Phytotoxic and Genotoxic Effects of Copper Nanoparticles in Coriander (Coriandrum sativum—Apiaceae)
Abstract
:1. Introduction
2. Results
2.1. CuNP Decreased C. sativum Root Length, Biomass, and Chlorophyll Content
2.2. CuNP Caused Membrane Damage on C. sativum Plants
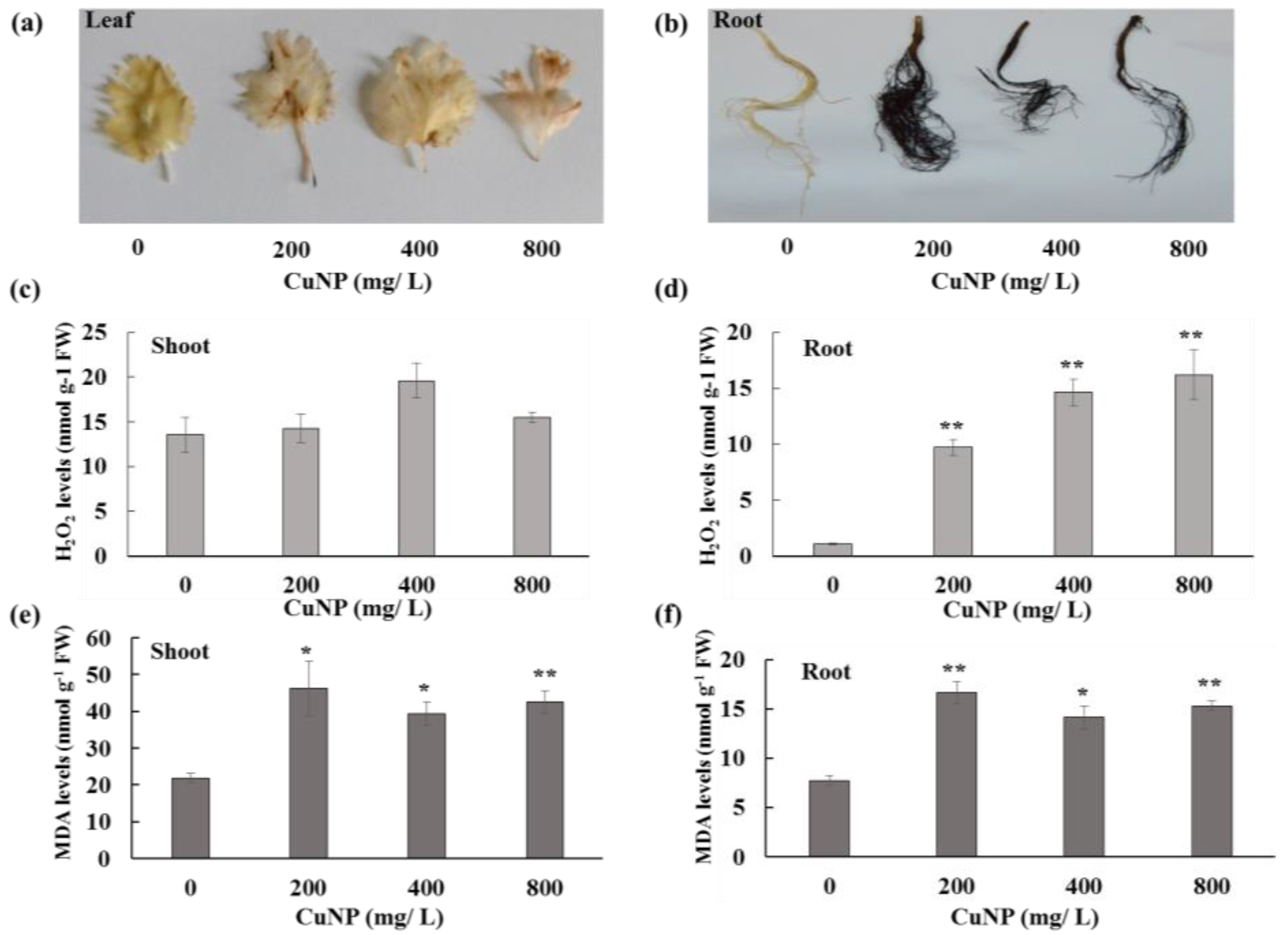
2.3. CuNP Increased Hydrogen Peroxide and Malondialdehyde Content in C. sativum Plants
2.4. C. sativum Plants Accumulated more CuNP in Root Tissues
2.5. CuNP Induced Genotoxicity in C. sativum Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Nanoparticle Treatment
4.2. Measurement of Biomass, Root Length, and Chlorophyll Contents
4.3. Root Membrane Integrity
4.4. Lipid per Oxidation and H2O2 Determination
4.5. X-ray Fluorescence (XRF) Analysis of CuNP in Plant Tissues
4.6. DNA Extraction and RAPD Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khot, L.R.; Sankaran, S.; Maja, J.M.; Ehsani, R.; Schuster, E.W. Applications of nanomaterials in agricultural production and crop protection: A review. Crop Prot. 2012, 35, 64–70. [Google Scholar] [CrossRef]
- Guo, H.; Barnard, A.S. Naturally occurring iron oxide nanoparticles: Morphology, surface chemistry and environmental stability. J. Mater. Chem. A 2013, 1, 27–42. [Google Scholar] [CrossRef]
- Kumar, P.; Kumar, A.; Lead, J.R. Nanoparticles in the Indian Environment: Known, Unknowns and Awareness. Environ. Sci. Technol. 2012, 46, 7071–7072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatt, I.; Tripathi, B.N. Interaction of engineered nanoparticles with various components of the environment and possible strategies for their risk assessment. Chemosphere 2011, 82, 308–317. [Google Scholar] [CrossRef]
- Fabrega, J.; Luoma, S.N.; Tyler, C.R.; Galloway, T.S.; Lead, J.R. Silver nanoparticles: Behaviour and effects in the aquatic environment. Environ. Int. 2011, 37, 517–531. [Google Scholar] [CrossRef]
- Bystrzejewska-Piotrowska, G.; Golimowski, J.; Urban, P.L. Nanoparticles: Their potential toxicity, waste and environmental management. Waste Manag. 2009, 29, 2587–2595. [Google Scholar] [CrossRef] [PubMed]
- Athanassiou, E.K.; Grass, R.N.; Stark, W.J. Large-scale production of carbon-coated copper nanoparticles for sensor applications. Nanotechnology 2006, 17, 1668. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, J.R.; Lee, K.J.; Stott, N.E.; Kim, D. Large-scale synthesis of copper nanoparticles by chemically controlled reduction for applications of inkjet-printed electronics. Nanotechnology 2008, 19, 415604. [Google Scholar] [CrossRef]
- Oberdürster, G. Toxicology of ultrafine particles: In vivo studies. Philos. Trans. R. Soc. Lond. Ser. A 2000, 358, 2719. [Google Scholar] [CrossRef]
- Yruela, I. Copper in plants. Braz. J. Plant Physiol. 2005, 17, 145–156. [Google Scholar] [CrossRef]
- Sommer, A.L. Copper as an essential for plant growth. Plant Physiol. 1931, 6, 339. [Google Scholar] [CrossRef] [PubMed]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 2010, 8, 199–216. [Google Scholar] [CrossRef]
- Tripathi, D.K.; Singh, S.; Singh, S.; Pandey, R.; Singh, V.P.; Sharma, N.C.; Prasad, S.M.; Dubey, N.K.; Chauhan, D.K. An overview on manufactured nanoparticles in plants: Uptake, translocation, accumulation and phytotoxicity. Plant Physiol. Biochem. 2017, 110, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Faisal, M.; Saquib, Q.; Alatar, A.A.; Al-Khedhairy, A.A. Phytotoxicity of Nanoparticles; Springer: Berlin, Germany, 2018. [Google Scholar]
- Ravishankar Rai, V.; Jamuna Bai, A. Nanoparticles and Their Potential Application as Antimicrobials; Méndez-Vilas, A., Ed.; Formatex: Mysore, India, 2011. [Google Scholar]
- Palza, H. Antimicrobial Polymers with Metal Nanoparticles. Int. J. Mol. Sci. 2015, 16, 2099–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiaune, L.; Singhasemanon, N. Pesticidal copper (I) oxide: Environmental fate and aquatic toxicity. Rev. Environ. Contam. Toxicol. 2011, 213, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Ponmurugan, P.; Manjukarunambika, K.; Elango, V.; Gnanamangai, B.M. Antifungal activity of biosynthesised copper nanoparticles evaluated against red root-rot disease in tea plants. J. Exp. Nanosci. 2016, 11, 1019–1031. [Google Scholar] [CrossRef]
- Stampoulis, D.; Sinha, S.K.; White, J.C. Assay-Dependent Phytotoxicity of Nanoparticles to Plants. Environ. Sci. Technol. 2009, 43, 9473–9479. [Google Scholar] [CrossRef]
- Shi, J.; Peng, C.; Yang, Y.; Yang, J.; Zhang, H.; Yuan, X.; Chen, Y.; Hu, T. Phytotoxicity and accumulation of copper oxide nanoparticles to the Cu-tolerant plant Elsholtzia splendens. Nanotoxicology 2014, 8, 179–188. [Google Scholar] [CrossRef]
- Nair, P.M.G.; Chung, I.M. Study on the correlation between copper oxide nanoparticles induced growth suppression and enhanced lignification in Indian mustard (Brassica juncea L.). Ecotoxicol. Environ. Saf. 2015, 113, 302–313. [Google Scholar] [CrossRef]
- Mosa, K.A.; El-Naggar, M.; Ramamoorthy, K.; Alawadhi, H.; Elnaggar, A.; Wartanian, S.; Ibrahim, E.; Hani, H. Copper Nanoparticles Induced Genotoxicity, Oxidative Stress, and Changes in Superoxide Dismutase (SOD) Gene Expression in Cucumber (Cucumis sativus) Plants. Front. Plant Sci. 2018, 9, 872. [Google Scholar] [CrossRef]
- Atha, D.H.; Wang, H.; Petersen, E.J.; Cleveland, D.; Holbrook, R.D.; Jaruga, P.; Dizdaroglu, M.; Xing, B.; Nelson, B.C. Copper Oxide Nanoparticle Mediated DNA Damage in Terrestrial Plant Models. Environ. Sci. Technol. 2012, 46, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Hocking, B.; Dayod, M.; Xu, B.; Athman, A.; Henderson, S.; Aukett, L.; Conn, V.; Shearer, M.K.; Fuentes, S.; et al. Protocol: Optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 2013, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Kivuti, N.M. Using Cilantro Leaves and Stems to Remove Lead, Cadmium and Turbidity from Contaminated Water; Kenyatta University: Kenya, Nairobi, 2017. [Google Scholar]
- Gaur, N.; Kukreja, A.; Yadav, M.; Tiwari, A. Assessment of phytoremediation ability of Coriander sativum for soil and water co-contaminated with lead and arsenic: A small-scale study. 3 Biotech 2017, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, M.V.J.; Sharma, P.K. Effect of copper oxide nanoparticles on growth, morphology, photosynthesis, and antioxidant response in Oryza sativa. Photosynthetica 2016, 54, 110–119. [Google Scholar] [CrossRef]
- Hong, J.; Rico, C.M.; Zhao, L.; Adeleye, A.S.; Keller, A.A.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Toxic effects of copper-based nanoparticles or compounds to lettuce (Lactuca sativa) and alfalfa (Medicago sativa). Environ. Sci. Process. Impacts 2015, 17, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.M.G.; Chung, I.M. Impact of copper oxide nanoparticles exposure on Arabidopsis thaliana growth, root system development, root lignificaion, and molecular level changes. Environ. Sci. Pollut. Res. 2014, 21, 12709–12722. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.K.; Hossain, Z. Impact of nano-CuO stress on rice (Oryza sativa L.) seedlings. Chemosphere 2013, 93, 906–915. [Google Scholar] [CrossRef]
- Shaw, A.K.; Ghosh, S.; Kalaji, H.M.; Bosa, K.; Brestic, M.; Zivcak, M.; Hossain, Z. Nano-CuO stress induced modulation of antioxidative defense and photosynthetic performance of Syrian barley (Hordeum vulgare L.). Environ. Exp. Bot. 2014, 102, 37–47. [Google Scholar] [CrossRef]
- Dingle, T.C.; MacCannell, D.R. Chapter 9—Molecular Strain Typing and Characterisation of Toxigenic Clostridium difficile. In Methods in Microbiology; Sails, A., Tang, Y.-W., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 42, pp. 329–357. [Google Scholar]
- Gui, X.; Zhang, Z.; Liu, S.; Ma, Y.; Zhang, P.; He, X.; Li, Y.; Zhang, J.; Li, H.; Rui, Y.; et al. Fate and Phytotoxicity of CeO2 Nanoparticles on Lettuce Cultured in the Potting Soil Environment. PLoS ONE 2015, 10, e0134261. [Google Scholar] [CrossRef]
- De Filippis, L.F.; Ziegler, H. Effect of Sublethal Concentrations of Zinc, Cadmium and Mercury on the Photosynthetic Carbon Reduction Cycle of Euglena. J. Plant Physiol. 1993, 142, 167–172. [Google Scholar] [CrossRef]
- Chandra, R.; Kang, H. Mixed heavy metal stress on photosynthesis, transpiration rate, and chlorophyll content in poplar hybrids. For. Sci. Technol. 2016, 12, 55–61. [Google Scholar] [CrossRef]
- Lequeux, H.; Hermans, C.; Lutts, S.; Verbruggen, N. Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 2010, 48, 673–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feigl, G.; Kumar, D.; Lehotai, N.; Tugyi, N.; Molnar, A.; Ordog, A.; Szepesi, A.; Gemes, K.; Laskay, G.; Erdei, L.; et al. Physiological and morphological responses of the root system of Indian mustard (Brassica juncea L. Czern.) and rapeseed (Brassica napus L.) to copper stress. Ecotoxicol. Environ. Saf. 2013, 94, 179–189. [Google Scholar] [CrossRef]
- Hernandez-Viezcas, J.A.; Castillo-Michel, H.; Andrews, J.C.; Cotte, M.; Rico, C.; Peralta-Videa, J.R.; Ge, Y.; Priester, J.H.; Holden, P.A.; Gardea-Torresdey, J.L. In Situ Synchrotron X-ray Fluorescence Mapping and Speciation of CeO2 and ZnO Nanoparticles in Soil Cultivated Soybean (Glycine max). ACS Nano 2013, 7, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, Y.; Hernandez-Viezcas, J.A.; Hong, J.; Majumdar, S.; Niu, G.; Duarte-Gardea, M.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Monitoring the environmental effects of CeO2 and ZnO nanoparticles through the life cycle of corn (Zea mays) plants and in situ mu-XRF mapping of nutrients in kernels. Environ. Sci. Technol. 2015, 49, 2921–2928. [Google Scholar] [CrossRef]
- Larue, C.; Laurette, J.; Herlin-Boime, N.; Khodja, H.; Fayard, B.; Flank, A.-M.; Brisset, F.; Carriere, M. Accumulation, translocation and impact of TiO2 nanoparticles in wheat (Triticum aestivum spp.): Influence of diameter and crystal phase. Sci. Total Environ. 2012, 431, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Leisner, S.M.; Frantz, J. Alleviation of copper toxicity in Arabidopsis thaliana by silicon addition to hydroponic solutions. J. Am. Soc. Hort. Sci. 2008, 133, 670–677. [Google Scholar]
- Moreno-Olivas, F.; Gant, V.U.; Johnson, K.L.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Random amplified polymorphic DNA reveals that TiO2 nanoparticles are genotoxic to Cucurbita pepo. J. Zhejiang Univ. Sci. A 2014, 15, 618–623. [Google Scholar] [CrossRef]
- Mattiello, A.; Filippi, A.; Pošćić, F.; Musetti, R.; Salvatici, M.C.; Giordano, C.; Vischi, M.; Bertolini, A.; Marchiol, L. Evidence of Phytotoxicity and Genotoxicity in Hordeum vulgare L. Exposed to CeO2 and TiO2 Nanoparticles. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Lee, S.; Chung, H.; Kim, S.; Lee, I. The Genotoxic Effect of ZnO and CuO Nanoparticles on Early Growth of Buckwheat, Fagopyrum Esculentum. Water Air Soil Pollut. 2013, 224, 1668. [Google Scholar] [CrossRef]
- López-Moreno, M.L.; de la Rosa, G.; Hernández-Viezcas, J.Á.; Castillo-Michel, H.; Botez, C.E.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Evidence of the Differential Biotransformation and Genotoxicity of ZnO and CeO2 Nanoparticles on Soybean (Glycine max) Plants. Environ. Sci. Technol. 2010, 44, 7315–7320. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591. [Google Scholar] [CrossRef]
- Liu, X.; Huang, B. Cytokinin Effects on Creeping Bentgrass Response to Heat Stress. Crop Sci. 2002, 42, 466–472. [Google Scholar] [CrossRef]
- Zanardo, D.I.L.; Lima, R.B.; Ferrarese, M.d.L.L.; Bubna, G.A.; Ferrarese-Filho, O. Soybean root growth inhibition and lignification induced by p-coumaric acid. Environ. Exp. Bot. 2009, 66, 25–30. [Google Scholar] [CrossRef]
- Zhou, W.; Leul, M. Uniconazole-induced alleviation of freezing injury in relation to changes in hormonal balance, enzyme activities and lipid peroxidation in winter rape. Plant Growth Regul. 1998, 26, 41–47. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—Powdery mildew interaction. Plant J. 2002, 11, 1187–1194. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
AlQuraidi, A.O.; Mosa, K.A.; Ramamoorthy, K. Phytotoxic and Genotoxic Effects of Copper Nanoparticles in Coriander (Coriandrum sativum—Apiaceae). Plants 2019, 8, 19. https://doi.org/10.3390/plants8010019
AlQuraidi AO, Mosa KA, Ramamoorthy K. Phytotoxic and Genotoxic Effects of Copper Nanoparticles in Coriander (Coriandrum sativum—Apiaceae). Plants. 2019; 8(1):19. https://doi.org/10.3390/plants8010019
Chicago/Turabian StyleAlQuraidi, Alya O., Kareem A. Mosa, and Kalidoss Ramamoorthy. 2019. "Phytotoxic and Genotoxic Effects of Copper Nanoparticles in Coriander (Coriandrum sativum—Apiaceae)" Plants 8, no. 1: 19. https://doi.org/10.3390/plants8010019




