Assessing and Resolving Model Misspecifications in Metabolic Flux Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Metabolic Flux Analysis
2.2. Model Misspecification
2.2.1. Effects of Missing Reactions
2.2.2. Model Misspecification Tests
2.2.3. Resolving Model Misspecification
- Given the exchange fluxes vE, the stoichiometric matrices SE and SI, and the possible missing reaction stoichiometric matrix SA, we formulate the linear least square regression problem with , , and .
- Compute using Z constructed from every k-tuple combination of the columns (reactions) of SA.
- Identify the k-tuple combination(s) satisfying and move the corresponding columns from SA to SI.
- Repeat steps 2 to 3 until no more reactions can be moved from SA to SI, that is, until the remaining set of k-tuple reaction combinations satisfying is empty.
2.3. In Silico Metabolic Network Models and Data Generation
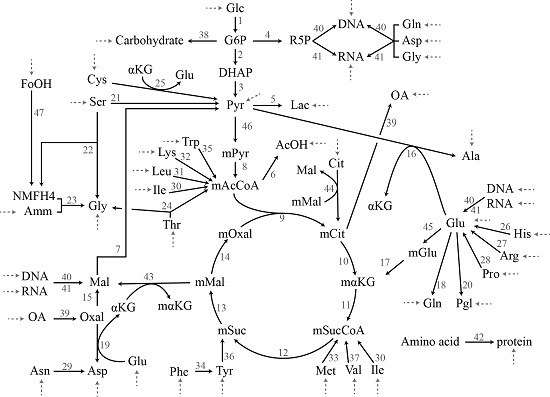
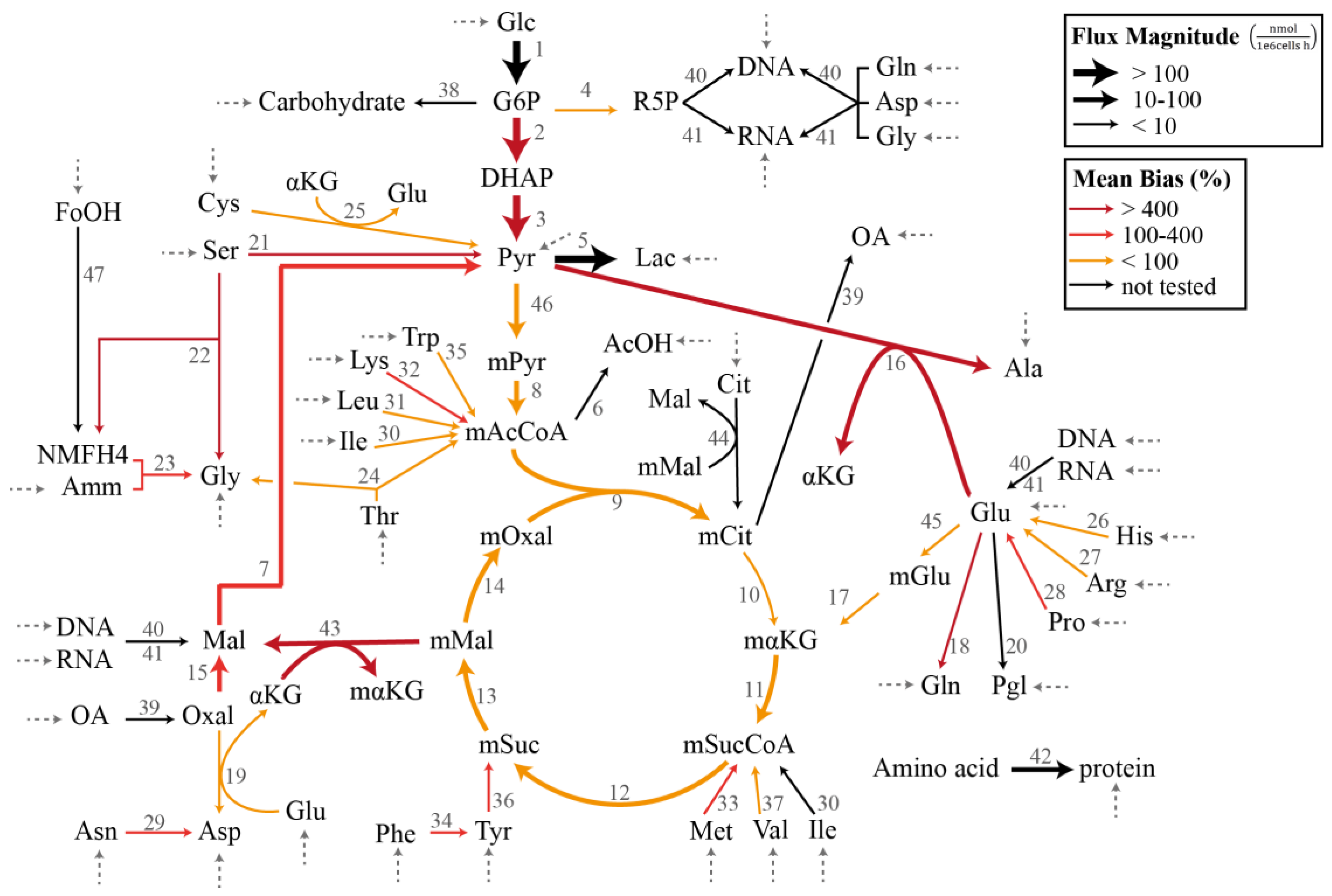
2.3.1. Chinese Hamster Ovary Model
2.3.2. Random Metabolic Models
3. Results
3.1. Case Study I: Specification Bias
3.2. Case Study II: Stoichiometric Model Misspecification Tests
3.3. Case Study III: Resolving Model Misspecification
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stephanopoulos, G. Metabolic fluxes and metabolic engineering. Metab. Eng. 1999, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.E. Bioprocess Engineering. Adv. Chem. Eng. 1991, 16, 425–462. [Google Scholar]
- Lee, S.Y.; Park, J.M.; Kim, T.Y. Application of metabolic flux analysis in metabolic engineering. Methods Enzymol. 2011, 498, 67–93. [Google Scholar] [PubMed]
- Lewis, N.E.; Nagarajan, H.; Palsson, B.O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012, 10, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Crown, S.B.; Antoniewicz, M.R. Publishing 13C metabolic flux analysis studies: A review and future perspectives. Metab. Eng. 2013, 20, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Naderi, S.; Meshram, M.; Wei, C.; Mcconkey, B.; Ingalls, B.; Budman, H.; Scharer, J. Development of a mathematical model for evaluating the dynamics of normal and apoptotic Chinese hamster ovary cells. Biotechnol. Prog. 2011, 27, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Nolan, R.P.; Lee, K. Dynamic model of CHO cell metabolism. Metab. Eng. 2011, 13, 108–124. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, R.T.J.M.; Heijnen, J.J.; Hellinga, C.; Romein, B.; Luyben, K.C.A.M. Linear constraint relations in biochemical reaction systems: I. Classification of the calculability and the balanceability of conversion rates. Biotechnol. Bioeng. 1994, 43, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.S.; Stephanopoulos, G. Application of Macroscopic Balances to the Identification of Gross Measurement Errors. Biotechnol. Bioeng. 1983, 25, 2177–2208. [Google Scholar] [CrossRef] [PubMed]
- Goudar, C.T.; Biener, R.K.; Piret, J.M.; Konstantinov, K.B. Metabolic Flux Estimation in Mammalian Cell Cultures. In Animal Cell Biotechnology: Methods and Protocols; Pörtner, R., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 193–209. [Google Scholar]
- Wiechert, W.; Siefke, C.; de Graaf, A.A.; Marx, A. Bidirectional reaction steps in metabolic networks: II. Flux estimation and statistical analysis. Biotechnol. Bioeng. 1997, 55, 118–135. [Google Scholar] [CrossRef]
- Goudar, C.T.; Biener, R.; Piret, J.M.; Konstantinov, K.B. Metabolic flux estimation in mammalian cell cultures. In Methods in Biotechnology; Springer: Berlin, Germany, 2007; Volume 24, pp. 301–317. [Google Scholar]
- Hädicke, O.; Lohr, V.; Genzel, Y.; Reichl, U.; Klamt, S. Evaluating differences of metabolic performances: Statistical methods and their application to animal cell cultivations. Biotechnol. Bioeng. 2013, 110, 2633–2642. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, R.T.J.M.; Romein, B.; Heijnen, J.J.; Hellinga, C.; Luyben, K.C.A.M. Linear constraint relations in biochemical reaction systems: II. Diagnosis and estimation of gross errors. Biotechnol. Bioeng. 1994, 43, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Quek, L.-E.; Dietmair, S.; Krömer, J.O.; Nielsen, L.K. Metabolic flux analysis in mammalian cell culture. Metab. Eng. 2010, 12, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Sokolenko, S.; Quattrociocchi, M.; Aucoin, M.G. Identifying model error in metabolic flux analysis—A generalized least squares approach. BMC Syst. Biol. 2016, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Erdrich, P.; Steuer, R.; Klamt, S. An algorithm for the reduction of genome-scale metabolic network models to meaningful core models. BMC Syst. Biol. 2015, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chipman, J.S. Gauss-Markov Theorem. In International Encyclopedia of Statistical Science; Springer: Berlin, Germany, 2014; pp. 577–582. [Google Scholar]
- Rao, P. Some Notes on Misspecification in Multiple Regressions. Am. Stat. 1971, 25, 37–39. [Google Scholar] [CrossRef]
- Long, J.S.; Trivedi, P.K. Some Specification Tests for the Linear Regression Model. Sociol. Methods Res. 1992, 21, 161–204. [Google Scholar] [CrossRef]
- Ramsey, J.B. Tests for Specification Errors in Classical Linear Least-Squares Regression Analysis. J. R. Stat. Soc. Ser. B 1969, 31, 350–371. [Google Scholar]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]
- Morgat, A.; Lombardot, T.; Axelsen, K.B.; Aimo, L.; Niknejad, A.; Hyka-Nouspikel, N.; Coudert, E.; Pozzato, M.; Pagni, M.; Moretti, S.; et al. Updates in rhea-an expert curated resource of biochemical reactions. Nucleic Acids Res. 2017, 45, D415–D418. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.V.; Arnold, K.J. Parameter Estimation in Engineering and Science; Wiley: Hoboken, NJ, USA, 1977. [Google Scholar]
- Davidson, R.; MacKinnon, J. Heteroskedasticity-Robust Tests in Regression Directions. Ann. Insee. 1985, 59/60, 183–218. [Google Scholar] [CrossRef]
- Altamirano, C.; Illanes, A.; Casablancas, A.; Gámez, X.; Cairó, J.J.; Gòdia, C. Analysis of CHO cells metabolic redistribution in a glutamate-based defined medium in continuous culture. Biotechnol. Prog. 2001, 17, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Aho, T.; Smolander, O.-P.; Niemi, J.; Yli-Harja, O. RMBNToolbox: Random models for biochemical networks. BMC Syst. Biol. 2007, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, D.C. Applied Statistics and Probability for Engineers, 6th ed.; Wiley: Hoboken, NJ, USA, 2003; Volume 37. [Google Scholar]
- Conover, W.J. Practical Nonparametric Statistics, 3rd ed.; Wiley: Hoboken, NJ, USA, 1999. [Google Scholar]
- Niklas, J.; Heinzle, E. Metabolic flux analysis in systems biology of mammalian cells. In Genomics and Systems Biology of Mammalian Cell Culture; Springer: Berlin, Germany, 2011; pp. 109–132. [Google Scholar]
| Reaction a | p Value c | Absolute Specification Bias (%) d | ||||
|---|---|---|---|---|---|---|
| Min | Median | Mean | Max | |||
| 25 | −0.02 | 0.00 ± 0.00 | 0.00 | 0.41 | 2.73 | 54.1 |
| 19 | 0.03 | 0.00 ± 0.00 | 0.00 | 0.39 | 2.48 | 48.8 |
| 10 | −1.46 | 0.00 ± 0.00 | 0.00 | 0.15 | 1.96 | 11.6 |
| 45 | −0.21 | 0.00 ± 0.00 | 0.00 | 2.04 | 18.3 | 269 |
| 17 | −0.21 | 0.00 ± 0.00 | 0.00 | 2.11 | 19.0 | 280 |
| 31 | −0.24 | 0.00 ± 0.00 | 0.00 | 2.83 | 24.9 | 361 |
| 27 | 0.34 | 0.00 ± 0.00 | 0.00 | 2.12 | 15.6 | 229 |
| 14 | 12.50 | 0.00 ± 0.00 | 0.00 | 1.31 | 33.3 | 855 |
| 9 | 12.50 | 0.00 ± 0.00 | 0.00 | 1.31 | 33.3 | 855 |
| 46 | 15.04 | 0.00 ± 0.00 | 0.00 | 0.88 | 38.1 | 1020 |
| 8 | 15.04 | 0.00 ± 0.00 | 0.00 | 0.88 | 38.1 | 1020 |
| 37 | 0.27 | 0.00 ± 0.00 | 0.00 | 5.86 | 54.1 | 753 |
| 12 | 17.42 | 0.00 ± 0.00 | 0.02 | 1.28 | 43.1 | 1190 |
| 11 | 17.84 | 0.00 ± 0.00 | 0.02 | 1.09 | 44.0 | 1220 |
| 13 | 18.06 | 0.00 ± 0.00 | 0.02 | 1.66 | 46.7 | 1230 |
| 30 | −0.27 | 0.00 ± 0.00 | 0.00 | 6.87 | 63.8 | 889 |
| 24 | −0.38 | 0.00 ± 0.00 | 0.00 | 6.67 | 60.9 | 860 |
| 26 | 0.27 | 0.00 ± 0.00 | 0.00 | 4.19 | 36.1 | 509 |
| 35 | 0.13 | 0.00 ± 0.00 | 0.00 | 3.12 | 28.8 | 399 |
| 33 | 0.22 | 0.00 ± 0.00 | 0.00 | 11.7 | 124 | 2060 |
| 32 b | −1.18 | 0.01 ± 0.01 | 0.00 | 21.0 | 196 | 2770 |
| 29 b | 0.99 | 0.01 ± 0.01 | 0.00 | 13.9 | 170 | 3840 |
| 34 b | 0.47 | 0.01 ± 0.01 | 0.00 | 16.2 | 152 | 2110 |
| 36 b | 0.87 | 0.02 ± 0.01 | 0.00 | 23.1 | 217 | 3020 |
| 23 b | −1.34 | 0.02 ± 0.01 | 0.00 | 17.1 | 177 | 2470 |
| 28 b | 1.03 | 0.02 ± 0.01 | 0.00 | 17.6 | 158 | 2220 |
| 15 | 12.31 | 0.05 ± 0.02 | 0.00 | 14.3 | 121 | 2210 |
| 4 | 1.24 | 0.05 ± 0.02 | 0.00 | 5.25 | 65.1 | 2420 |
| 21 | −6.81 | 0.13 ± 0.03 | 0.00 | 53.9 | 475 | 6980 |
| 16 | 19.26 | 0.15 ± 0.03 | 0.00 | 61.4 | 573 | 8100 |
| 18 | −21.53 | 0.20 ± 0.04 | 0.00 | 61.6 | 632 | 8830 |
| 43 | 19.52 | 0.46 ± 0.04 | 0.00 | 74.0 | 477 | 8050 |
| 22 | 7.24 | 0.47 ± 0.04 | 0.00 | 121 | 988 | 14,700 |
| 7 | 19.63 | 0.52 ± 0.04 | 0.00 | 21.8 | 114 | 2360 |
| 2 | 157.77 | 0.97 ± 0.01 | 0.00 | 1.28 | 803 | 21,600 |
| 3 | 315.55 | 0.97 ± 0.01 | 0.00 | 1.28 | 803 | 21,600 |
| m | CoV | RESET Test (p = 1) | RESET Test (p = 2) | F-Test | LM Test | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TN | TP | FN | FP | TN | TP | FN | FP | TN | TP | FN | FP | TN | |||||
| 100 | 60 | 50 | 2 | 0.01 | 0.18 | 0.82 | 0.56 | 0.44 | 0.33 | 0.67 | 0.75 | 0.25 | 0.86 | 0.14 | 0.09 | 0.91 | 0.68 | 0.32 | 0.11 | 0.89 |
| 0.05 | 0.28 | 0.72 | 0.57 | 0.43 | 0.44 | 0.56 | 0.78 | 0.22 | 0.82 | 0.19 | 0.09 | 0.91 | 0.67 | 0.33 | 0.14 | 0.86 | ||||
| 0.1 | 0.32 | 0.69 | 0.58 | 0.42 | 0.51 | 0.49 | 0.76 | 0.24 | 0.82 | 0.19 | 0.10 | 0.90 | 0.66 | 0.34 | 0.16 | 0.84 | ||||
| 0.2 | 0.42 | 0.58 | 0.56 | 0.44 | 0.69 | 0.31 | 0.81 | 0.19 | 0.71 | 0.29 | 0.08 | 0.92 | 0.60 | 0.41 | 0.18 | 0.82 | ||||
| 5 | 0.01 | 0.11 | 0.89 | 0.57 | 0.43 | 0.33 | 0.67 | 0.76 | 0.25 | 0.99 | 0.01 | 0.14 | 0.87 | 0.71 | 0.29 | 0.07 | 0.93 | |||
| 0.05 | 0.12 | 0.88 | 0.54 | 0.46 | 0.34 | 0.67 | 0.73 | 0.27 | 0.98 | 0.02 | 0.12 | 0.88 | 0.73 | 0.27 | 0.06 | 0.94 | ||||
| 0.1 | 0.19 | 0.81 | 0.54 | 0.46 | 0.41 | 0.59 | 0.75 | 0.25 | 0.97 | 0.03 | 0.13 | 0.87 | 0.71 | 0.29 | 0.11 | 0.90 | ||||
| 0.2 | 0.29 | 0.71 | 0.55 | 0.45 | 0.58 | 0.42 | 0.82 | 0.19 | 0.93 | 0.07 | 0.11 | 0.89 | 0.70 | 0.30 | 0.12 | 0.88 | ||||
| 10 | 0.01 | 0.11 | 0.89 | 0.57 | 0.43 | 0.40 | 0.60 | 0.73 | 0.27 | 1.00 | 0.00 | 0.11 | 0.89 | 0.47 | 0.53 | 0.00 | 1.00 | |||
| 0.05 | 0.13 | 0.87 | 0.57 | 0.43 | 0.42 | 0.58 | 0.76 | 0.24 | 1.00 | 0.00 | 0.10 | 0.90 | 0.48 | 0.52 | 0.01 | 0.99 | ||||
| 0.1 | 0.16 | 0.84 | 0.54 | 0.46 | 0.47 | 0.53 | 0.75 | 0.26 | 1.00 | 0.00 | 0.13 | 0.87 | 0.48 | 0.52 | 0.01 | 0.99 | ||||
| 0.2 | 0.26 | 0.74 | 0.57 | 0.43 | 0.57 | 0.43 | 0.79 | 0.21 | 0.99 | 0.01 | 0.12 | 0.88 | 0.44 | 0.56 | 0.01 | 0.99 | ||||
| m | CoV | TP | FN | FP | TN | |||
|---|---|---|---|---|---|---|---|---|
| 50 | 30 | 25 | 2 | 0.01 | 0.86 | 0.14 | 0.11 | 0.89 |
| 0.05 | 0.82 | 0.18 | 0.10 | 0.90 | ||||
| 0.1 | 0.75 | 0.25 | 0.09 | 0.91 | ||||
| 0.2 | 0.69 | 0.31 | 0.09 | 0.91 | ||||
| 5 | 0.01 | 0.99 | 0.01 | 0.10 | 0.90 | |||
| 0.05 | 0.98 | 0.02 | 0.10 | 0.90 | ||||
| 0.1 | 0.97 | 0.03 | 0.10 | 0.90 | ||||
| 0.2 | 0.92 | 0.08 | 0.11 | 0.89 | ||||
| 10 | 0.01 | 1.00 | 0.00 | 0.10 | 0.90 | |||
| 0.05 | 1.00 | 0.00 | 0.09 | 0.91 | ||||
| 0.1 | 1.00 | 0.00 | 0.09 | 0.91 | ||||
| 0.2 | 0.99 | 0.02 | 0.11 | 0.90 | ||||
| 200 | 120 | 100 | 2 | 0.01 | 0.76 | 0.24 | 0.11 | 0.89 |
| 0.05 | 0.73 | 0.27 | 0.10 | 0.90 | ||||
| 0.1 | 0.67 | 0.33 | 0.07 | 0.93 | ||||
| 0.2 | 0.58 | 0.42 | 0.10 | 0.90 | ||||
| 5 | 0.01 | 0.97 | 0.03 | 0.16 | 0.84 | |||
| 0.05 | 0.95 | 0.05 | 0.11 | 0.89 | ||||
| 0.1 | 0.94 | 0.07 | 0.13 | 0.87 | ||||
| 0.2 | 0.88 | 0.12 | 0.13 | 0.88 | ||||
| 10 | 0.01 | 1.00 | 0.00 | 0.15 | 0.85 | |||
| 0.05 | 0.99 | 0.01 | 0.16 | 0.84 | ||||
| 0.1 | 1.00 | 0.01 | 0.13 | 0.87 | ||||
| 0.2 | 0.98 | 0.02 | 0.15 | 0.85 | ||||
| 20 | 0.01 | 1.00 | 0.00 | 0.14 | 0.86 | |||
| 0.05 | 1.00 | 0.00 | 0.14 | 0.86 | ||||
| 0.1 | 1.00 | 0.00 | 0.15 | 0.86 | ||||
| 0.2 | 1.00 | 0.00 | 0.14 | 0.86 |
| k | nextra | nomit | Number of Remaining Reactions a | |
|---|---|---|---|---|
| Extra Reactions | Omitted Reactions | |||
| 1 | 3 | 3 | 2.82 ± 0.38 | 0.99 ± 0.10 |
| 5 | 5 | 4.13 ± 0.63 | 1.34 ± 0.46 | |
| 8 | 8 | 5.89 ± 0.83 | 2.21 ± 0.48 | |
| 1 then 2 | 5 | 5 | 3.66 ± 0.59 | 0.97 ± 0.17 |
| 8 | 8 | 5.03 ± 0.70 | 1.00 ± 0.29 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gunawan, R.; Hutter, S. Assessing and Resolving Model Misspecifications in Metabolic Flux Analysis. Bioengineering 2017, 4, 48. https://doi.org/10.3390/bioengineering4020048
Gunawan R, Hutter S. Assessing and Resolving Model Misspecifications in Metabolic Flux Analysis. Bioengineering. 2017; 4(2):48. https://doi.org/10.3390/bioengineering4020048
Chicago/Turabian StyleGunawan, Rudiyanto, and Sandro Hutter. 2017. "Assessing and Resolving Model Misspecifications in Metabolic Flux Analysis" Bioengineering 4, no. 2: 48. https://doi.org/10.3390/bioengineering4020048
APA StyleGunawan, R., & Hutter, S. (2017). Assessing and Resolving Model Misspecifications in Metabolic Flux Analysis. Bioengineering, 4(2), 48. https://doi.org/10.3390/bioengineering4020048