally, there was substantial interobserver agreement (IOA) among external nCLE experts in detecting HGD-Ca, with a κ value of 0.61 for epithelial width and 0.55 for darkness [
48].
3.2. CNN-AI Algorithm for nCLE Analysis
By employing accurate AI-driven image analysis and recognition, the need for time-consuming manual quantification of papillary epithelial parameters by endomicroscopy specialists can be bypassed. In a recent post hoc analysis of video frames from the INDEX study, which included patients with histopathologically proven IPMNs, two CNN-based algorithms were developed to detect HGD-Ca in the lesions [
45]. The first algorithm utilizes an instance-segmentation-based model, specifically Mask R-CNN, to detect and segment papillary structures. It then measures papillary epithelial thickness and darkness and employs these features for diagnosing HGD-Ca. This model achieved an accuracy of 82.9%. The second algorithm applies a CNN model, namely VGGNet, to directly extract features from the holistic nCLE video frames for risk stratification. This approach yielded an accuracy of 85.7%. In comparison, the accuracy of current society guidelines (AGA and Fukuoka) reached only 68.6% and 74.3%, respectively. These findings highlight the potential of AI-based approaches in improving diagnostic accuracy and outperforming existing guidelines in the assessment of IPMNs.
3.3. Improving and Prospectively Evaluating the Single-Center Algorithm
In addition to high accuracy, our single-center algorithm must also demonstrate the ability to incorporate new patient data. Many patients with PCLs require EUS with or without nCLE, with big data being stored as large video files. The mean duration of the unedited EUS-nCLE videos in our previous studies was approximately seven minutes, and the videos were manually shortened to under three minutes of high-yield portions [
45,
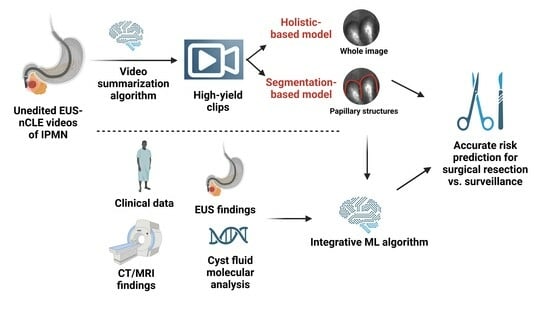
48]. Both algorithms in our pilot study used pre-edited videos where frames with artifact, blurring, or redundancy were removed by an nCLE expert. An algorithm that can be directly applied to unedited videos, without the need for manual editing, will greatly increase model efficiency, applicability, and generalizability.
Our future plans involve the development of a video summarization AI algorithm that will convert unedited nCLE videos into shorter, high-yield video clips, which will, in turn, improve the performance of our CNN-based algorithm [
49,
50,
51]. This approach aims to streamline the diagnostic process for HGD-Ca detection in three steps. First, a CNN-based classifier will be used to classify edited video frames into “high-risk” and “low-risk” images based on the presence or absence of papillary structures (as an indicator of potential dysplasia) [
52]. Second, an instance-segmentation-based algorithm will be employed to segment papillae and measure papillary epithelial thickness and darkness, enabling the grading of dysplasia. Concurrently, a holistic-based algorithm will utilize a CNN-based model to extract features from nCLE images, focusing on identifying HGD-Ca. The outputs of these two algorithms will be appropriately fused to leverage their complementary abilities and reduce uncertainty in the diagnostic process. Finally, image-level predictions from the entire edited video will be aggregated using a majority voting approach to diagnose HGD-Ca in each subject. By implementing these steps and integrating the video summarization algorithm, we aim to enhance the accuracy and efficiency of our diagnostic approach for HGD-Ca detection in PCLs.
3.4. Creating an Integrative Predictive Algorithm
While EUS-nCLE provides valuable information for PCL diagnosis, it represents only one source of patient data. Multiple clinical, demographic, genomic, and radiographic data points have been identified as significant predictors of HGD-Ca; incorporating all of these data into an integrative predictive algorithm will likely significantly improve accuracy. It is important to acknowledge that existing predictive models and expert consensus-led guidelines may oversimplify the impact of each variable considered. In routine medical practice, clinicians take into account patient demographics, apply guidelines such as Fukuoka-ICG, and often engage in multidisciplinary team discussions to assess the risk of malignancy associated with PCLs [
13]. By leveraging an ML-powered integrative framework, we can potentially optimize diagnostic accuracy by incorporating and analyzing the available data in a more comprehensive manner. This approach aims to combine the expertise of clinicians with the analytical power of ML algorithms to improve the accuracy of diagnosing PCLs and risk stratifying them accordingly.
Molecular analysis using NGS so far has been shown to more reliably predict HGD-Ca in IPMNs as compared to the standard of care. In a 2015 composite analysis, the combination of genetic mutations in SMAD4, RNF43, TP53, and aneuploidy predicted HGD-Ca in IPMNs with a sensitivity of 75% and a specificity of 92% [
52]. Another study showed that the presence of KRAS/GNAS along with additional mutations in TP53, PIK3CA, and PTEN produced 88% sensitivity and 97% specificity for BD-IPMNs with HGD-Ca [
53].
Standard-of-care variables include clinical characteristics (age, gender, onset of diabetes, family history symptoms, pancreatitis history), serum CA 19-9, cyst fluid analysis (glucose, CEA, cytology) as well as cyst and pancreas morphology, as detected via CT/MRI/EUS (factors, such as size, wall, thickness, mural nodules, growth rate, and pancreatic duct diameter). We intend to integrate four sources: cyst fluid NGS, standard-of-care variables in expert consensus guidelines (Fukuoka-ICG 2017 version), manual/human EUS-nCLE results, and CNN-AI results [
13].
The first component of our vision involves creating a logistic regression model that integrates the four elements mentioned to predict HGD-Ca or LGD in BD-IPMNs. The contribution of each variable will be assessed, and our sample size is expected to include at least 300 BD-IPMNs, with approximately 50% being HGD-Ca cases.
The second component aims to develop an ML-based approach for integrating the four data sources. Decision trees are chosen as the ML algorithm of choice due to their ability to handle both continuous and categorical variables seamlessly. Additionally, decision trees provide human-understandable explanations of the decision-making process, enabling experts to verify and interpret the learned decision trees. This feature facilitates the integration of the model into future management guidelines. Suitable options for doing this include using random forests and XGBoost, both of which are well-known ensemble methods over decisions trees that offer easy ways to control the model complexity to overcome over/under-fitting by controlling the number of input variables, tree depths/widths, and number of trees [
54,
55,
56]. These enhanced models have the potential to diagnose HGD-Ca in BD-IPMNs with optimized diagnostic accuracy. Furthermore, by identifying the most significant contributors from the data sources, these models can potentially inform new clinical practice guidelines and improve the risk stratification of BD-IPMNs.
3.5. Prior Integrative Algorithms
Integration of multiple data sources into AI algorithms to solve clinical problems related to PCLs and pancreatic malignancy have shown promising results, as illustrated in
Table 2 [
52,
57,
58,
59]. In 2015, Springer et al. developed an algorithm that analyzed multi-parametric features (known as Multivariate Organization of Combinatorial Alterations or MOCA) to identify PCLs requiring resection [
52]. The researchers combined composite clinical markers (such as age, presence of abdominal symptoms, and specific imaging results) and composite molecular markers (including aneuploidy and various gene mutations) in their MOCA algorithm. When evaluating the markers individually, the composite clinical marker and the composite molecular marker showed sensitivities of 75% and 77%, respectively, in identifying cysts that required resection. However, when used together, their sensitivity increased to 89%. Furthermore, the combination of molecular and clinical markers in the study resulted in a sensitivity of 94% for detecting IPMNs, an increase from the 76% sensitivity observed with the composite molecular marker alone. Subsequently, the same group developed CompCyst in 2019, which is a test built on the MOCA algorithm. This test categorizes patients with PCLs into three management groups (surgery, surveillance, or discharge) based on the evaluation of various clinical and molecular markers [
58]. Overall, these advancements in combining molecular and clinical markers, as demonstrated by the MOCA algorithm and CompCyst, offer improved stratification and management of patients with PCLs. The test integrated three data elements: presence of VHL mutation but absence of GNAS mutation, decreased expression of a VEGF-A protein, and a combination of factors (solid component of cyst seen on imaging, aneuploidy, and presence of mutations in certain genes). Clinical, imaging, and molecular data were integrated in their test, and they produced higher accuracy compared to conventional clinical and imaging criteria. The authors estimated that widespread use of CompCyst may reduce the number of unnecessary surgical resections by 60%.
Other recent studies have also leveraged ML to perform integrative analyses to manage other malignancies. One study used a deep-learning-based stacked ensemble model to predict the prognosis of breast cancer from available multi-modal cancer datasets, including genetic data (gene expression, copy number variation) and clinical data (age, subjective timing of menstruation and menopause, timing of pregnancy and others) [
64]. Integrative approaches have also been applied to determine prognosis of clear cell renal cell carcinoma and lung adenocarcinoma [
65,
66]. Additionally, it is well established in the literature that deep learning methods utilizing multiple modalities of input data sources (multimodal) outperform methods with a single source of input data (unimodal) [
67,
68,
69]. Given the promising evidence regarding the success of previous integrative AI algorithms in addressing both clinical and non-clinical challenges, we anticipate that our next step, involving an integrative approach with an ML-powered model, will contribute to optimizing diagnostic accuracy and further enhancing the ability to predict HGD-Ca in branch-duct IPMNs.
4. Discussion
In this review, we present some existing applications of AI in diagnosing and predicting malignant potential of PCLs. We propose a comprehensive plan for designing an accurate, integrative, and scalable predictive algorithm for predicting the progression of branch-duct IPMNs using a combination of EUS-nCLE findings, clinical data, fluid analysis, and radiographic imaging. To reduce the workload of the time-consuming pre-processing of videos by specialists, we will develop a video summarization algorithm capable of automating the editing of nCLE videos. This algorithm will use AI to identify high-yield video clips that contain relevant information for analysis. Next, we will employ two models to risk stratify the IPMNs based on the selected video clips. These models will utilize advanced ML techniques to assess and predict the likelihood of neoplasia progression. Subsequently, we will develop an integrative algorithm that incorporates standard-of-care variables, nCLE videos, and NGS in cyst fluid to further enhance the accuracy of risk stratification. By combining multiple data sources and leveraging the power of AI, we aim to improve the predictive capabilities of our algorithm. By implementing these steps, our goal is to develop an advanced AI-powered algorithm that can accurately predict the malignant progression of IPMNs, providing clinicians with valuable insights for personalized patient management and decision making.
There are several foreseeable limitations to this model. Firstly, EUS-nCLE is typically only available at large academic institutions and referral centers. Patients who undergo these procedures usually reside in metropolitan regions or possess the resources to seek treatment at such institutions. This is in contrast to non-invasive imaging modalities, like CT and MRI or even routine cyst fluid analysis, which are more widely accessible. Consequently, the preference for using data from patients who have undergone EUS-nCLE may introduce a selection bias into the final algorithm.
Another source of selection bias arises from the necessity for surgical histopathology to establish a formal diagnosis. As a result, the data used for training the models must come from patients who have already been identified as high risk and have undergone surgical resection.
Additionally, the costs and resources required for implementing the AI algorithm, including software development, maintenance, and staff training, could pose barriers to widespread adoption, especially in lower-resource regions. As is the case with all applications of AI in medicine, our algorithm should be viewed as an adjunct to assist in decision making rather than as a replacement for clinical expertise. Lastly, our plan to enhance generalizability by utilizing data from multiple hospitals across the U.S. presents unique challenges related to patient privacy.
Multi-Center Collaboration
To ensure the generalizability of our algorithm to a larger patient population, it is crucial to validate it in a multi-center cohort. Our aim is to include patients from at least ten tertiary care centers, and the development and validation of the multi-center algorithm will be carried out concurrently with the design of our improved CNN and integrative models. Combining data from multiple centers presents several challenges, including increased variability, domain discrepancies, and differences in devices and favored imaging modalities. Furthermore, the importance of data privacy, protection, and ownership has become increasingly recognized in recent years.
Even when transmitting anonymous data between centers, it is widely acknowledged that simply removing a patient’s name or medical record number is insufficient to protect their privacy. Ideally, collaboration and algorithm updates should be achieved without the need to share patient data across centers. This would require implementing privacy-preserving techniques and secure protocols that allow for the exchange of knowledge and model updates while maintaining data confidentiality. Addressing these challenges will be essential in order to ensure effective collaboration and advancements in algorithm development without compromising patient privacy.
To address these challenges, we intend to utilize federated learning (FL) and domain adaptation (DA) techniques. Traditionally, data used in ML algorithms are collected from various sources and processed in a centralized manner at a single site. ML models are then trained using these centralized data. In contrast, FL aims to train AI models while keeping the data decentralized at multiple data collection sites [
70,
71]. In the mainstream FL framework, a centralized server coordinates the model training process. Initially, the server broadcasts the ML model to each data collection site, known as a client. These client devices, which are secure servers located in their respective hospitals, then train the models using their local data. Subsequently, the updated model parameters are sent back to the server from each client. The server aggregates these client model parameters into a single ML model. This approach enables collaborative model development and adaptation among multiple hospitals without the need to share any patient data [
71,
72,
73,
74,
75,
76,
77]. FL has demonstrated reliable results when applied to clinical information, PET images, and multimodal data. Building upon our prior work, we will develop an FL framework that leverages unlabeled data at our center to enhance the aggregation of clients’ models. This approach will further improve the performance and adaptability of the AI model while maintaining data privacy and security [
78,
79,
80].
In addition to FL, we will also employ DA techniques to address the data domain discrepancy across different centers. It is widely recognized in the ML community that such domain discrepancy can negatively impact the accuracy of ML models. Traditional DA methods typically involve sharing data across different sources or centers to quantify the differences in data distributions and then applying transformations to reduce the domain discrepancy [
81,
82,
83]. To better protect patient privacy, recent advancements have introduced source-free DA algorithms, where the learned model only has access to unlabeled target data during the adaptation process, bypassing the need for data sharing [
84,
85]. This approach allows for the development of models that can adapt to different domains without compromising patient data security. Integrating these source-free DA techniques into our FL framework will enable us to adapt the ML model to different hospitals without the requirement of sharing patient data (particularly all 18 HIPAA identifiers for protected health information), ultimately improving the overall accuracy of the model. By combining FL and DA techniques, we aim to address both the challenges of data decentralization and domain discrepancy, ensuring the development of accurate and robust ML models while safeguarding patient data privacy (
Figure 2).
5. Conclusions
AI is increasingly being adopted in the field of medicine to derive actionable insights from extensive patient data and alleviate the workload of specialists. Shortcomings in current models utilizing nCLE for risk stratification of PCLs include the necessity for manual image pre-processing and the exclusion of pertinent data with significant predictive value. Our research group has previously developed nCLE-based CNN models for risk stratification of BD-IPMNs, which exhibited diagnostic accuracy surpassing that of standard-of-care guidelines, albeit using expert-edited endomicroscopy videos. We now present our strategy for enhancing the model’s accuracy and generalizability, which encompasses a video summarization algorithm, an integrated ML algorithm that harnesses a vast amount of data, and the validation of our predictive model within a multi-center cohort. While ML and AI require extensive imaging and datasets, the aspiration of this methodology review is to encourage researchers worldwide to collaborate in enhancing the management of patients with pancreatic cystic lesions.
Author Contributions
Conceptualization, W.-L.C. and S.G.K.; resources, J.J., T.C. and S.G.K.; writing—original draft preparation, J.J., T.C., T.M. and S.G.K.; writing—review and editing, all authors; supervision, S.G.K. All authors have read and agreed to the published version of the manuscript.
Funding
Research by S.G.K was supported by a grant from the National Institutes of Health, the National Cancer Institute (NIH/NCI) (R01CA279965).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of The Ohio State University.
Conflicts of Interest
S.G.K—research grant support (investigator-initiated studies) from Mauna Kea Technologies, Paris, France, and Taewoong Medical, USA.
References
- Zerboni, G.; Signoretti, M.; Crippa, S.; Falconi, M.; Arcidiacono, P.G.; Capurso, G. Systematic review and meta-analysis: Prevalence of incidentally detected pancreatic cystic lesions in asymptomatic individuals. Pancreatology 2019, 19, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, F.; Davis, A.M.; Chapman, C.G. Pancreatic Cysts-An Overview and Summary of Society Guidelines, 2021. JAMA 2021, 325, 391–392. [Google Scholar] [CrossRef] [PubMed]
- Elta, G.H.; Enestvedt, B.K.; Sauer, B.G.; Lennon, A.M. ACG Clinical Guideline: Diagnosis and Management of Pancreatic Cysts. Am. J. Gastroenterol. 2018, 113, 464–479. [Google Scholar] [CrossRef]
- Kaimakliotis, P.; Riff, B.; Pourmand, K.; Chandrasekhara, V.; Furth, E.E.; Siegelman, E.S.; Drebin, J.; Vollmer, C.M.; Kochman, M.L.; Ginsberg, G.G.; et al. Sendai and Fukuoka consensus guidelines identify advanced neoplasia in patients with suspected mucinous cystic neoplasms of the pancreas. Clin. Gastroenterol. Hepatol. 2015, 13, 1808–1815. [Google Scholar] [CrossRef]
- Sakorafas, G.H.; Sarr, M.G.; van de Velde, C.J.; Peros, G. Intraductal papillary mucinous neoplasms of the pancreas: A surgical perspective. Surg. Oncol. 2005, 14, 155–178. [Google Scholar] [CrossRef] [PubMed]
- Castellano-Megías, V.M.; Andrés, C.I.; López-Alonso, G.; Colina-Ruizdelgado, F. Pathological features and diagnosis of intraductal papillary mucinous neoplasm of the pancreas. World J. Gastrointest. Oncol. 2014, 6, 311–324. [Google Scholar] [CrossRef]
- Machado, N.O.; Al Qadhi, H.; Al Wahibi, K. Intraductal Papillary Mucinous Neoplasm of Pancreas. N. Am. J. Med. Sci. 2015, 7, 160–175. [Google Scholar] [CrossRef]
- Scheiman, J.M.; Hwang, J.H.; Moayyedi, P. American gastroenterological association technical review on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015, 148, 824–848.e22. [Google Scholar] [CrossRef]
- Marchegiani, G.; Pollini, T.; Andrianello, S.; Tomasoni, G.; Biancotto, M.; Javed, A.A.; Kinny-Köster, B.; Amini, N.; Han, Y.; Kim, H.; et al. Progression vs cyst stability of branch-duct intraductal papillary mucinous neoplasms after observation and surgery. JAMA Surg. 2021, 156, 654–661. [Google Scholar] [CrossRef]
- Crist, D.W.; Sitzmann, J.V.; Cameron, J.L. Improved hospital morbidity, mortality, and survival after the Whipple procedure. Ann. Surg. 1987, 206, 358–365. [Google Scholar] [CrossRef]
- Chierici, A.; Intotero, M.; Granieri, S.; Paleino, S.; Flocchini, G.; Germini, A.; Cotsoglou, C. Timely synergic surgical and radiological aggressiveness improves perioperative mortality after hemorrhagic complication in Whipple procedure. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 387–390. [Google Scholar] [CrossRef]
- Tanaka, M. Intraductal papillary mucinous neoplasm of the pancreas as the main focus for early detection of pancreatic adenocarcinoma. Pancreas 2018, 47, 544–550. [Google Scholar] [CrossRef]
- Tanaka, M.; Fernández-del Castillo, C.; Kamisawa, T.; Jang, J.Y.; Levy, P.; Ohtsuka, T.; Salvia, R.; Shimizu, Y.; Tada, M.; Wolfgang, C.L. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017, 17, 738–753. [Google Scholar] [CrossRef]
- Sharib, J.M.; Fonseca, A.L.; Swords, D.S.; Jaradeh, K.; Bracci, P.M.; Firpo, M.A.; Hatcher, S.; Scaife, C.L.; Wang, H.; Kim, G.E.; et al. Surgical overtreatment of pancreatic intraductal papillary mucinous neoplasms: Do the 2017 International Consensus Guidelines improve clinical decision making? Surgery 2018, 164, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Dbouk, M.; Brewer Gutierrez, O.I.; Lennon, A.M.; Chuidian, M.; Shin, E.J.; Kamel, I.R.; Fishman, E.K.; He, J.; Burkhart, R.A.; Wolfgang, C.L.; et al. Guidelines on management of pancreatic cysts detected in high-risk individuals: An evaluation of the 2017 Fukuoka guidelines and the 2020 International Cancer of the Pancreas Screening (CAPS) consortium statements. Pancreatology 2021, 21, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Heckler, M.; Michalski, C.W.; Schaefle, S.; Kaiser, J.; Büchler, M.W.; Hackert, T. The Sendai and Fukuoka consensus criteria for the management of branch duct IPMN-A meta-analysis on their accuracy. Pancreatology 2017, 17, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Takasu, N.; Watanabe, T.; Fukumoto, T.; Okazaki, S.; Tezuka, K.; Sugawara, S.; Hirai, I.; Kimura, W. Validation of the 2012 Fukuoka Consensus Guideline for Intraductal Papillary Mucinous Neoplasm of the Pancreas From a Single Institution Experience. Pancreas 2017, 46, 936–942. [Google Scholar] [CrossRef] [PubMed]
- David, G.K.; Gail, D.M.; Klein, M. Logistic Regression; Springer: New York, NY, USA, 2002. [Google Scholar]
- Kotsiantis, S.B. Decision trees: A recent overview. Artif. Intell. Rev. 2013, 39, 261–283. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. TIST 2011, 3, 2. [Google Scholar] [CrossRef]
- Thomas, H.; Schölkopf, B.; Smola, A.J. Kernel methods in machine learning. Ann. Statist. 2008, 36, 1171–1220. [Google Scholar]
- Yann, L.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 7553, 436–444. [Google Scholar]
- Ian, G.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Yann, L.; Bengio, Y. Convolutional networks for images, speech, and time series. In The handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 1995; Volume 10, p. 3361. [Google Scholar]
- Alex, K.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 6, 84–90. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. arXiv 2015, arXiv:1506.01497. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 4, 834–848. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Si, K.; Xue, Y.; Yu, X.; Zhu, X. Fully end-to-end deep-learning-based diagnosis of pancreatic tumors. Theranostics 2021, 11, 1982–1990. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, Y.; Yue, X.; Ma, C.; Liu, X.; Yang, P.; Lu, J. Deep pancreas segmentation with uncertain regions of shadowed sets. Magn. Reson. Imaging 2020, 68, 45–52. [Google Scholar] [CrossRef]
- Wei, R.; Lin, K.; Yan, W.; Guo, Y.; Wang, Y.; Li, J.; Zhu, J. Computer-Aided Diagnosis of Pancreas Serous Cystic Neoplasms: A Radiomics Method on Preoperative MDCT Images. Technol. Cancer Res. Treat. 2019, 18, 1533033818824339. [Google Scholar] [CrossRef]
- Chakraborty, J.; Midya, A.; Gazit, L.; Attiyeh, M.; Langdon-Embry, L.; Allen, P.J.; Do, R.K.G.; Simpson, A.L. CT radiomics to predict high-risk intraductal papillary mucinous neoplasms of the pancreas. Med. Phys. 2018, 45, 5019–5029. [Google Scholar] [CrossRef]
- Chu, L.C.; Park, S.; Soleimani, S.; Fouladi, D.F.; Shayesteh, S.; He, J.; Javed, A.A.; Wolfgang, C.L.; Vogelstein, B.; Kinzler, K.W. Classification of pancreatic cystic neoplasms using radiomic feature analysis is equivalent to an experienced academic radiologist: A step toward computer-augmented diagnostics for radiologists. Abdom. Radiol. 2022, 47, 4139–4150. [Google Scholar] [CrossRef]
- Liang, W.; Tian, W.; Wang, Y.; Wang, P.; Wang, Y.; Zhang, H.; Ruan, S.; Shao, J.; Zhang, X.; Huang, D.; et al. Classification prediction of pancreatic cystic neoplasms based on radiomics deep learning models. BMC Cancer 2022, 22, 1237. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Heilmaier, M.; Phillip, V.; Treiber, M.; Mayr, U.; Lahmer, T.; Mueller, J.; Demir, I.E.; Friess, H.; Reichert, M.; et al. Accurate prediction of histological grading of intraductal papillary mucinous neoplasia using deep learning. Endoscopy 2023, 55, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Javed, S.; Qureshi, T.A.; Deng, Z.; Wachsman, A.; Raphael, Y.; Gaddam, S.; Xie, Y.; Pandol, S.J.; Li, D. Segmentation of Pancreatic Subregions in Computed Tomography Images. J. Imaging 2022, 8, 195. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-H.; Kim, Y.J.; Park, Y.-H.; Kim, D.; Kim, K.G.; Lee, D.-H. Automated pancreas segmentation and volumetry using deep neural network on computed tomography. Sci. Rep. 2022, 12, 4075. [Google Scholar] [CrossRef]
- Kamboj, A.K.; Modi, R.M.; Swanson, B.; Conwell, D.L.; Krishna, S.G. A comprehensive examination of the novel techniques used for in vivo and ex vivo confocal laser endomicroscopy of pancreatic cystic lesions. VideoGIE 2016, 1, 6–7. [Google Scholar] [CrossRef]
- Nakai, Y.; Iwashita, T.; Park, D.H.; Samarasena, J.B.; Lee, J.G.; Chang, K.J. Diagnosis of pancreatic cysts: EUS-guided, through-the-needle confocal laser-induced endomicroscopy and cystoscopy trial: DETECT study. Gastrointest. Endosc. 2015, 81, 1204–1214. [Google Scholar] [CrossRef]
- Machicado, J.D.; Chao, W.L.; Carlyn, D.E.; Pan, T.Y.; Poland, S.; Alexander, V.L.; Maloof, T.G.; Dubay, K.; Ueltschi, O.; Middendorf, D.M.; et al. High performance in risk stratification of intraductal papillary mucinous neoplasms by confocal laser endomicroscopy image analysis with convolutional neural networks (with video). Gastrointest. Endosc. 2021, 94, 78–87.e2. [Google Scholar] [CrossRef]
- Krishna, S.G.; Hart, P.A.; DeWitt, J.M.; DiMaio, C.J.; Kongkam, P.; Napoleon, B.; Othman, M.O.; Tan, D.M.Y.; Strobel, S.G.; Stanich, P.P.; et al. EUS-guided confocal laser endomicroscopy: Prediction of dysplasia in intraductal papillary mucinous neoplasms (with video). Gastrointest. Endosc. 2020, 91, 551–563.e5. [Google Scholar] [CrossRef]
- Coban, S.; Brugge, W. EUS-guided confocal laser endomicroscopy: Can we use thick and wide for diagnosis of early cancer? Gastrointest. Endosc. 2020, 91, 564–567. [Google Scholar] [CrossRef]
- Krishna, S.G.; Hart, P.A.; Malli, A.; Kruger, A.J.; McCarthy, S.T.; El-Dika, S.; Walker, J.P.; Dillhoff, M.E.; Manilchuk, A.; Schmidt, C.R.; et al. Endoscopic Ultrasound-Guided Confocal Laser Endomicroscopy Increases Accuracy of Differentiation of Pancreatic Cystic Lesions. Clin. Gastroenterol. Hepatol. 2020, 18, 432–440.e6. [Google Scholar] [CrossRef]
- Ruder, S. An overview of multi-task learning in deep neural networks. arXiv 2017, arXiv:1706.05098. [Google Scholar]
- Zhang, K.; Chao, W.-L.; Sha, F.; Grauman, K. Video summarization with long short-term memory. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Gong, B.; Chao, W.-L.; Grauman, K.; Sha, F. Diverse sequential subset selection for supervised video summarization. Adv. Neural Inf. Process. Syst. 2014, 27, 2069–2077. [Google Scholar]
- Springer, S.; Wang, Y.; Dal Molin, M.; Masica, D.L.; Jiao, Y.; Kinde, I.; Blackford, A.; Raman, S.P.; Wolfgang, C.L.; Tomita, T.; et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015, 149, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; McGrath, K.; E Brand, R.; Khalid, A.; Zeh, H.J.; Chennat, J.S.; E Fasanella, K.; I Papachristou, G.; Slivka, A.; Bartlett, D.L.; et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2018, 67, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Li, W.; Yin, Y.; Quan, X.; Zhang, H. Gene expression value prediction based on XGBoost algorithm. Front. Genet. 2019, 10, 1077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qian, L.; Mao, B.; Huang, C.; Huang, B.; Si, Y. A data-driven design for fault detection of wind turbines using random forests and XGboost. IEEE Access 2018, 6, 21020–21031. [Google Scholar] [CrossRef]
- Rangwani, S.; Ardeshna, D.R.; Rodgers, B.; Melnychuk, J.; Turner, R.; Culp, S.; Chao, W.-L.; Krishna, S.G. Application of Artificial Intelligence in the Management of Pancreatic Cystic Lesions. Biomimetics 2022, 7, 79. [Google Scholar] [CrossRef]
- Springer, S.; Masica, D.L.; Dal Molin, M.; Douville, C.; Thoburn, C.J.; Afsari, B.; Li, L.; Cohen, J.D.; Thompson, E.; Allen, P.J.; et al. A multimodality test to guide the management of patients with a pancreatic cyst. Sci. Transl. Med. 2019, 11, eaav4772. [Google Scholar] [CrossRef]
- Tang, B.; Chen, Y.; Wang, Y.; Nie, J. A Wavelet-Based Learning Model Enhances Molecular Prognosis in Pancreatic Adenocarcinoma. BioMed. Res. Int. 2021, 2021, 7865856. [Google Scholar] [CrossRef]
- Permuth, J.B.; Choi, J.; Balarunathan, Y.; Kim, J.; Chen, D.-T.; Chen, L.; Orcutt, S.; Doepker, M.P.; Gage, K.; Zhang, G.; et al. Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget 2016, 7, 85785–85797. [Google Scholar] [CrossRef] [PubMed]
- Kurita, Y.; Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Koda, H.; Tajika, M.; Shimizu, Y.; et al. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci. Rep. 2019, 9, 6893. [Google Scholar] [CrossRef] [PubMed]
- Blyuss, O.; Zaikin, A.; Cherepanova, V.; Munblit, D.; Kiseleva, E.M.; Prytomanova, O.M.; Duffy, S.W.; Crnogorac-Jurcevic, T. Development of PancRISK, a urine biomarker-based risk score for stratified screening of pancreatic cancer patients. Br. J. Cancer 2020, 122, 692–696. [Google Scholar] [CrossRef]
- Hernandez-Barco, Y.G.; Daye, D.; Castillo, C.F.F.-D.; Parker, R.F.; Casey, B.W.; Warshaw, A.L.; Ferrone, C.R.; Lillemoe, K.D.; Qadan, M. IPMN-LEARN: A linear support vector machine learning model for predicting low-grade intraductal papillary mucinous neoplasms. Ann. Hepatobiliary Pancreat. Surg. 2023, 27, 195–200. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Puppala, M.; Ezeana, C.F.; Huang, Y.-S.; Chou, P.-H.; Yu, X.; Chen, S.; Wang, L.; Yin, Z.; Danforth, R.L.; et al. A deep learning–based decision support tool for precision risk assessment of breast cancer. JCO Clin. Cancer Inform. 2019, 3, 1–12. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, J.; Han, Y.; Wang, X.; Ye, X.; Meng, Y.; Parwani, A.; Han, Z.; Feng, Q.; Huang, K. Integrative analysis of histopathological images and genomic data predicts clear cell renal cell carcinoma prognosis. Cancer Res. 2017, 77, e91–e100. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, D.; Gu, J.; Xu, F.; Zhu, Q.; Lu, C. A large cohort study identifying a novel prognosis prediction model for lung adenocarcinoma through machine learning strategies. BMC Cancer 2019, 19, 886. [Google Scholar] [CrossRef]
- Alleman, K.; Knecht, E.; Huang, J.; Zhang, L.; Lam, S.; DeCuypere, M. Multimodal Deep Learning-Based Prognostication in Glioma Patients: A Systematic Review. Cancers 2023, 15, 545. [Google Scholar] [CrossRef]
- Soto, J.T.; Hughes, J.W.; Sanchez, P.A.; Perez, M.; Ouyang, D.; A Ashley, E. Multimodal deep learning enhances diagnostic precision in left ventricular hypertrophy. Eur. Heart J. Digit. Health 2022, 3, 380–389. [Google Scholar] [CrossRef]
- Zhao, D.; Homayounfar, M.; Zhen, Z.; Wu, M.-Z.; Yu, S.Y.; Yiu, K.-H.; Vardhanabhuti, V.; Pelekos, G.; Jin, L.; Koohi-Moghadam, M. A Multimodal Deep Learning Approach to Predicting Systemic Diseases from Oral Conditions. Diagnostics 2022, 12, 3192. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, C.; Zhu, M.; Peng, Z.; Yuan, Y. Personalized Retrogress-Resilient Federated Learning Toward Imbalanced Medical Data. IEEE Trans. Med. Imaging 2022, 41, 3663–3674. [Google Scholar] [CrossRef] [PubMed]
- Wicaksana, J.; Yan, Z.; Yang, X.; Liu, Y.; Fan, L.; Cheng, K.-T. Customized Federated Learning for Multi-Source Decentralized Medical Image Classification. IEEE J. Biomed. Health Inform. 2022, 26, 5596–5607. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.; Guo, P. Federated learning in cloud-edge collaborative architecture: Key technologies, applications and challenges. J. Cloud Comput. Heidelb. 2022, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Shiri, I.; Sadr, A.V.; Akhavan, A.; Salimi, Y.; Sanaat, A.; Amini, M.; Razeghi, B.; Saberi, A.; Arabi, H.; Ferdowsi, S.; et al. Decentralized collaborative multi-institutional PET attenuation and scatter correction using federated deep learning. Eur. J. Nucl. Med. Mol. Imaging 2022, 50, 1034–1050. [Google Scholar] [CrossRef] [PubMed]
- Kidd, B.; Wang, K.; Xu, Y.; Ni, Y. Federated Learning for Sparse Bayesian Models with Applications to Electronic Health Records and Genomics. Pac. Symp. Biocomput. 2023, 28, 484–495. [Google Scholar]
- Alam, M.U.; Rahmani, R. FedSepsis: A Federated Multi-Modal Deep Learning-Based Internet of Medical Things Application for Early Detection of Sepsis from Electronic Health Records Using Raspberry Pi and Jetson Nano Devices. Sensors 2023, 23, 970. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Tardy, M.; Ballester, M.A.G.; Mateus, D.; Piella, G. Memory-aware curriculum federated learning for breast cancer classification. Comput. Methods Programs Biomed. 2022, 229, 107318. [Google Scholar] [CrossRef]
- Ogier du Terrail, J.; Leopold, A.; Joly, C.; Béguier, C.; Andreux, M.; Maussion, C.; Schmauch, B.; Tramel, E.W.; Bendjebbar, E.; Zaslavskiy, M.; et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat. Med. 2023, 29, 135–146. [Google Scholar] [CrossRef]
- Han, S.; Ding, H.; Zhao, S.; Ren, S.; Wang, Z.; Lin, J.; Zhou, S. Practical and Robust Federated Learning With Highly Scalable Regression Training. IEEE Trans. Neural. Netw. Learn Syst. 2023; advance online Publication. [Google Scholar] [CrossRef]
- Kwak, L.; Bai, H. The Role of Federated Learning Models in Medical Imaging. Radiol. Artif. Intell. 2023, 5, e230136. [Google Scholar] [CrossRef]
- Ullah, F.; Srivastava, G.; Xiao, H.; Ullah, S.; Lin, J.C.-W.; Zhao, Y. A Scalable Federated Learning Approach for Collaborative Smart Healthcare Systems with Intermittent Clients using Medical Imaging. IEEE J. Biomed. Health Inform. 2023, 1–13. [Google Scholar] [CrossRef]
- Goel, P.; Ganatra, A. Unsupervised Domain Adaptation for Image Classification and Object Detection Using Guided Transfer Learning Approach and JS Divergence. Sensors 2023, 23, 4436. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zong, Y.; He, Y.; Shi, G.; Jiang, C. Domain Adaptation-Based Automated Detection of Retinal Diseases from Optical Coherence Tomography Images. Curr. Eye Res. 2023, 48, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Chen, H.; Xu, Y.; Chen, H.; Liu, Y.; Tan, H.; Yan, Y.; Yu, H. Multicomponent Adversarial Domain Adaptation: A General Framework. IEEE Trans. Neural. Netw. Learn Syst. 2023, 34, 6824–6838. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Hu, D.; Feng, J. Do we really need to access the source data? source hypothesis transfer for unsupervised domain adaptation. In Proceedings of the International Conference on Machine Learning, Vienna, Austria, 12–18 July 2020; pp. 6028–6039. [Google Scholar]
- Yang, S.; Wang, Y.; Van De Weijer, J.; Herranz, L.; Jui, S. Generalized source-free domain adaptation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 8978–8987. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).