Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis
Abstract
:1. Introduction
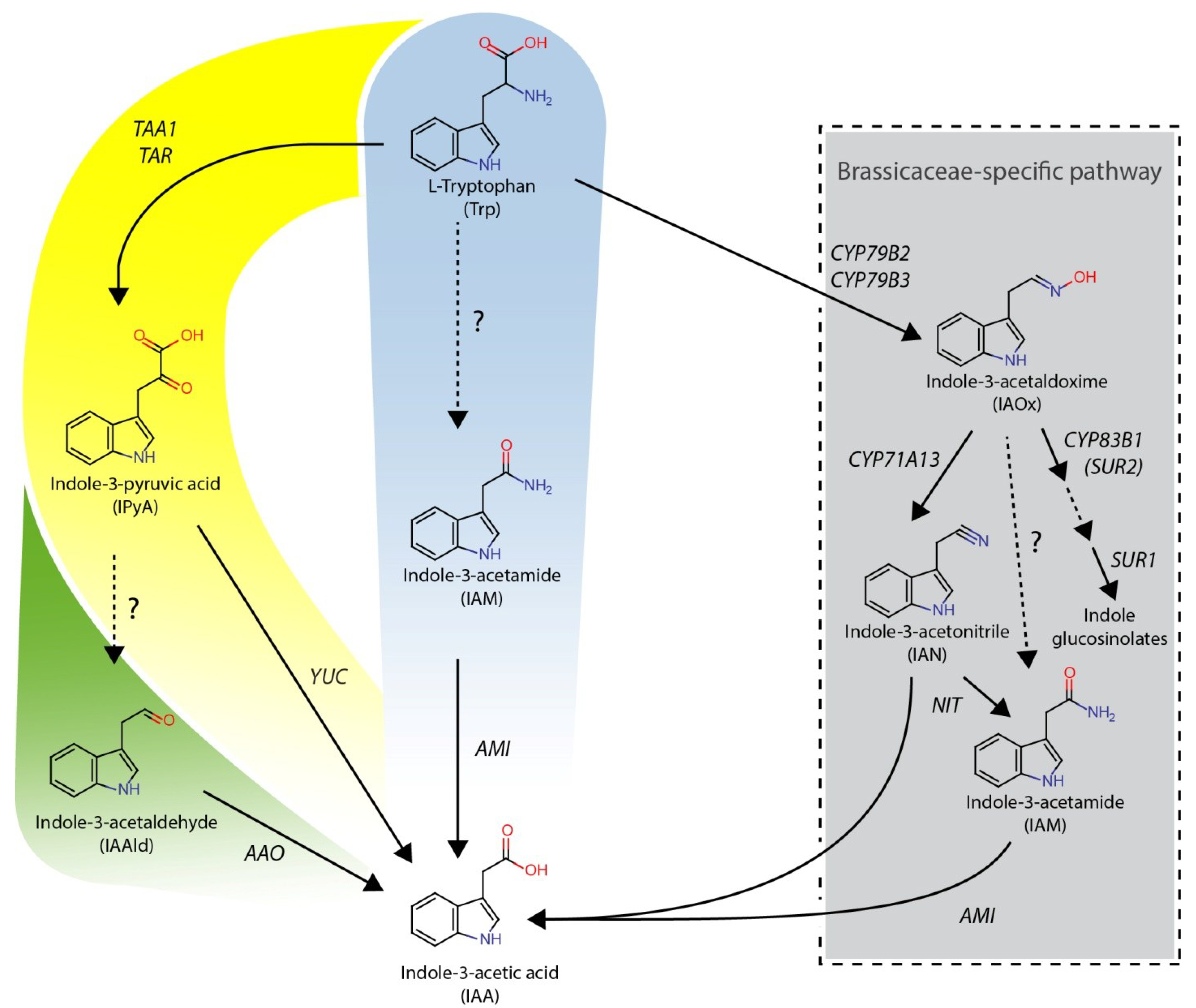
2. Results and Discussion
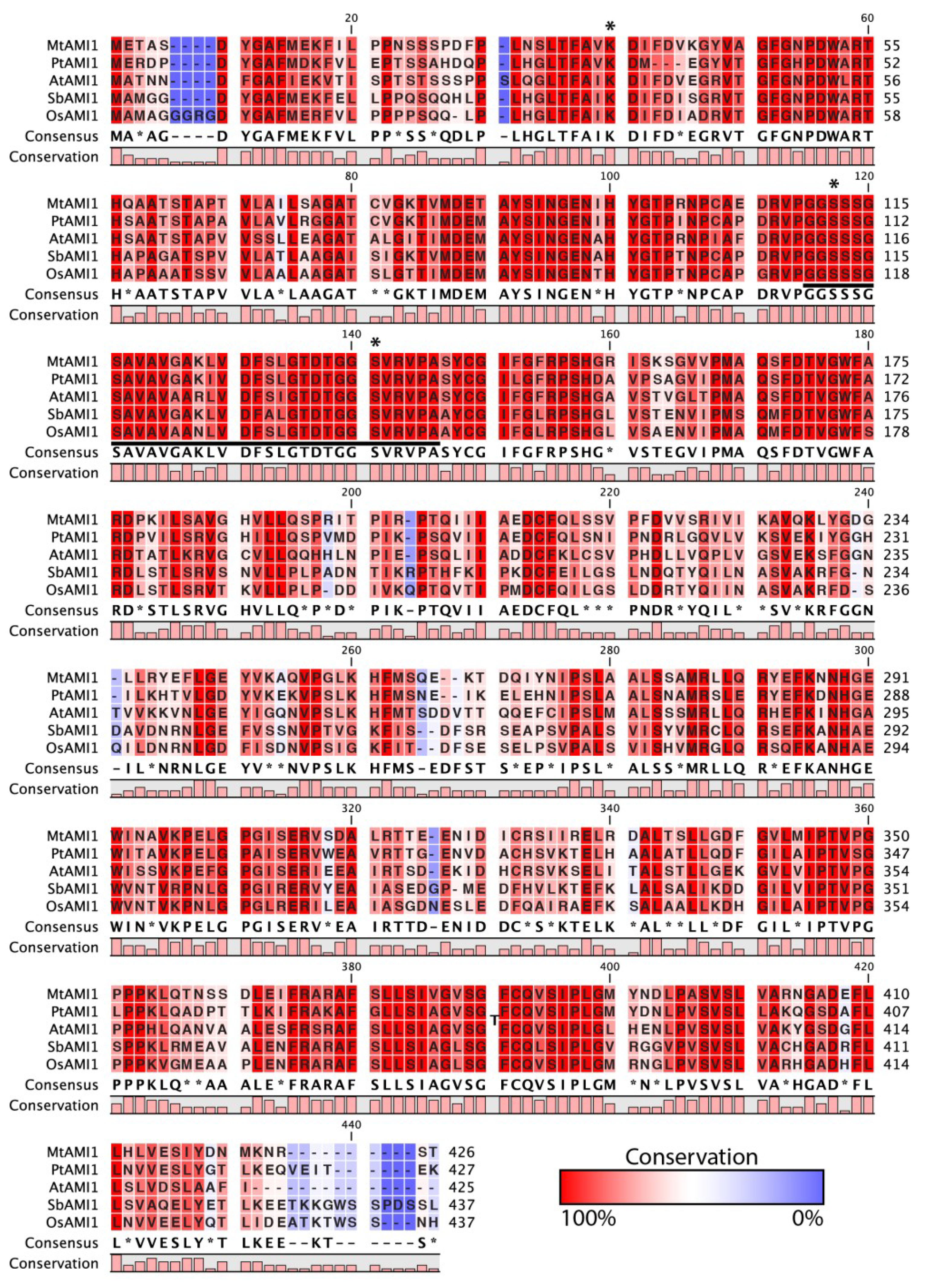
2.1. Identification of AMI1-like Proteins in Plant Genomes

2.2. Subcellular Localization of Selected Plant Amidases
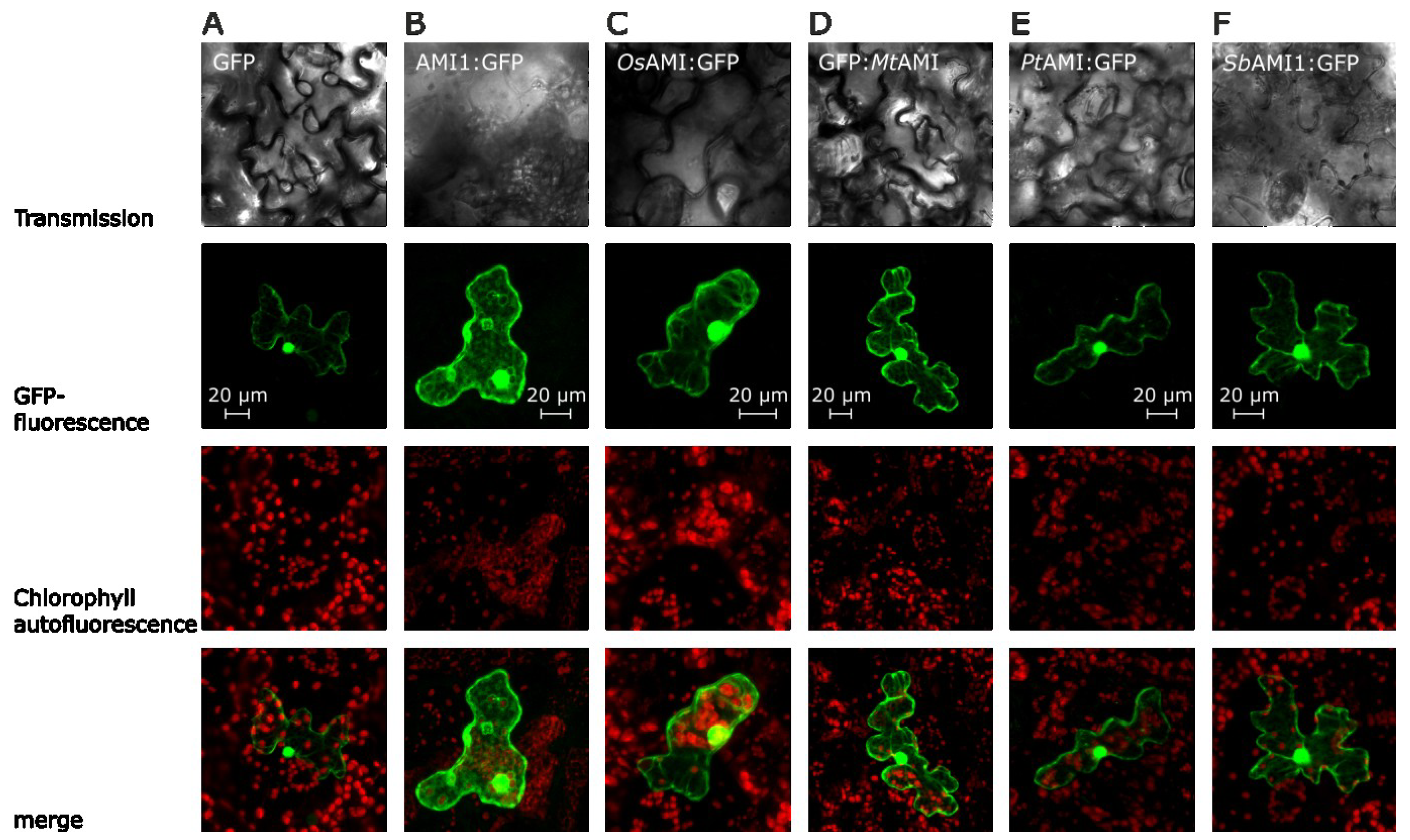
2.3. Functional Analysis of the Selected Plant Amidases
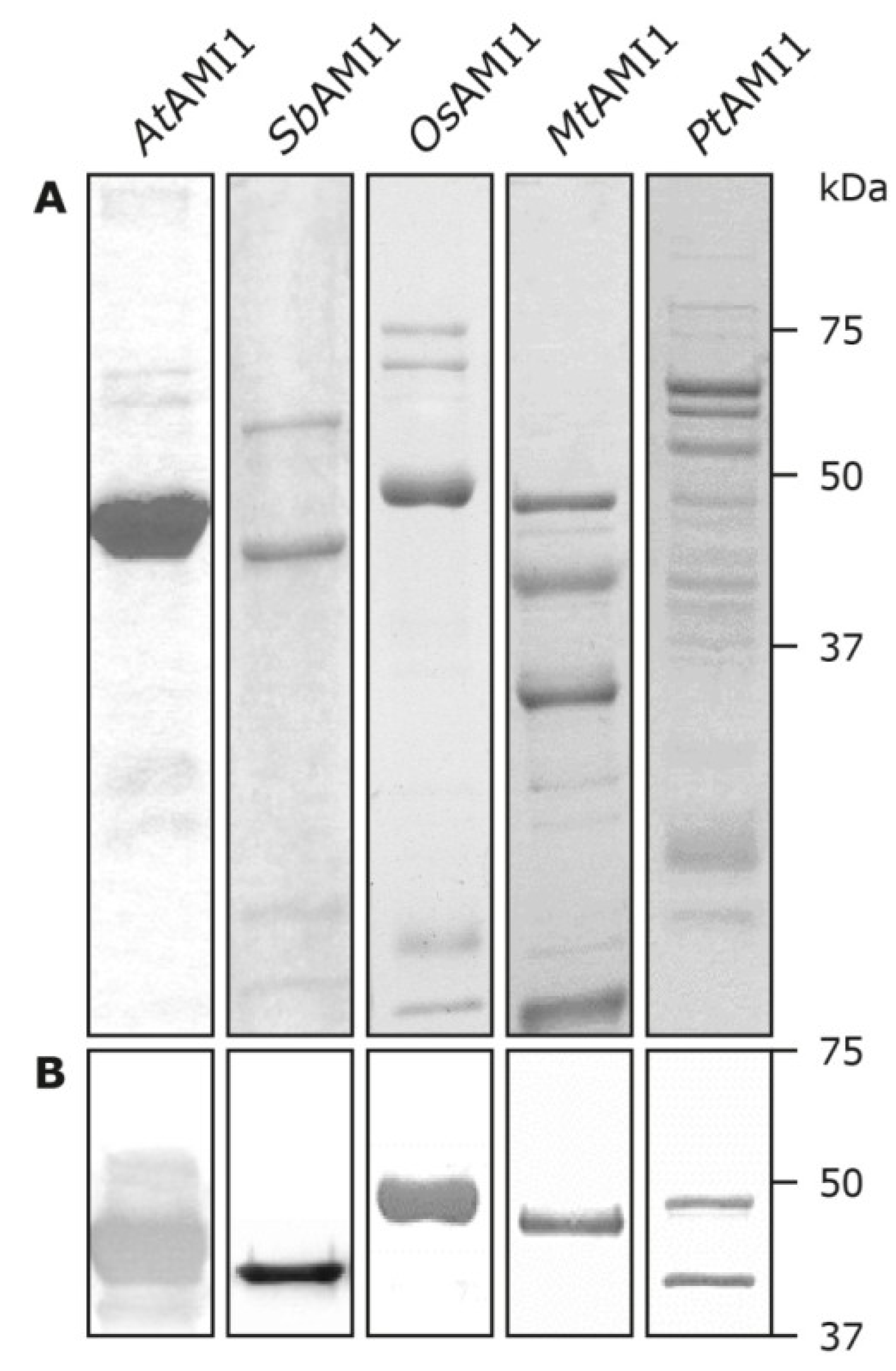
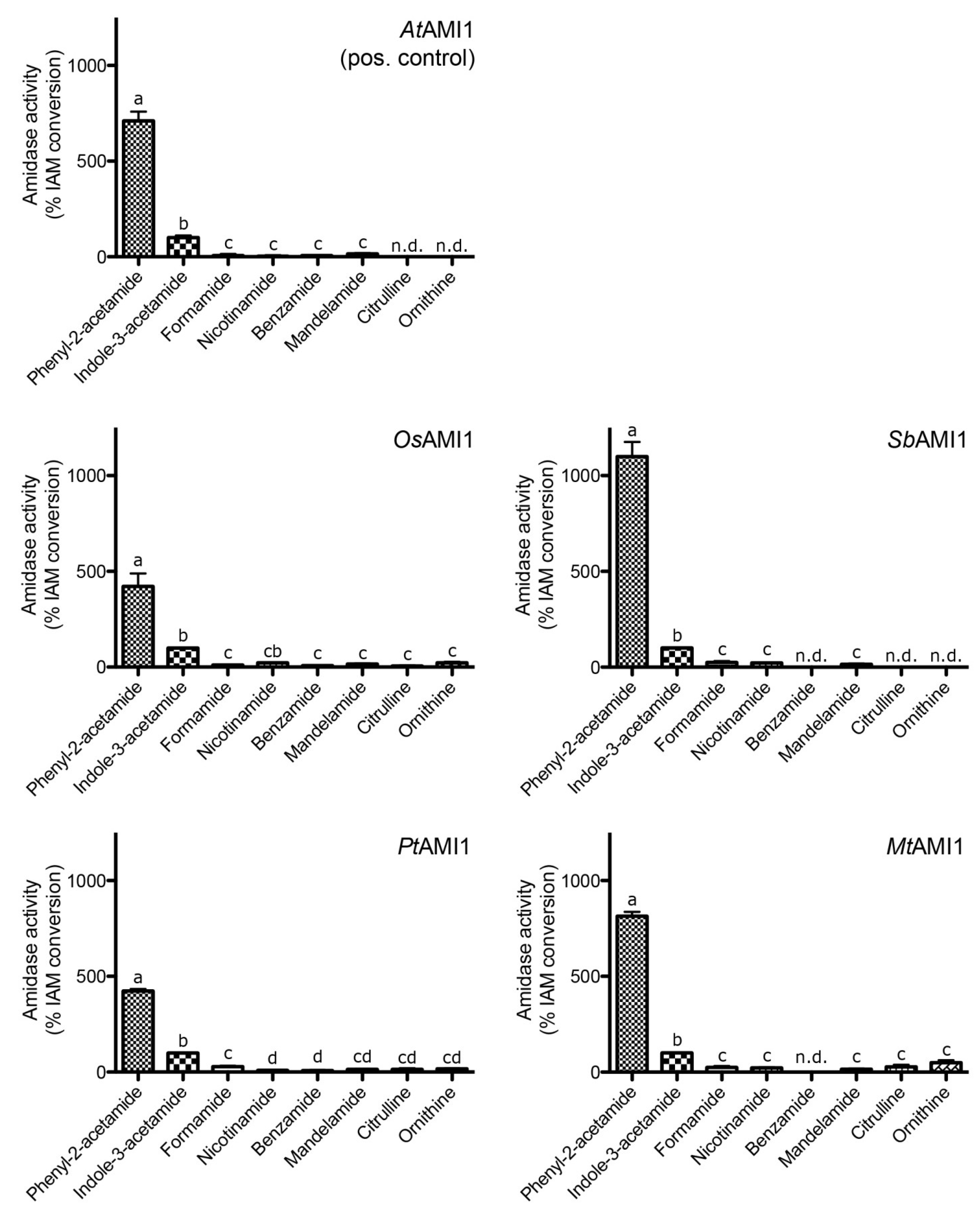
| Parameter | AtAMI1 | SbAMI1 | OsAMI1 | PtAMI1 | MtAMI1 |
|---|---|---|---|---|---|
| Amino acid identity to AtAMI1 [%] | 100 | 58 | 57 | 64 | 66 |
| Intracellular localization | cytoplasm/nucleoplasm | cytoplasm/nucleoplasm | cytoplasm/nucleoplasm | cytoplasm/nucleoplasm | cytoplasm/nucleoplasm |
| Specific activity (IAM) [pkat mg−1] | 3070 ± 520 | 2378 ± 324 | 375 ± 52 | 329 ± 22 | 37 ± 3 |
| Temperature optimum [°C] | 37 | 45 | 27 | 37 | 35 |
| pH optimum | 7.5 | 6 | 7.5 | 7.5 | 7.5 |
| Calculated molecular weight [kDa] | 46 | 45 | 50 | 53 | 57 |
2.4. Occurrence and Auxin Activity of Phenyl-2-acetic Acid in Arabidopsis
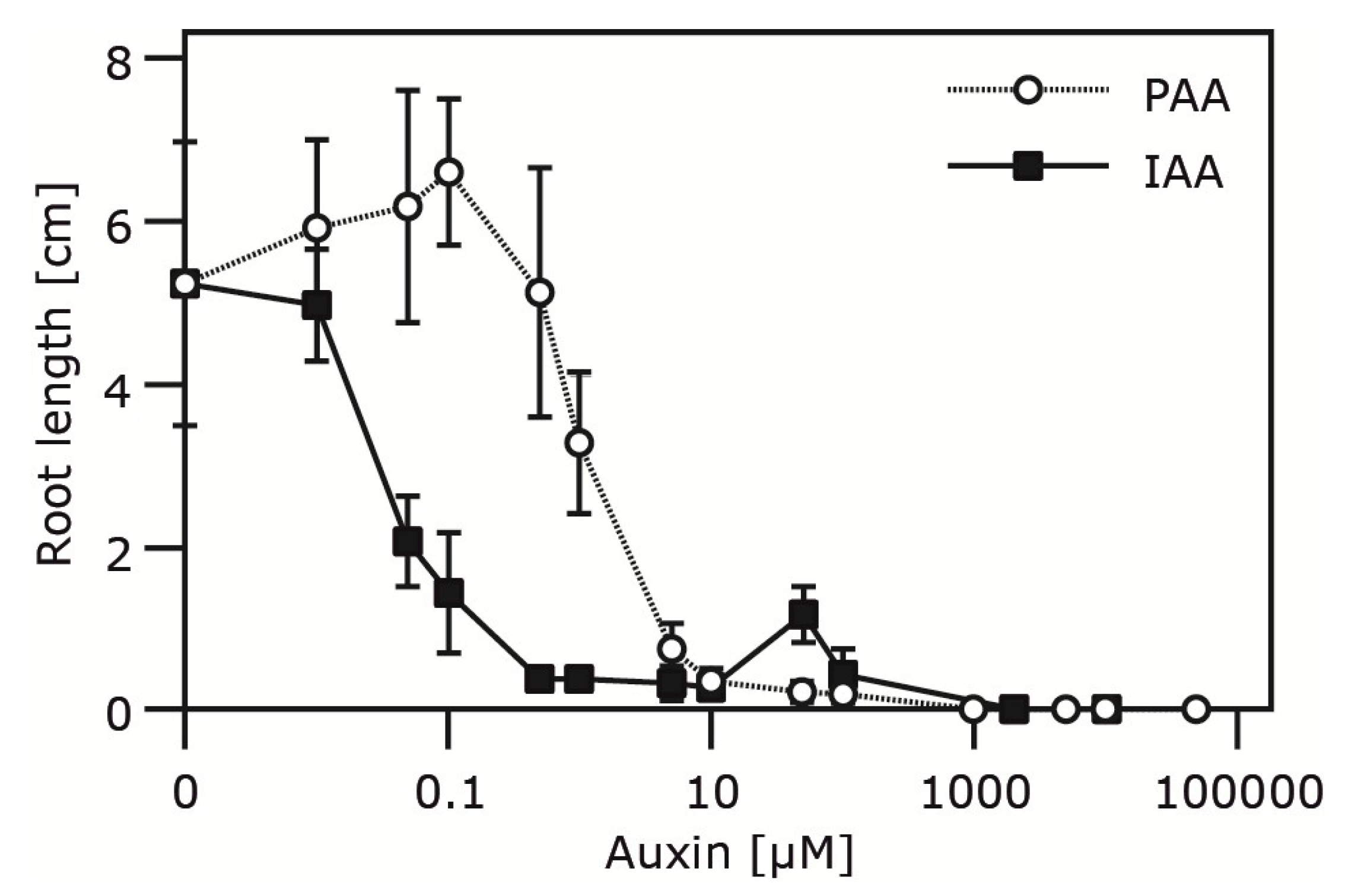
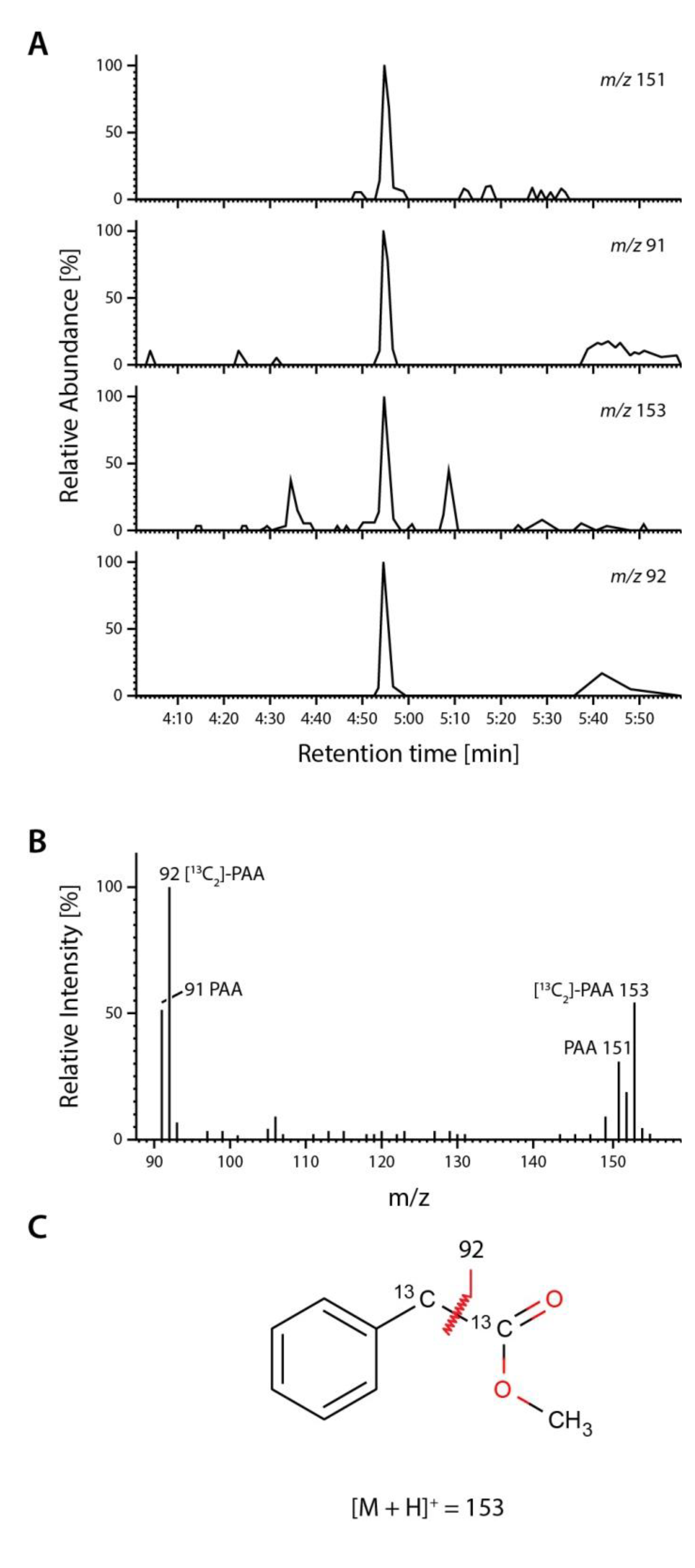
| Plant Age (Weeks) | Free PAA pmol (g FW)−1 | Free IAA pmol (g FW)−1 |
|---|---|---|
| Two weeks | 98 ± 6.4 | 35 ± 2.7 |
| Six weeks | 23 ± 2 | 15 ± 1 |
3. Experimental Section
3.1. Plant Material and Plant Growth Conditions
3.2. RNA Isolation and RT-PCR
| Primer Name | Sequence (5'–3') |
|---|---|
| OsAMI1-SacI-His2C-For | 5'-TATGAGCTCTATGGCGATGGCGGGTGGAG-3' |
| OsAMI1-KpnI-His2C-Rev | 5'-TATGGTACCGTGATTGGAGGACCAAGTTTTAG-3' |
| SbAMI1-SpeI-pUC-For | 5'-TATACTAGTATGGCGATGGGCGGCGATTAC-3' |
| SbAMI1-SmaI-pUC-Rev | 5'-TATCCCGGGGAGAGAGGAGTCTGGTGAGC-3' |
| MtAMI1-XhoI-His2B-For | 5'-TATCTCGAGATGGAAACAGCCTCAGACTATG-3' |
| MtAMI1-XbaI-His2B-Rev | 5'-TATTCTAGATATTTTTCAATGTTATCATAAATACTC-3' |
| PtAMI1-XhoI-His2B-For | 5'-TATCTCGAGATGGAACGAGACCCGGATTATG-3' |
| PtAMI1-XbaI-His2B-Rev | 5'-TATTCTAGATATTTTTCAGTGATCTCAACCTG-3' |
| MtAMI1-BglII-pUC-For | 5'-TATAGATCTATGGAAACAGCCTCAGACTATG-3' |
| MtAMI1-SalI-pUC-Rev | 5'-TATGTCGACCTATTTTTCAATGTTATCATAAATACTC-3' |
3.3. Generation of Bacterial Expression Constructs
3.4. Preparation of GFP Amidase Fusion Constructs
3.5. Heterologous Expression of Recombinant Amidases
3.6. Transient Expression in Plants and Confocal Laser Scanning Microscopy
3.7. Assay for Amidase Activity
3.8. Root Growth Assay
3.9. Auxin Extraction and Purification
3.10. Quantification of Endogenous Phenyl-2-Acetic Acid and Indole-3-Acetic Acid from Arabidopsis thaliana
3.11. Gel Electrophoresis and Immunoblotting
3.12. Phylogenetic Analysis
3.13. Statistic Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Thimann, K.V. Hormone Action in the Whole Life of Plants; Univ of Massachusetts Press: Amherst, MA, USA, 1977. [Google Scholar]
- Davies, P.J. Plant Hormones. Biosynthesis, Signal Transduction, Action! Revised 3rd ed.; Kluwer Academic Publishers: London, UK, 2010. [Google Scholar]
- Slovin, J.P.; Bandurski, R.S.; Cohen, J.D. Auxin. In Biochemistry and Molecular Biology of Plant Hormones; Hooykaas, P.J.J., Hall, M.A., Libbenga, K.R., Eds.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 115–140. [Google Scholar]
- Wightman, F.; Lighty, D.L. Identification of phenylacetic acid as a natural auxin in the shoots of higher plants. Physiol. Plant. 1982, 55, 17–24. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Cohen, J.D. Identification and quantification of three active auxins in different tissues of Tropaeolum majus. Physiol. Plant. 2002, 115, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, C.H.; Wain, R.L.; Wightman, F. The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proc. R. Soc. Lond. B Biol. Sci. 1960, 152, 231–254. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E.; Chen, K.-H.; Cohen, J. Identification of indole-3-butyric acid as an endogenous constituent of maize kernels and leaves. Plant Growth Regul. 1989, 8, 215–223. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Epstein, E. Occurrence and in vivo biosynthesis of indole-3-butyric acid in corn (Zea mays L.). Plant Physiol. 1991, 97, 765–770. [Google Scholar]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Müller, A.; Weiler, E.W. Many roads lead to “auxin”: Of nitrilases, synthases, and amidases. Plant Biol. 2006, 8, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, T.; Hoffmann, M.; Hentrich, M.; Pollmann, S. Indole-3-acetamide-dependent auxin biosynthesis: A widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 2010, 89, 895–905. [Google Scholar]
- Brumos, J.; Alonso, J.M.; Stepanova, A.N. Genetic aspects of auxin biosynthesis and its regulation. Physiol. Plant. 2014, 151, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.O. Genes specifying auxin and cytokinin biosynthesis in phytopathogens. Ann. Rev. Plant Physiol. 1986, 37, 509–538. [Google Scholar] [CrossRef]
- Manulis, S.; Valinski, L.; Gafni, Y.; Hershenhorn, J. Indole-3-acetic acid biosynthetic pathways in Erwinia herbicola in relation to pathogenicity on Gypsophila paniculata. Physiol. Mol. Plant Pathol. 1991, 39, 161–171. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yamaguchi, I.; Kõno, T.; Igoshi, M.; Hirose, K.; Suzuki, K. Characterization of plant growth substances in Citrus unshiu and their change in fruit development. Plant Cell Physiol. 1975, 16, 1101–1111. [Google Scholar]
- Saotome, M.; Shirahata, K.; Nishimura, R.; Yahaba, M.; Kawaguchi, M.; Syõno, K.; Kitsuwa, T.; Ishii, Y.; Nakamura, T. The identification of indole-3-acetic acid and indole-3-acetamide in the hypocotyls of japanese cherry. Plant Cell Physiol. 1993, 34, 157–159. [Google Scholar]
- Rajagopal, R.; Tsurusaki, K.I.; Kannangara, G.; Kuraishi, S.; Sakurai, N. Natural occurrence of indoleacetamide and amidohydrolase activity in etiolated aseptically-grown squash seedlings. Plant Cell Physiol. 1994, 35, 329–339. [Google Scholar]
- Pollmann, S.; Müller, A.; Piotrowski, M.; Weiler, E.W. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 2002, 216, 155–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Kobayashi, M.; Sakurai, A.; Syõno, K. The presence of an enzyme that converts indoIe-3-acetamide into IAA in wild and cultivated rice. Plant Cell Physiol. 1991, 32, 143–149. [Google Scholar]
- Arai, Y.; Kawaguchi, M.; Syono, K.; Ikuta, A. Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J. Plant Res. 2004, 117, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Fujioka, S.; Sakurai, A.; Yamaki, Y.T.; Syõno, K. Presence of a pathway for the biosynthesis of auxin via indole-3-acetamide in Trifoliata orange. Plant Cell Physiol. 1993, 34, 121–128. [Google Scholar]
- Pollmann, S.; Neu, D.; Weiler, E.W. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 2003, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, K.; Hara, M.; Suzuki, M.; Seki, H.; Muranaka, T.; Mano, Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 2009, 583, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Schröder, G.; Waffenschmidt, S.; Weiler, E.W.; Schröder, J. The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur. J. Biochem. 1984, 138, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Palm, C.J.; Brooks, B.; Kosuge, T. Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc. Natl. Acad. Sci. USA 1985, 82, 6522–6526. [Google Scholar] [CrossRef] [PubMed]
- Neu, D.; Lehmann, T.; Elleuche, S.; Pollmann, S. Arabidopsis amidase 1, a member of the amidase signature family. FEBS J. 2007, 274, 3440–3451. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Neu, D.; Lehmann, T.; Berkowitz, O.; Schäfer, T.; Weiler, E.W. Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 2006, 224, 1241–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronsson, H.; Boij, P.; Patel, R.; Wardle, A.; Töpel, M.; Jarvis, P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007, 52, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Expósito-Rodríguez, M.; Borges, A.; Borges-Pérez, A.; Hernández, M.; Pérez, J. Cloning and biochemical characterization of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan-dependent auxin biosynthesis pathway. J. Plant Growth Regul. 2007, 26, 329–340. [Google Scholar] [CrossRef]
- LeClere, S.; Schmelz, E.A.; Chourey, P.S. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010, 153, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Tobeña-Santamaria, R.; Bliek, M.; Ljung, K.; Sandberg, G.; Mol, J.N.; Souer, E.; Koes, R. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002, 16, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Davies, N.W.; Molesworth, P.P.; Davidson, S.E.; Smith, J.A.; Lowe, E.K.; Reid, J.B.; Ross, J.J. Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol. 2010, 154, 1957–1965. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Janowitz, T.; Trompetter, I.; Piotrowski, M. Evolution of nitrilases in glucosinolate-containing plants. Phytochemistry 2009, 70, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Cokol, M.; Nair, R.; Rost, B. Finding nuclear localization signals. EMBO Rep. 2000, 1, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Isogai, Y.; Okamoto, T.; Koizumi, T. Isolation of 2-phenylacetamide, indole-3-acetamide, and indole-3-carboxaldehyde from etiolated seedling of phaseolus. Chem. Pharm. Bull. 1963, 11, 1217–1218. [Google Scholar] [CrossRef] [PubMed]
- Isogai, Y.; Okamoto, T.; Koizumi, T. Studies on plant growth regulators. I. Isolation of indole-3-acetamide, 2-phenylacetamide, and indole-3-carboxaldehyde from etiolated seedlings of phaseolus. Chem. Pharm. Bull. 1967, 15, 151–158. [Google Scholar]
- Wittstock, U.; Halkier, B.A. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. Catalyzes the conversion of l-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 2000, 275, 14659–14666. [Google Scholar]
- Okamoto, T.; Koizumi, T.; Isogai, Y. Studies on plant growth regulators. II. Isolation of indole-3-acetic acid, phenylacetic acid, and several plant growth inhibitors from etiolated seedlings of phaseolus. Chem. Pharm. Bull. 1967, 15, 159–163. [Google Scholar]
- Yokohari, M.; Brown, R.D.; Kato, Y.; Yamamoto, S. The cooling effect of paddy fields on summertime air temperature in residential Tokyo, Japan. Landsc. Urban Plan. 2001, 53, 17–27. [Google Scholar] [CrossRef]
- Koepfli, J.B.; Thimann, K.V.; Went, F.W. Phytohormones: Structure and physiological activity. I. J. Biol. Chem. 1938, 122, 763–780. [Google Scholar]
- Muir, R.M.; Fujita, T.; Hansch, C. Structure-activity relationship in the auxin activity of mono-substituted phenylacetic acids. Plant Physiol. 1967, 42, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Weiler, E.W. IAA-synthase, an enzyme complex from Arabidopsis thaliana catalyzing the formation of indole-3-acetic acid from (S)-tryptophan. Biol. Chem. 2000, 381, 679–686. [Google Scholar] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Lehmann, T.; Pollmann, S. Gene expression and characterization of a stress-induced tyrosine decarboxylase from Arabidopsis thaliana. FEBS Lett. 2009, 583, 1895–1900. [Google Scholar] [CrossRef] [PubMed]
- Finer, J.J.; Vain, P.; Jones, M.W.; McMullen, M.D. Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 1992, 11, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, P.W.; Hitchcock, A.E. Substituted phenoxy and benzoic acid growth substances and the relation of structure to physiological activity. Contrib. Boyce Thompson Inst. 1942, 12, 321–343. [Google Scholar]
- Müller, A.; Düchting, P.; Weiler, E.W. A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 2002, 216, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Düchting, P.; Weiler, E.W. Hormone profiling in Arabidopsis. Methods Mol. Biol. 2006, 323, 449–457. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Parets-Soler, A.; Pardo, J.M.; Serrano, R. Immunocytolocalization of plasma membrane H-ATPase. Plant Physiol. 1990, 93, 1654–1658. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- Haagen-Smit, A.J.; Went, F.W. A physiological analysis of the growth substance. Proc. K. Ned. Akad. Wet. 1935, 38, 852–857. [Google Scholar]
- Zimmerman, P.W.; Wilcoxon, F. Several chemical growth substances which cause initiation of roots and other responses in plants. Contrib. Boyce Thompson Inst. 1935, 7, 209–229. [Google Scholar]
- Thimann, K.V.; Schneider, C.L. The relative activities of different auxins. Am. J. Bot. 1939, 26, 328–333. [Google Scholar] [CrossRef]
- Leuba, V.; LeTourneau, D. Auxin activity of phenylacetic acid in tissue culture. J. Plant Growth Regul. 1990, 9, 71–76. [Google Scholar] [CrossRef]
- Wightman, F.; Schneider, E.A.; Thimann, K.V. Hormonal factors controlling the initiation and development of lateral roots. Physiol. Plant. 1980, 49, 304–314. [Google Scholar] [CrossRef]
- Burkhead, K.D.; Slininger, P.J.; Schisler, D.A. Biological control bacterium Enterobacter cloacae S11:T:07 (NRRL B-21050) produces the antifungal compound phenylacetic acid in Sabouraud maltose broth culture. Soil Biol. Biochem. 1998, 30, 665–667. [Google Scholar] [CrossRef]
- Hwang, B.K.; Lim, S.W.; Kim, B.S.; Lee, J.Y.; Moon, S.S. Isolation and in vivo and in vitro antifungal activity of phenylacetic acid and sodium phenylacetate from Streptomyces humidus. Appl. Environ. Microbiol. 2001, 67, 3739–3745. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cho, J.Y.; Kuk, J.H.; Moon, J.H.; Cho, J.I.; Kim, Y.C.; Park, K.H. Identification and antimicrobial activity of phenylacetic acid produced by Bacillus licheniformis isolated from fermented soybean, Chungkook-Jang. Curr. Microbiol. 2004, 48, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J.; Horsch, R.B.; Hinchee, M.A.; Hein, M.B.; Hoffmann, N.L. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1987, 1, 86–96. [Google Scholar] [CrossRef]
- Inzé, D.; Follin, A.; Velten, J.; Velten, L.; Prinsen, E.; Rüdelsheim, P.; van Onckelen, H.; Schell, J.; van Montagu, M. The Pseudomonas savastanoi tryptophan-2-mono-oxygenase is biologically active in Nicotiana tabacum. Planta 1987, 172, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Mashiguchi, K.; Chen, Q.; Kasahara, H.; Kamiya, Y.; Ojha, S.; Dubois, J.; Ballou, D.; Zhao, Y. The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
Appendix
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sánchez-Parra, B.; Frerigmann, H.; Alonso, M.-M.P.; Loba, V.C.; Jost, R.; Hentrich, M.; Pollmann, S. Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis. Plants 2014, 3, 324-347. https://doi.org/10.3390/plants3030324
Sánchez-Parra B, Frerigmann H, Alonso M-MP, Loba VC, Jost R, Hentrich M, Pollmann S. Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis. Plants. 2014; 3(3):324-347. https://doi.org/10.3390/plants3030324
Chicago/Turabian StyleSánchez-Parra, Beatriz, Henning Frerigmann, Marta-Marina Pérez Alonso, Víctor Carrasco Loba, Ricarda Jost, Mathias Hentrich, and Stephan Pollmann. 2014. "Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis" Plants 3, no. 3: 324-347. https://doi.org/10.3390/plants3030324





