2,4-Thiazolidinedione in Well-Fed Lactating Dairy Goats: I. Effect on Adiposity and Milk Fat Synthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Animal Management
2.2. Treatments
2.3. Measurements, Sample Collection, and Blood Metabolites
2.4. Adipose Biopsy and Histological Analysis, and Mammary Epithelial Cells Isolation
2.5. Reverse Transcriptase Quantitative Polymerase Chain Reaction (RT-qPCR)
2.6. Statistical Analysis
3. Results
3.1. Animal Performance
3.2. Blood Metabolic Parameters
3.3. Adipocytes Size Distribution
3.4. Gene Expression
3.4.1. Subcutaneous Adipose Tissue
3.4.2. Mammary Epithelial Cells
3.5. Fatty Acid Profile in Milk
4. Discussion
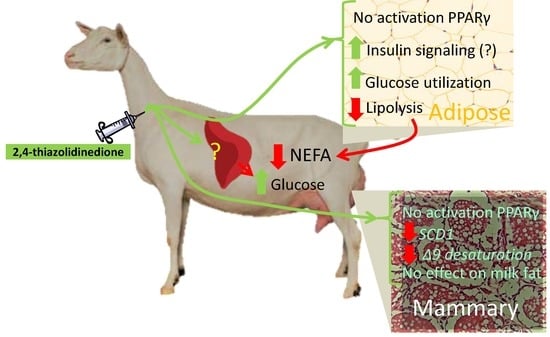
4.1. 2,4-TZD Induces Greater Glucose Utilization in the Adipose Tissue but Did Not Activate PPARγ
4.2. 2,4-TZD Does Not Affect Milk Fat Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Supplementary Materials and Methods
Appendix A.1. Animal Management
Appendix A.2. Dry Matter Intake, Energy Corrected Milk (ECM), Dairy Efficiency (DE), and Body Condition Score (BCS)
Appendix A.3. Adipose Biopsy
Appendix A.4. Histological Analysis of the Adipose Tissue
Appendix A.5. Mammary Epithelial Cells Isolation
Appendix A.6. RT-qPCR
Appendix A.7. Milking and Milk Sample Analysis
Appendix A.8. Milk Fatty Acid Analysis
Appendix A.9. Blood Metabolites
Appendix A.10. Statistical Analysis
References
- Bionaz, M.; Chen, S.; Khan, M.J.; Loor, J.J. Functional Role of PPARs in Ruminants: Potential Targets for Fine-Tuning Metabolism during Growth and Lactation. PPAR Res. 2013, 2013, 684159. [Google Scholar] [CrossRef]
- Bionaz, M.; Osorio, J.; Loor, J.J. TRIENNIAL LACTATION SYMPOSIUM: Nutrigenomics in dairy cows: Nutrients, transcription factors, and techniques. J. Anim. Sci. 2015, 93, 5531–5553. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, J.; Ma, L.; Wang, H.; Cao, W.; Xu, H.; Zhu, J.; Sun, Y.; Li, J.; Yao, D.; et al. MiR130b-Regulation of PPARgamma Coactivator- 1alpha Suppresses Fat Metabolism in Goat Mammary Epithelial Cells. PLoS ONE 2015, 10, e0142809. [Google Scholar] [CrossRef]
- Shi, H.B.; Zhao, W.S.; Zhang, C.H.; Shahzad, K.; Luo, J.; Loor, J.J. Transcriptome-Wide Analysis Reveals the Role of PPAR gamma Controlling the Lipid Metabolism in Goat Mammary Epithelial Cells. PPAR Res. 2016, 2016. [Google Scholar] [CrossRef]
- Liu, L.; Lin, Y.; Liu, L.; Wang, L.; Bian, Y.; Gao, X.; Li, Q. Regulation of peroxisome proliferator-activated receptor gamma on milk fat synthesis in dairy cow mammary epithelial cells. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 1044–1059. [Google Scholar] [CrossRef]
- Smith, K.L.; Butler, W.R.; Overton, T.R. Effects of prepartum 2,4-thiazolidinedione on metabolism and performance in transition dairy cows. J. Dairy Sci. 2009, 92, 3623–3633. [Google Scholar] [CrossRef]
- Yousefi, A.R.; Kohram, H.; Shahneh, A.Z.; Zamiri, M.J.; Fouladi-Nashta, A.A. Effects of dietary supplementation of pioglitazone on metabolism, milk yield, and reproductive performance in transition dairy cows. Theriogenology 2016, 85, 1540–1548. [Google Scholar] [CrossRef] [Green Version]
- Palmquist, D.L. Milk Fat: Origin of Fatty Acids and Influence of Nutritional Factors Thereon. In Advanced Dairy Chemistry; Fox, P.F., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2006; Volume 2. [Google Scholar]
- Rosa, F.; Osorio, J.S.; Trevisi, E.; Yanqui-Rivera, F.; Estill, C.T.; Bionaz, M. 2,4-Thiazolidinedione Treatment Improves the Innate Immune Response in Dairy Goats with Induced Subclinical Mastitis. PPAR Res. 2017, 2017, 7097450. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg, K.M.; Perfield, K.L.; Farney, J.K.; Bradford, B.J.; Boisclair, Y.R.; Overton, T.R. Effects of prepartum 2,4-thiazolidinedione on insulin sensitivity, plasma concentrations of tumor necrosis factor-alpha and leptin, and adipose tissue gene expression. J. Dairy Sci. 2011, 94, 5523–5532. [Google Scholar] [CrossRef]
- Hosseini, A.; Tariq, M.R.; da Rosa, F.T.; Kesser, J.; Iqbal, Z.; Mora, O.; Sauerwein, H.; Drackley, J.K.; Trevisi, E.; Loor, J.J. Insulin Sensitivity in Adipose and Skeletal Muscle Tissue of Dairy Cows in Response to Dietary Energy Level and 2,4-Thiazolidinedione (TZD). PLoS ONE 2015, 10, e0142633. [Google Scholar] [CrossRef]
- Sandri, E.C.; Camera, M.; Sandri, E.M.; Harvatine, K.J.; De Oliveira, D.E. Peroxisome proliferator-activated receptor gamma (PPARgamma) agonist fails to overcome trans-10, cis-12 conjugated linoleic acid (CLA) inhibition of milk fat in dairy sheep. Anim. Int. J. Anim. Biosci. 2017. [Google Scholar] [CrossRef]
- Rosa, F.; Moridi, M.; Osorio, J.; Lohakare, J.; Trevisi, E.; Filley, S.; Estill, C.T.; Bionaz, M. 2,4-thiazolidinedione in well-fed lactating dairy goats: II. Response to intramammary infection. Vet. Sci. 2019, submitted. [Google Scholar]
- National Research Council. Nutrient Requirements of Small Ruminants: Sheep, Goats, Cervids, and New World Camelids; National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Quinn, C.E.; Hamilton, P.K.; Lockhart, C.J.; McVeigh, G.E. Thiazolidinediones: Effects on insulin resistance and the cardiovascular system. Br. J. Pharmacol. 2008, 153, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.S.; Vora, D.K.; Ramaa, C.S. Thiazolidine-2,4-diones: Progress towards multifarious applications. Bioorg. Med. Chem. 2013, 21, 1599–1620. [Google Scholar] [CrossRef]
- Calamari, L.; Ferrari, A.; Minuti, A.; Trevisi, E. Assessment of the main plasma parameters included in a metabolic profile of dairy cow based on Fourier Transform mid-infrared spectroscopy: Preliminary results. BMC Vet. Res. 2016, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Trevisi, E.; Calamari, L.; Librandi, F.; Ferrari, A.; Bertoni, G. Plasma paraoxonase, health, inflammatory conditions, and liver function in transition dairy cows. J. Dairy Sci. 2007, 90, 1740–1750. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.T.; Xing, T.Y.; Chu, Y.K.; Li, H.; Wang, N. Genetic and epigenetic regulation of PPARgamma during adipogenesis. Yi Chuan Hereditas 2017, 39, 1066–1077. [Google Scholar] [CrossRef]
- Boden, G.; Cheung, P.; Mozzoli, M.; Fried, S.K. Effect of thiazolidinediones on glucose and fatty acid metabolism in patients with type 2 diabetes. Metabolism 2003, 52, 753–759. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 2011, 1812, 1007–1022. [Google Scholar] [CrossRef]
- Smith, K.L.; Stebulis, S.E.; Waldron, M.R.; Overton, T.R. Prepartum 2,4-thiazolidinedione alters metabolic dynamics and dry matter intake of dairy cows. J. Dairy Sci. 2007, 90, 3660–3670. [Google Scholar] [CrossRef]
- Guo, S. Insulin signaling, resistance, and the metabolic syndrome: Insights from mouse models into disease mechanisms. J. Endocrinol. 2014, 220, T1–T23. [Google Scholar] [CrossRef]
- Alexander, M.C.; Lomanto, M.; Nasrin, N.; Ramaika, C. Insulin Stimulates Glyceraldehyde-3-Phosphate Dehydrogenase Gene-Expression through Cis-Acting DNA-Sequences. Proc. Natl. Acad. Sci. USA 1988, 85, 5092–5096. [Google Scholar] [CrossRef]
- Nafikov, R.A.; Beitz, D.C. Carbohydrate and lipid metabolism in farm animals. J. Nutr. 2007, 137, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Adewuyi, A.A.; Gruys, E.; van Eerdenburg, F.J. Non esterified fatty acids (NEFA) in dairy cattle. A review. Vet. Q. 2005, 27, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Dugail, I.; Quignard-Boulange, A.; Bazin, R.; Le Liepvre, X.; Lavau, M. Adipose-tissue-specific increase in glyceraldehyde-3-phosphate dehydrogenase activity and mRNA amounts in suckling pre-obese Zucker rats. Effect of weaning. Biochem. J. 1988, 254, 483–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, R.W.; Reshef, L. Glyceroneogenesis revisited. Biochimie 2003, 85, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Tordjman, J.; Khazen, W.; Antoine, B.; Chauvet, G.; Quette, J.; Fouque, F.; Beale, E.G.; Benelli, C.; Forest, C. Regulation of glyceroneogenesis and phosphoenolpyruvate carboxykinase by fatty acids, retinoic acids and thiazolidinediones: Potential relevance to type 2 diabetes. Biochimie 2003, 85, 1213–1218. [Google Scholar] [CrossRef]
- Schoenberg, K.M.; Overton, T.R. Effects of plane of nutrition and 2,4-thiazolidinedione on insulin responses and adipose tissue gene expression in dairy cattle during late gestation. J. Dairy Sci. 2011, 94, 6021–6035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10. [Google Scholar] [CrossRef]
- Lohakare, J.; Osorio, J.S.; Bionaz, M. Peroxisome proliferator-activated receptor beta/delta does not regulate glucose uptake and lactose synthesis in bovine mammary epithelial cells cultivated in vitro. J. Dairy Res. 2018, 85, 295–302. [Google Scholar] [CrossRef]
- Yu, Y.H.; Wang, P.H.; Cheng, W.T.K.; Mersmann, H.J.; Wu, S.C.; Ding, S.T. Porcine peroxisome proliferator-activated receptor delta mediates the lipolytic effects of dietary fish oil to reduce body fat deposition. J. Anim. Sci. 2010, 88, 2009–2018. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, D.L.; Spagnolo, A.; Akar, C.; Weinberg, G.; Murphy, P.; Gavrilyuk, V.; Russo, C.D. Receptor-independent actions of PPAR thiazolidinedione agonists: Is mitochondrial function the key? Biochem. Pharmacol. 2005, 70, 177–188. [Google Scholar] [CrossRef]
- Miller, P.S.; Reis, B.L.; Calvert, C.C.; DePeters, E.J.; Baldwin, R.L. Patterns of nutrient uptake by the mammary glands of lactating dairy cows. J. Dairy Sci. 1991, 74, 3791–3799. [Google Scholar] [CrossRef]
- Mensink, R.P. Effects of stearic acid on plasma lipid and lipoproteins in humans. Lipids 2005, 40, 1201–1205. [Google Scholar] [CrossRef]
- Moreira, D.K.T.; Santos, P.S.; Gambero, A.; Macedo, G.A. Evaluation of structured lipids with behenic acid in the prevention of obesity. Food Res. Int. 2017, 95, 52–58. [Google Scholar] [CrossRef]
- Shabbir, M.A.; Khan, M.R.; Saeed, M.; Pasha, I.; Khalil, A.A.; Siraj, N. Punicic acid: A striking health substance to combat metabolic syndromes in humans. Lipids Health Dis. 2017, 16, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mozaffarian, D.; Otto, M.C.D.; Lemaitre, R.N.; Fretts, A.M.; Hotamisligil, G.; Tsai, M.Y.; Siscovick, D.S.; Nettleton, J.A. trans-Palmitoleic acid, other dairy fat biomarkers, and incident diabetes: The Multi-Ethnic Study of Atherosclerosis (MESA). Am. J. Clin. Nutr. 2013, 97, 854–861. [Google Scholar] [CrossRef]
- Merry, T.L.; Ristow, M. Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J. Physiol. 2016, 594, 5195–5207. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, W.; Li, G.; Chen, J.; Guan, X.; Chen, X.; Guan, Z. Neuroprotective Effect and Mechanism of Thiazolidinedione on Dopaminergic Neurons In Vivo and In Vitro in Parkinson’s Disease. PPAR Res. 2017, 2017, 4089214. [Google Scholar] [CrossRef]
- Griinari, J.M.; Corl, B.A.; Lacy, S.H.; Chouinard, P.Y.; Nurmela, K.V.; Bauman, D.E. Conjugated linoleic acid is synthesized endogenously in lactating dairy cows by Delta(9)-desaturase. J. Nutr. 2000, 130, 2285–2291. [Google Scholar] [CrossRef] [PubMed]
- Sjaunja, L.O.; Baevre, L.; Junkkarinene, L.; Pedersen, J.; Setala, J. A Nordic proposal for an energy corrected milk (ECM) formula. In Proceedings of the 27th Session of the International Commission for Breeding and Productivity of Milk Animals, Paris, France, 2–6 July 1990. [Google Scholar]
- Villaquiran, M.; Gipson, T.A.; Merkel, R.C.; Goetsch, A.L.; Sahlu, T. Body Condition Scores in Goats. Am. Inst. Goat Res. 2004, 1–8. [Google Scholar]
- Kamentsky, L.; Jones, T.R.; Fraser, A.; Bray, M.A.; Logan, D.J.; Madden, K.L.; Ljosa, V.; Rueden, C.; Eliceiri, K.W.; Carpenter, A.E. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics 2011, 27, 1179–1180. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genom. 2007, 29, 312–319. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Oeffner, S.P.; Qu, Y.; Just, J.; Quezada, N.; Ramsing, E.; Keller, M.; Cherian, G.; Goddick, L.; Bobe, G. Effect of flaxseed supplementation rate and processing on the production, fatty acid profile, and texture of milk, butter, and cheese. J. Dairy Sci. 2013, 96, 1177–1188. [Google Scholar] [CrossRef]




| Parameter | Group | Day of Treatment | SEM | p-Value 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 5 | 15 | 16 | 17 | 20 | 21 | 22 | Z | Time | Z × T | |||
| Protein % | CTR | 2.32 | 2.29 | 2.22 | 2.32 | 2.35 | 2.28 | 2.41 | 2.41 | 2.39 | 0.09 | 0.30 | 0.10 | 0.78 |
| TZD | 2.32 | 2.26 | 2.19 | 2.18 | 2.18 | 2.17 | 2.21 | 2.21 | 2.19 | 0.09 | ||||
| Protein g 2 | CTR | 18.3 | 19.4 | 18.2 | 20.3 | 18.2 | 17.0 | 19.2 | 18.4 | 18.1 | 2.5 | 0.98 | <0.05 | 0.76 |
| TZD | 18.3 | 20.4 | 17.9 | 18.9 | 18.2 | 17.5 | 18.4 | 19.2 | 19.0 | 2.5 | ||||
| Lactose % | CTR | 4.45 | 4.47 | 4.42 | 4.38 | 4.40 | 4.35 | 4.34 | 4.35 | 4.31 | 0.07 | 0.45 | <0.05 | 0.23 |
| TZD | 4.45 | 4.68 | 4.51 | 4.33 | 4.37 | 4.41 | 4.42 | 4.39 | 4.37 | 0.07 | ||||
| Lactose g 2 | CTR | 34.9 | 37.2 | 36.0 | 37.8 | 33.7 | 32.1 | 34.2 | 32.7 | 32.2 | 4.3 | 0.61 | <0.05 | 0.61 |
| TZD | 34.9 | 40.3 | 36.6 | 37.3 | 36.4 | 35.5 | 36.9 | 38.1 | 37.9 | 4.3 | ||||
| SNF 3% | CTR | 7.66 | 7.61 | 7.55 | 7.53 | 7.51 | 7.45 | 7.48 | 7.51 | 7.46 | 0.31 | 0.98 | 0.44 | 0.63 |
| TZD | 7.66 | 7.87 | 7.61 | 7.42 | 7.44 | 7.46 | 7.46 | 7.43 | 7.40 | 0.31 | ||||
| SNF g 2 | CTR | 60.2 | 64.6 | 61.5 | 65.4 | 57.8 | 55.1 | 59.2 | 56.8 | 56.0 | 7.6 | 0.69 | <0.05 | 0.67 |
| TZD | 60.2 | 69.3 | 62.0 | 64.1 | 62.1 | 60.2 | 62.1 | 64.5 | 64.2 | 7.6 | ||||
| Parameter | Group | Day of Treatment | SEM | p-Value 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 9 | 15 | 16 | 17 | 18 | 21 | 26 | Z | Time | Z × T | |||
| Cholesterol, mM | CTR | 2.55 | 2.30 | 2.27 | 2.18 | 2.28 | 2.31 | 2.33 | 2.40 | 2.32 | 0.24 | 0.38 | 0.18 | 0.70 |
| TZD | 2.55 | 2.73 | 2.56 | 2.40 | 2.51 | 2.61 | 2.61 | 2.62 | 2.61 | 0.24 | ||||
| ∆FA, % 3 | CTR | 0.0 | 5.1 | −6.8 | 5.0 | 3.8 | 0.8 | 9.1 | 7.7 | −6.6 | 8.5 | 0.06 | 0.88 | 0.90 |
| TZD | 0.0 | −13.2 | −14.5 | −9.4 | −4.9 | −8.1 | −9.7 | −17.2 | −12.0 | 8.5 | ||||
| TAG, mM | CTR | 0.14 | 0.15 | 0.16 | 0.19 | 0.17 | 0.18 | 0.19 | 0.19 | 0.13 | 0.02 | 0.89 | 0.30 | 0.82 |
| TZD | 0.14 | 0.14 | 0.15 | 0.19 | 0.19 | 0.19 | 0.18 | 0.16 | 0.15 | 0.02 | ||||
| Creatinine, µM | CTR | 71.9 | 77.3 | 77.1 | 71.9 | 72.6 | 70.2 | 70.4 | 72.2 | 75.8 | 2.7 | 0.98 | <0.05 | 0.94 |
| TZD | 71.9 | 76.6 | 77.6 | 70.7 | 72.2 | 71.7 | 70.6 | 72.4 | 76.1 | 2.7 | ||||
| Urea, mM | CTR | 6.73 | 6.32 | 4.59 | 5.97 | 6.81 | 5.61 | 6.54 | 6.45 | 8.05 | 0.45 | 0.25 | <0.05 | 0.19 |
| TZD | 6.73 | 6.40 | 5.16 | 5.29 | 6.08 | 5.08 | 5.61 | 5.71 | 6.22 | 0.45 | ||||
| Gene | Group | Time | SEM 1 | p-Value 2 | ||||
|---|---|---|---|---|---|---|---|---|
| −1 | 13 | 26 | Z | Time | Z × T | |||
| Fatty Acids Import and De Novo Fatty Acid Synthesis | ||||||||
| CD36 | CTR | 7169.8 | 6987.6 | 6233.3 | 681.3 | 0.03 | 0.03 | 0.03 |
| TZD | 7169.8 | 3362.5 | 5781.9 | |||||
| LPL | CTR | 122.2 | 130.5 | 244.6 | 52.1 | 0.62 | 0.04 | 0.90 |
| TZD | 122.2 | 118.0 | 206.7 | |||||
| SLC27A6 | CTR | 0.09 | 0.12 | 0.11 | 0.03 | 0.84 | 0.15 | 0.94 |
| TZD | 0.09 | 0.14 | 0.11 | |||||
| VLDLR | CTR | 2.3 | 2.0 | 1.0 | 0.9 | 0.67 | 0.48 | 0.25 |
| TZD | 2.3 | 1.1 | 2.7 | |||||
| ACACA | CTR | 37.6 | 26.6 | 48.5 | 24.3 | 0.33 | 0.51 | 0.14 |
| TZD | 37.6 | 88.6 | 45.6 | |||||
| FASN | CTR | 473.3 | 47.7 | 58.6 | 439.9 | 0.13 | 0.29 | 0.18 |
| TZD | 473.3 | 163.1 | 1328.2 | |||||
| SCD1 | CTR | 275.2 | 500.8 | 410.9 | 333.4 | 0.67 | 0.16 | 0.54 |
| TZD | 275.2 | 917.0 | 329.3 | |||||
| Insulin Signal and Glucose Metabolism | ||||||||
| GAPDH | CTR | 159.2 | 125.6 | 91.2 | 41.7 | 0.01 | 0.25 | 0.08 |
| TZD | 159.2 | 271.2 | 195.9 | |||||
| INSR | CTR | 7.8 | 5.2 | 3.7 | 1.9 | 0.12 | 0.37 | 0.21 |
| TZD | 7.8 | 5.8 | 9.4 | |||||
| IRS1 | CTR | 9.2 | 6.8 | 4.6 | 2.0 | 0.04 | 0.42 | 0.05 |
| TZD | 9.2 | 7.4 | 12.5 | |||||
| SLC2A4 | CTR | 4.5 | 5.6 | 5.1 | 1.6 | 0.09 | 0.10 | 0.40 |
| TZD | 4.5 | 8.9 | 7.3 | |||||
| YWHAZ | CTR | 167.5 | 191.8 | 149.0 | 23.9 | 0.07 | 0.50 | 0.04 |
| TZD | 167.5 | 101.4 | 150.1 | |||||
| Glyceroneogenesis | ||||||||
| PCK1 | CTR | 8.0 | 9.8 | 15.0 | 4.2 | 0.67 | 0.01 | 0.47 |
| TZD | 8.0 | 8.7 | 21.0 | |||||
| PDK4 | CTR | 1.8 | 1.2 | 1.1 | 0.4 | 0.66 | 0.03 | 0.08 |
| TZD | 1.8 | 0.8 | 2.1 | |||||
| GPD1 | CTR | 71.5 | 109.1 | 90.8 | 33.9 | 0.45 | 0.07 | 0.71 |
| TZD | 71.5 | 145.8 | 116.4 | |||||
| Transcription Regulation | ||||||||
| PPARG | CTR | 43.5 | 41.7 | 49.5 | 9.6 | 0.81 | 0.19 | 0.70 |
| TZD | 43.5 | 37.3 | 58.3 | |||||
| SREBF1 | CTR | 16.1 | 19.3 | 23.0 | 16.7 | 0.61 | 0.38 | 0.61 |
| TZD | 16.1 | 14.8 | 44.7 | |||||
| Gene | Group | Cell 1 | SEM | p-Value 2 | |||
|---|---|---|---|---|---|---|---|
| Neg | Pos | Z | Cell | Z × Cell | |||
| Mammary Epithelial-Specific Genes | |||||||
| CSN3 | CTR | 6324 | 3877 | 875 | 0.46 | <0.05 | 0.42 |
| TZD | 5050 | 3326 | 911 | ||||
| FABP3 | CTR | 20.0 | 8.5 | 3.0 | 0.57 | <0.05 | 0.41 |
| TZD | 16.1 | 7.8 | 3.2 | ||||
| LALBA | CTR | 1341 | 578 | 233.4 | 0.56 | <0.05 | 0.56 |
| TZD | 1065 | 501 | 251.7 | ||||
| KRT8 (Ln) 3 | CTR | 1.78 | 2.50 | 0.27 | 0.30 | <0.05 | 0.64 |
| TZD | 1.48 | 2.03 | 0.29 | ||||
| Fatty Acid Transport and Synthesis | |||||||
| LPL | CTR | 9.68 | 4.87 | 1.52 | 0.70 | <0.05 | 0.77 |
| TZD | 10.8 | 5.19 | 1.68 | ||||
| CD36 | CTR | 1300 | 979 | 104 | 0.30 | <0.05 | 0.87 |
| TZD | 1445 | 1094 | 115 | ||||
| VLDLR (Ln) | CTR | 0.57 | 0.84 | 0.13 | 0.21 | <0.05 | 0.31 |
| TZD | 0.43 | 0.53 | 0.14 | ||||
| ACACA | CTR | 0.29 | 0.58 | 0.09 | 0.99 | 0.09 | 0.10 |
| TZD | 0.43 | 0.43 | 0.10 | ||||
| SCD1 | CTR | 5.84 | 5.03 | 0.68 | 0.06 | 0.02 | 0.53 |
| TZD | 4.19 | 2.89 | 0.71 | ||||
| Transcriptional Regulation | |||||||
| NFE2L2 | CTR | 51.2 | 62.0 | 12.3 | 0.30 | <0.05 | 0.50 |
| TZD | 66.4 | 83.7 | 12.6 | ||||
| NRF1 | CTR | 0.15 | 0.17 | 0.03 | 0.69 | 0.50 | 0.91 |
| TZD | 0.17 | 0.18 | 0.03 | ||||
| PPARG (Ln) | CTR | -2.73 | −1.94 | 0.33 | 0.38 | <0.05 | 0.50 |
| TZD | -2.20 | −1.71 | 0.35 | ||||
| SREBF1 | CTR | 0.85 | 1.17 | 0.13 | 0.45 | <0.05 | 0.79 |
| TZD | 0.74 | 1.01 | 0.14 | ||||
| Other | |||||||
| GAPDH (Ln) | CTR | 3.63 | 4.07 | 0.31 | 0.39 | <0.05 | 0.47 |
| TZD | 3.85 | 4.56 | 0.33 | ||||
| IL8 (Ln) | CTR | 4.8 | 4.9 | 0.6 | 0.31 | 0.17 | 0.40 |
| TZD | 5.5 | 5.9 | 0.6 | ||||
| YWHAZ | CTR | 31.6 | 40.7 | 8.3 | 0.64 | 0.19 | 0.61 |
| TZD | 39.1 | 43.3 | 8.3 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaaf, S.; Rosa, F.; Moridi, M.; Osorio, J.S.; Lohakare, J.; Trevisi, E.; Filley, S.; Cherian, G.; Estill, C.T.; Bionaz, M. 2,4-Thiazolidinedione in Well-Fed Lactating Dairy Goats: I. Effect on Adiposity and Milk Fat Synthesis. Vet. Sci. 2019, 6, 45. https://doi.org/10.3390/vetsci6020045
Jaaf S, Rosa F, Moridi M, Osorio JS, Lohakare J, Trevisi E, Filley S, Cherian G, Estill CT, Bionaz M. 2,4-Thiazolidinedione in Well-Fed Lactating Dairy Goats: I. Effect on Adiposity and Milk Fat Synthesis. Veterinary Sciences. 2019; 6(2):45. https://doi.org/10.3390/vetsci6020045
Chicago/Turabian StyleJaaf, Shana, Fernanda Rosa, Misagh Moridi, Johan S. Osorio, Jayant Lohakare, Erminio Trevisi, Shelby Filley, Gita Cherian, Charles T. Estill, and Massimo Bionaz. 2019. "2,4-Thiazolidinedione in Well-Fed Lactating Dairy Goats: I. Effect on Adiposity and Milk Fat Synthesis" Veterinary Sciences 6, no. 2: 45. https://doi.org/10.3390/vetsci6020045