Antisense Activity across the Nesp Promoter is Required for Nespas-Mediated Silencing in the Imprinted Gnas Cluster
Abstract
:1. Introduction
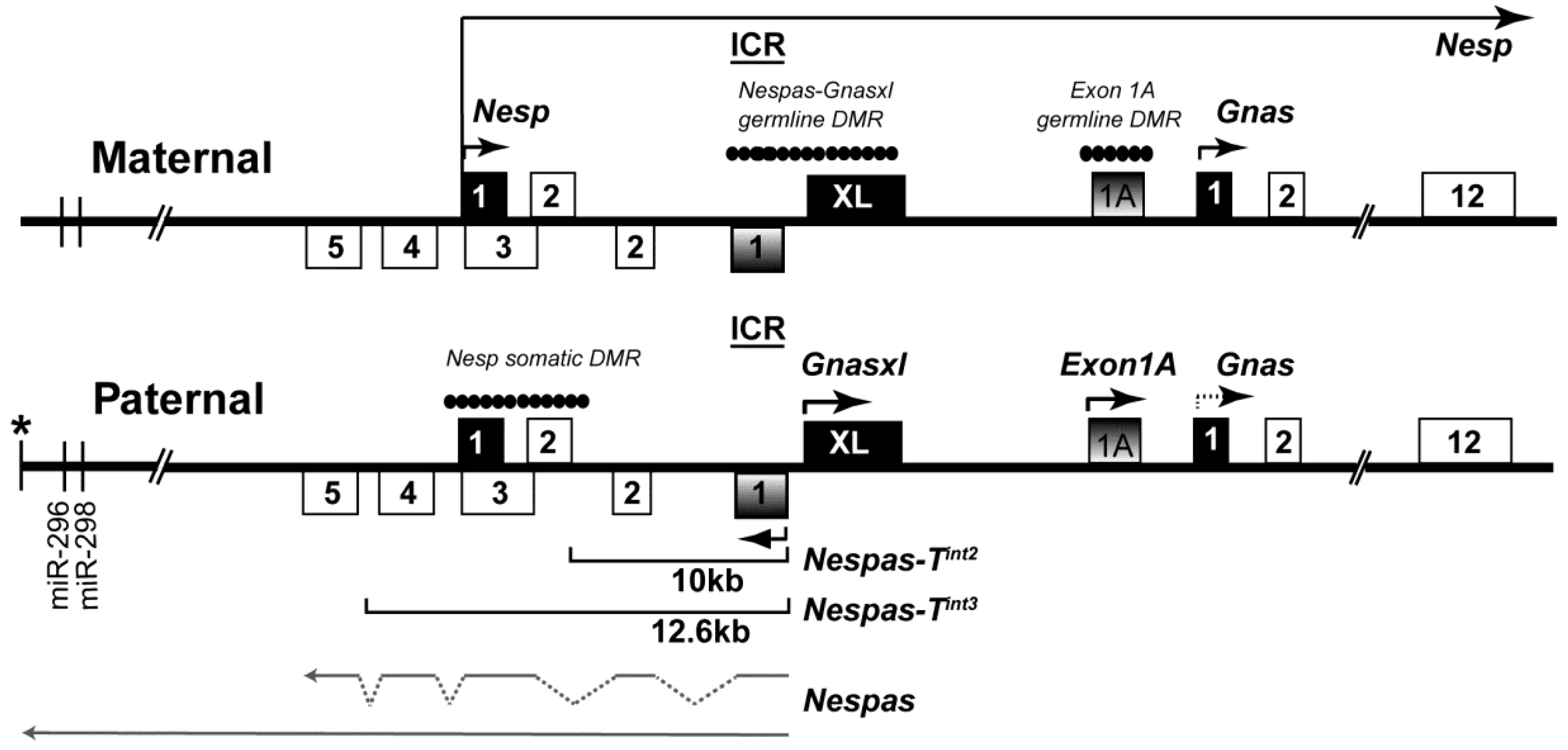
2. Results
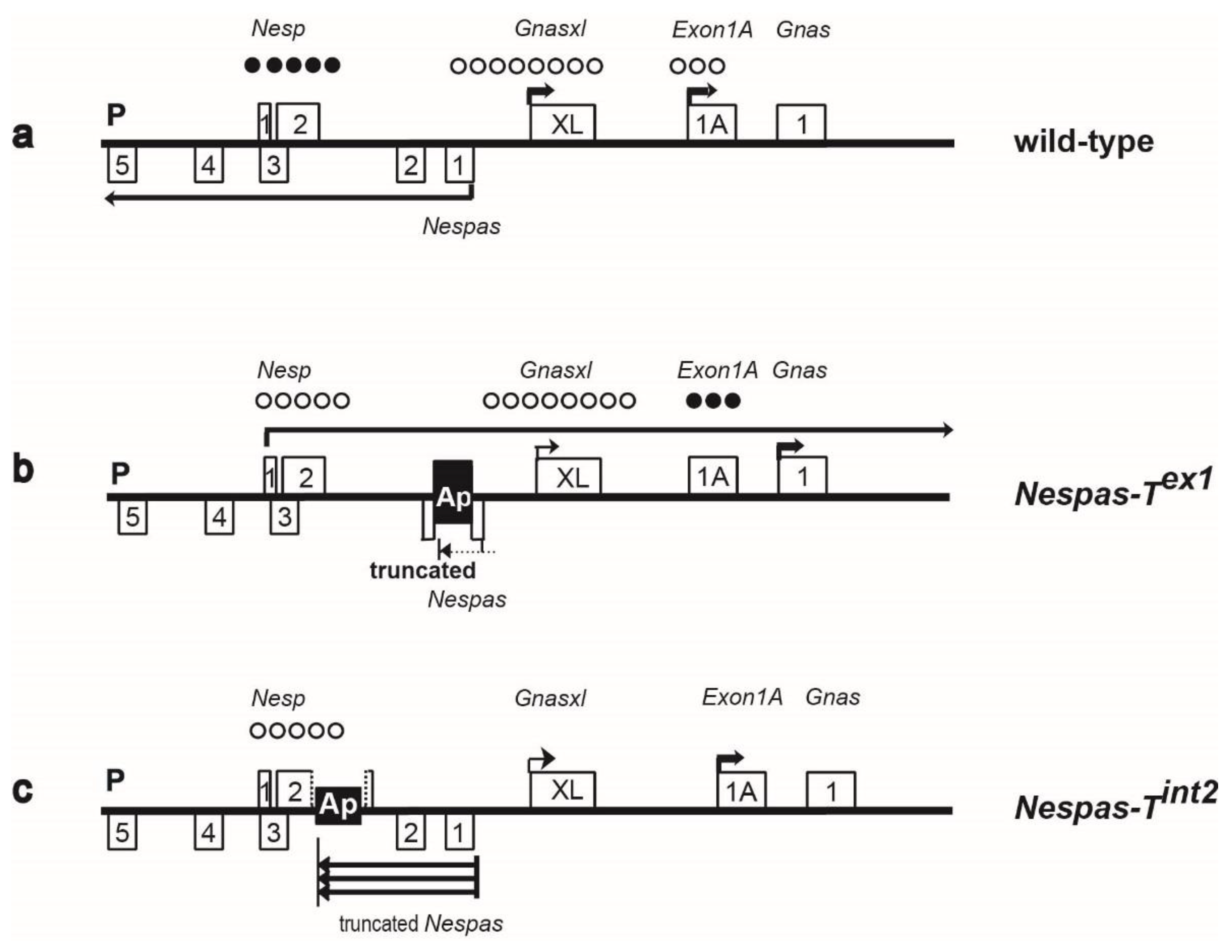
2.1. Nespas Transcription Extends over 30 kb
2.2. The Nesp DMR Is Unmethylated When Nespas Is Truncated before the Nesp Promoter

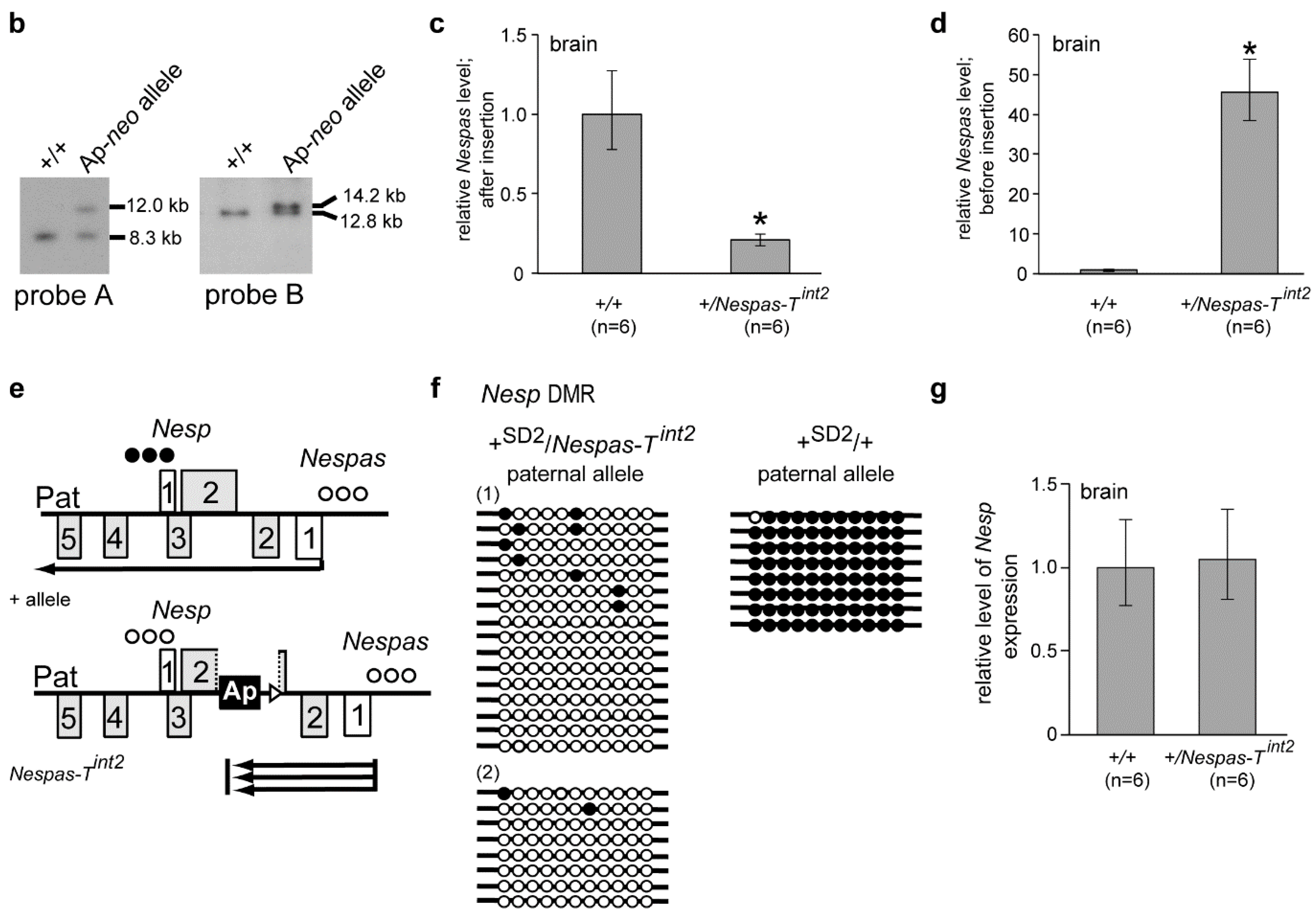
2.3. Lack of Methylation at the Nesp DMR Is Due to Truncation of Nespas


2.4. Truncation of Nespas after the Nesp Promoter
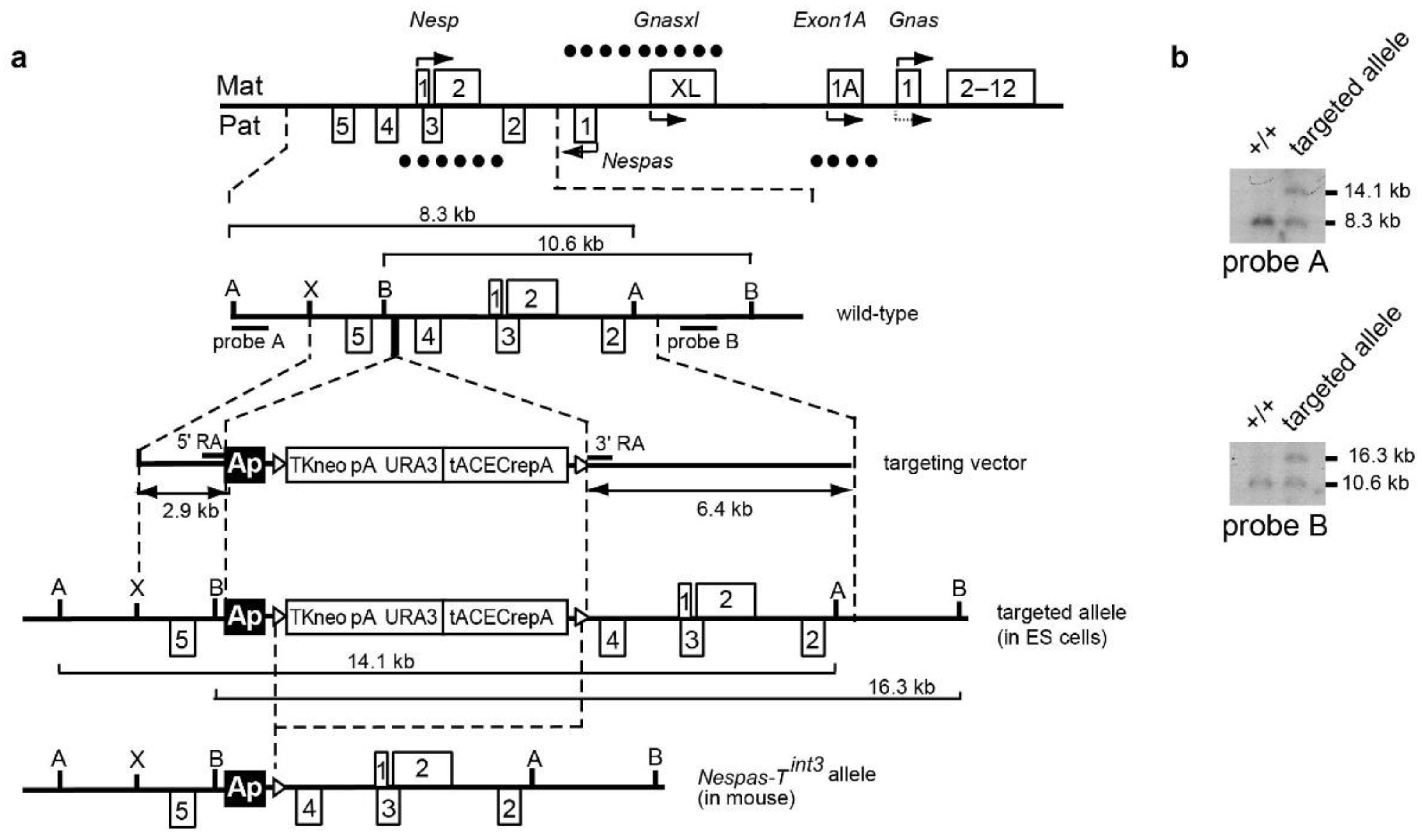
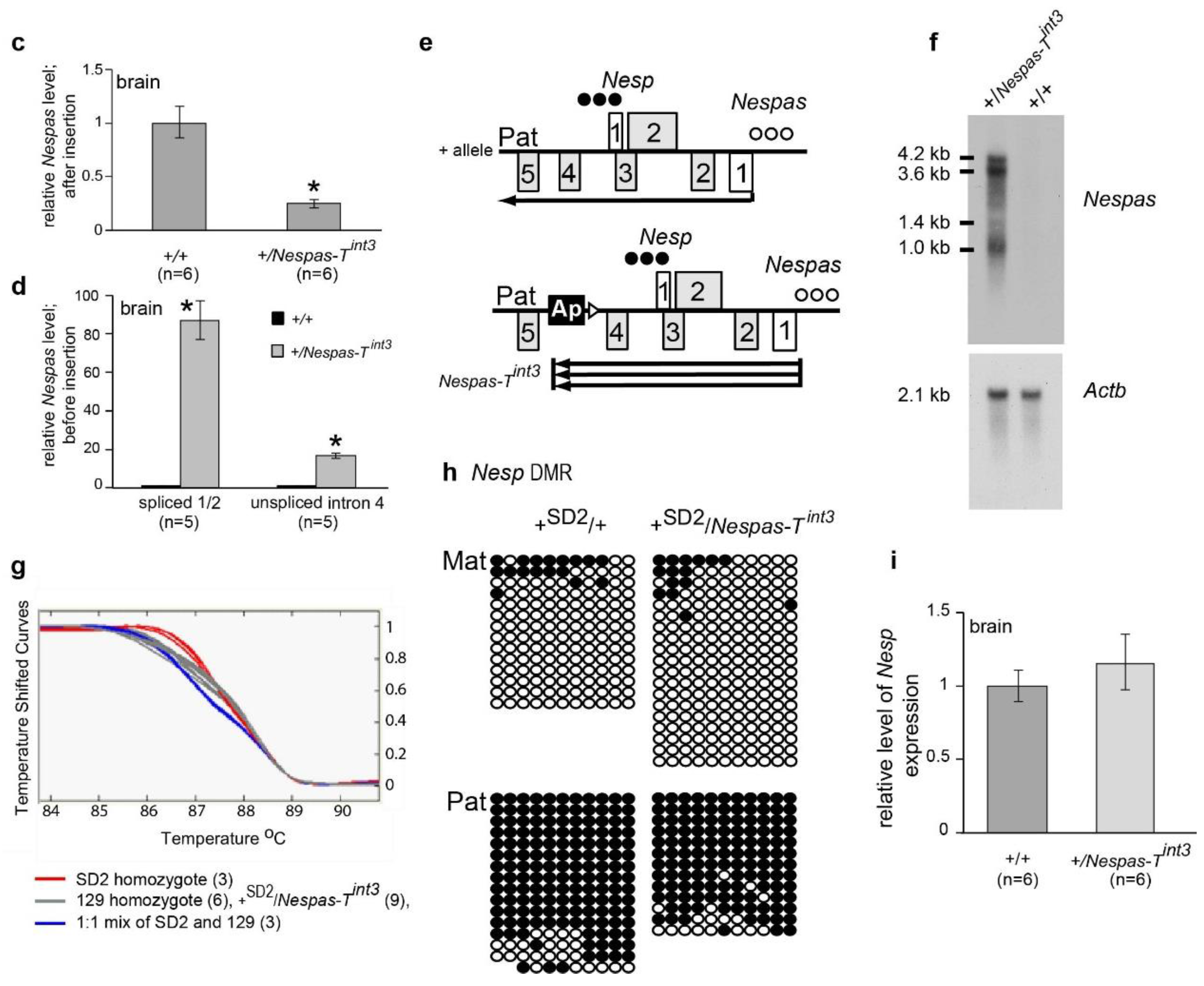
2.5. The Paternal Nesp DMR Is Methylated When Nespas Is Truncated after the Nesp Promoter
2.6. Stability of Truncated Nespas RNA in +/Nespas-Tint3

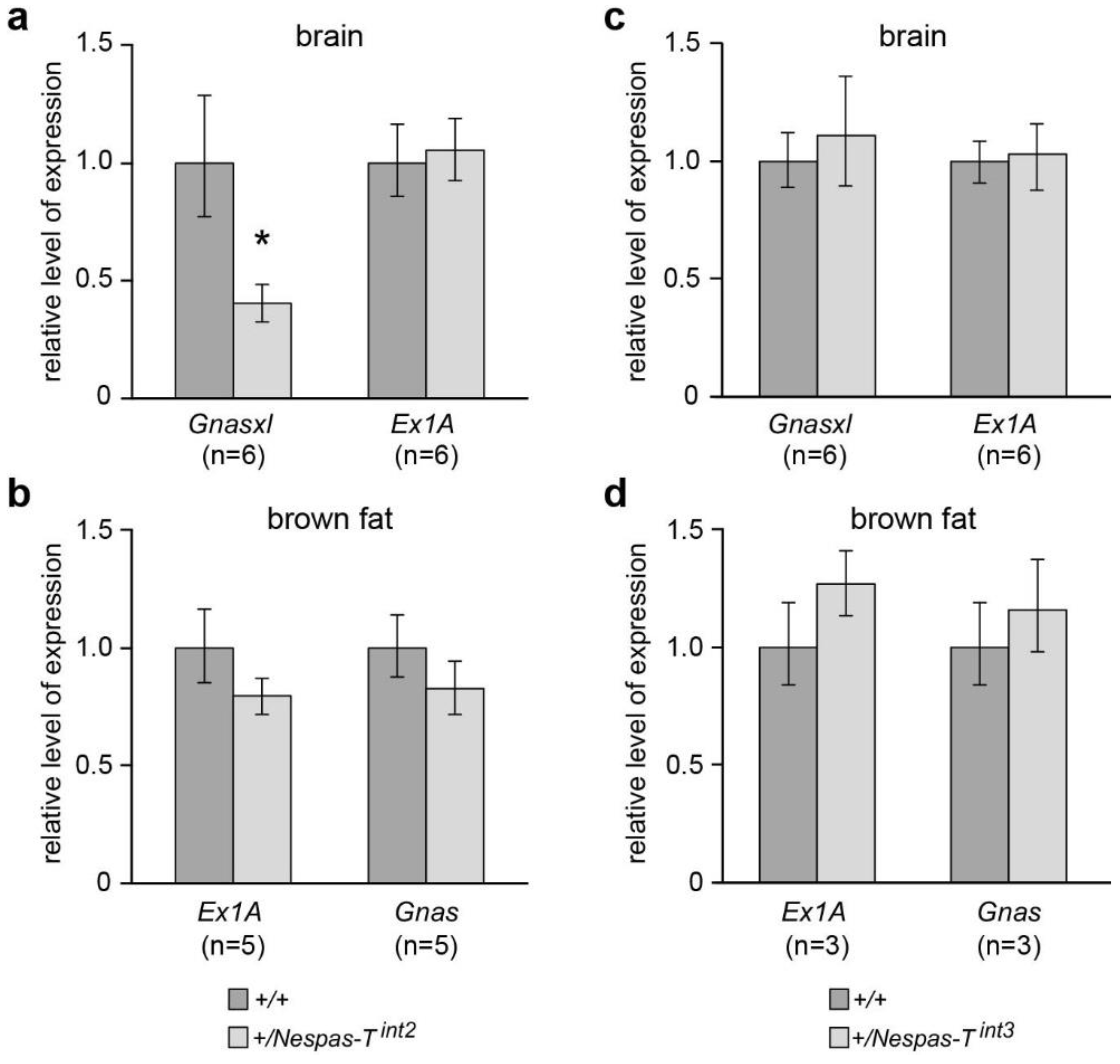
2.7. Reduced Level of Gnasxl on Paternal Transmission of Nespas-Tint2
3. Discussion

4. Material and Methods
4.1. GenBank Accession Number
4.2. Generation of Targeted Alleles
4.3. Genotyping Assay
4.4. Mouse Weight Data Analysis
4.5. Expression Analyses
4.6. PCR Product Melting Curve Analysis for Differentiating SNP Based Allelic Expression
4.7. Methylation Analysis
4.8. Luciferase Reporter Assay to Measure Promoter Activity
4.9. Transcription Inhibitor Assay
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Sleutels, F.; Zwart, R.; Barlow, D.P. The non-coding air rna is required for silencing autosomal imprinted genes. Nature 2002, 415, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Mancini-Dinardo, D.; Steele, S.J.; Levorse, J.M.; Ingram, R.S.; Tilghman, S.M. Elongation of the kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006, 20, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.M.; Ball, S.T.; Dawson, C.; Mehta, S.; Beechey, C.V.; Fray, M.; Teboul, L.; Dear, T.N.; Kelsey, G.; Peters, J. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted gnas cluster. PLoS Genet. 2011, 7, e1001347. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Person, R.E.; Beaudet, A.L. Ube3a-ats is an atypical rna polymerase ii transcript that represses the paternal expression of ube3a. Hum. Mol. Genet. 2012, 21, 3001–3012. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Person, R.E.; Huang, W.; Zhu, P.J.; Costa-Mattioli, M.; Beaudet, A.L. Truncation of ube3a-ats unsilences paternal ube3a and ameliorates behavioral defects in the angelman syndrome mouse model. PLoS Genet. 2013, 9, e1004039. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.; Williamson, C.M. Control of imprinting at the gnas cluster. Epigenetics 2007, 2, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Eaton, S.A.; Williamson, C.M.; Ball, S.T.; Beechey, C.V.; Moir, L.; Edwards, J.; Teboul, L.; Maconochie, M.; Peters, J. New mutations at the imprinted gnas cluster show gene dosage effects of gsalpha in postnatal growth and implicate xlalphas in bone and fat metabolism but not in suckling. Mol. Cell. Biol. 2012, 32, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Rebollo, E.; Maeda, A.; Reyes, M.; Turan, S.; Frohlich, L.F.; Plagge, A.; Kelsey, G.; Juppner, H.; Bastepe, M. Loss of xlalphas (extra-large alphas) imprinting results in early postnatal hypoglycemia and lethality in a mouse model of pseudohypoparathyroidism ib. Proc. Natl. Acad. Sci. USA 2012, 109, 6638–6643. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.T.; Kelly, M.L.; Robson, J.E.; Turner, M.D.; Harrison, J.; Jones, L.; Napper, D.; Beechey, C.V.; Hough, T.; Plagge, A.; et al. Gene dosage effects at the imprinted cluster. PLoS ONE 2013, 8, e65639. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.M.; Turner, M.D.; Ball, S.T.; Nottingham, W.T.; Glenister, P.; Fray, M.; Tymowska-Lalanne, Z.; Plagge, A.; Powles-Glover, N.; Kelsey, G.; et al. Identification of an imprinting control region affecting the expression of all transcripts in the gnas cluster. Nat. Genet. 2006, 38, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.M.; Skinner, J.A.; Kelsey, G.; Peters, J. Alternative non-coding splice variants of nespas, an imprinted gene antisense to nesp in the gnas imprinting cluster. Mamm. Genome 2002, 13, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.; Wroe, S.F.; Wells, C.A.; Miller, H.J.; Bodle, D.; Beechey, C.V.; Williamson, C.M.; Kelsey, G. A cluster of oppositely imprinted transcripts at the gnas locus in the distal imprinting region of mouse chromosome 2. Proc. Natl. Acad. Sci. USA 1999, 96, 3830–3835. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, G.; Bodle, D.; Miller, H.J.; Beechey, C.V.; Coombes, C.; Peters, J.; Williamson, C.M. Identification of imprinted loci by methylation-sensitive representational difference analysis: Application to mouse distal chromosome 2. Genomics 1999, 62, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, S.; Litman, D.; Chen, W.; Weinstein, L.S. Identification of a methylation imprint mark within the mouse gnas locus. Mol. Cell. Biol. 2000, 20, 5808–5817. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.; Williamson, C.; Peters, J.; Denny, P.; Wells, C.; Group, R.G.; Members, G.S.L. A comprehensive transcript map of the mouse gnas imprinted complex. Genome Res. 2003, 13, 1410–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robson, J.E.; Eaton, S.A.; Underhill, P.; Williams, D.; Peters, J. Micrornas 296 and 298 are imprinted and part of the gnas/gnas cluster and mir-296 targets ikbke and tmed9. Rna 2012, 18, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Barlow, D.P. Personal Communication; CeMM: Vienna, Austria, 2011. [Google Scholar]
- Chotalia, M.; Smallwood, S.A.; Ruf, N.; Dawson, C.; Lucifero, D.; Frontera, M.; James, K.; Dean, W.; Kelsey, G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009, 23, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Wroe, S.F.; Kelsey, G.; Skinner, J.A.; Bodle, D.; Ball, S.T.; Beechey, C.V.; Peters, J.; Williamson, C.M. An imprinted transcript, antisense to nesp, adds complexity to the cluster of imprinted genes at the mouse gnas locus. Proc. Natl. Acad. Sci. USA 2000, 97, 3342–3346. [Google Scholar] [CrossRef] [PubMed]
- Seidl, C.I.; Stricker, S.H.; Barlow, D.P. The imprinted air ncrna is an atypical rnapii transcript that evades splicing and escapes nuclear export. EMBO J. 2006, 25, 3565–3575. [Google Scholar] [CrossRef] [PubMed]
- Cattanach, B.M.; Peters, J.; Ball, S.; Rasberry, C. Two imprinted gene mutations: Three phenotypes. Hum. Mol. Genet. 2000, 9, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Plagge, A.; Gordon, E.; Dean, W.; Boiani, R.; Cinti, S.; Peters, J.; Kelsey, G. The imprinted signaling protein xl alpha s is required for postnatal adaptation to feeding. Nat. Genet. 2004, 36, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Williamson, C.M.; Ball, S.; Tibbit, C.; Beechey, C.; Fray, M.; Peters, J. Transcription driven somatic DNA methylation within the imprinted gnas cluster. PLoS ONE 2015, 10, e0117378. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.M.; Ball, S.T.; Nottingham, W.T.; Skinner, J.A.; Plagge, A.; Turner, M.D.; Powles, N.; Hough, T.; Papworth, D.; Fraser, W.D.; et al. A cis-acting control region is required exclusively for the tissue-specific imprinting of gnas. Nat. Genet. 2004, 36, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yu, D.; Lee, E.; Eckhaus, M.; Lee, R.; Corria, Z.; Accili, D.; Westphal, H.; Weinstein, L.S. Variable and tissue-specific hormone resistance in heterotrimeric gs protein alpha-subunit (gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc. Natl. Acad. Sci. USA 1998, 95, 8715–8720. [Google Scholar] [CrossRef] [PubMed]
- Stricker, S.H.; Steenpass, L.; Pauler, F.M.; Santoro, F.; Latos, P.A.; Huang, R.; Koerner, M.V.; Sloane, M.A.; Warczok, K.E.; Barlow, D.P. Silencing and transcriptional properties of the imprinted airn ncrna are independent of the endogenous promoter. EMBO J. 2008, 27, 3116–3128. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, M.; Deng, C.; Bourc'his, D.; Nealon, J.G.; Erlichman, B.; Bestor, T.H.; Weinstein, L.S. Identification of the control region for tissue-specific imprinting of the stimulatory g protein alpha-subunit. Proc. Natl. Acad. Sci. USA 2005, 102, 5513–5518. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S. Imprinting at the Mouse Gnas Cluster; University of Oxford: Oxford, UK, 2012. [Google Scholar]
- Ooi, S.K.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.P.; Allis, C.D.; et al. Dnmt3l connects unmethylated lysine 4 of histone h3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jurkowska, R.; Soeroes, S.; Rajavelu, A.; Dhayalan, A.; Bock, I.; Rathert, P.; Brandt, O.; Reinhardt, R.; Fischle, W.; et al. Chromatin methylation activity of dnmt3a and dnmt3a/3l is guided by interaction of the add domain with the histone h3 tail. Nucleic Acids Res. 2010, 38, 4246–4253. [Google Scholar] [CrossRef] [PubMed]
- Dhayalan, A.; Rajavelu, A.; Rathert, P.; Tamas, R.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. The dnmt3a pwwp domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010, 285, 26114–26120. [Google Scholar] [CrossRef] [PubMed]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Senergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncrna products, induces imprinted igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated rnas by rip-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.K.; Deaton, A.M.; Lee, J.T. A transient heterochromatic state in xist preempts x inactivation choice without rna stabilization. Mol. Cell 2006, 21, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Storck, T.; Kruth, U.; Kolhekar, R.; Sprengel, R.; Seeburg, P.H. Rapid construction in yeast of complex targeting vectors for gene manipulation in the mouse. Nucleic Acids Res. 1996, 24, 4594–4596. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A.; Rossant, J.; Nagy, R.; Abramow-Newerly, W.; Roder, J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 1993, 90, 8424–8428. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.P.; Beechey, C.V. Personal Communication; MRC Mammalian Genetics Unit: Harwell, Oxfordshire, UK, 2010. [Google Scholar]
- Lewandoski, M.; Meyers, E.N.; Martin, G.R. Analysis of fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997, 62, 159–168. [Google Scholar] [PubMed]
- Lewandoski, M.; Martin, G.R. Cre-mediated chromosome loss in mice. Nat. Genet. 1997, 17, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of pcr-quality mouse genomic DNA with hot sodium hydroxide and tris (hotshot). BioTechniques 2000, 29, 52, 54. [Google Scholar] [PubMed]
- Shimomura, K.; Galvanovskis, J.; Goldsworthy, M.; Hugill, A.; Kaizak, S.; Lee, A.; Meadows, N.; Quwailid, M.M.; Rydstrom, J.; Teboul, L.; et al. Insulin secretion from beta-cells is affected by deletion of nicotinamide nucleotide transhydrogenase. Methods Enzymol. 2009, 457, 451–480. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tibbit, C.J.; Williamson, C.M.; Mehta, S.; Ball, S.T.; Chotalia, M.; Nottingham, W.T.; Eaton, S.A.; Quwailid, M.M.; Teboul, L.; Kelsey, G.; et al. Antisense Activity across the Nesp Promoter is Required for Nespas-Mediated Silencing in the Imprinted Gnas Cluster. Non-Coding RNA 2015, 1, 246-265. https://doi.org/10.3390/ncrna1030246
Tibbit CJ, Williamson CM, Mehta S, Ball ST, Chotalia M, Nottingham WT, Eaton SA, Quwailid MM, Teboul L, Kelsey G, et al. Antisense Activity across the Nesp Promoter is Required for Nespas-Mediated Silencing in the Imprinted Gnas Cluster. Non-Coding RNA. 2015; 1(3):246-265. https://doi.org/10.3390/ncrna1030246
Chicago/Turabian StyleTibbit, Charlotte J., Christine M. Williamson, Stuti Mehta, Simon T. Ball, Mita Chotalia, Wade T. Nottingham, Sally A. Eaton, Mohamed M. Quwailid, Lydia Teboul, Gavin Kelsey, and et al. 2015. "Antisense Activity across the Nesp Promoter is Required for Nespas-Mediated Silencing in the Imprinted Gnas Cluster" Non-Coding RNA 1, no. 3: 246-265. https://doi.org/10.3390/ncrna1030246
APA StyleTibbit, C. J., Williamson, C. M., Mehta, S., Ball, S. T., Chotalia, M., Nottingham, W. T., Eaton, S. A., Quwailid, M. M., Teboul, L., Kelsey, G., & Peters, J. (2015). Antisense Activity across the Nesp Promoter is Required for Nespas-Mediated Silencing in the Imprinted Gnas Cluster. Non-Coding RNA, 1(3), 246-265. https://doi.org/10.3390/ncrna1030246





