SmRAV1, an AP2 and B3 Transcription Factor, Positively Regulates Eggplant’s Response to Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Sequence Analysis of SmRAV1
2.2. Expression Profiles of SmRAV1 under Different Stresses
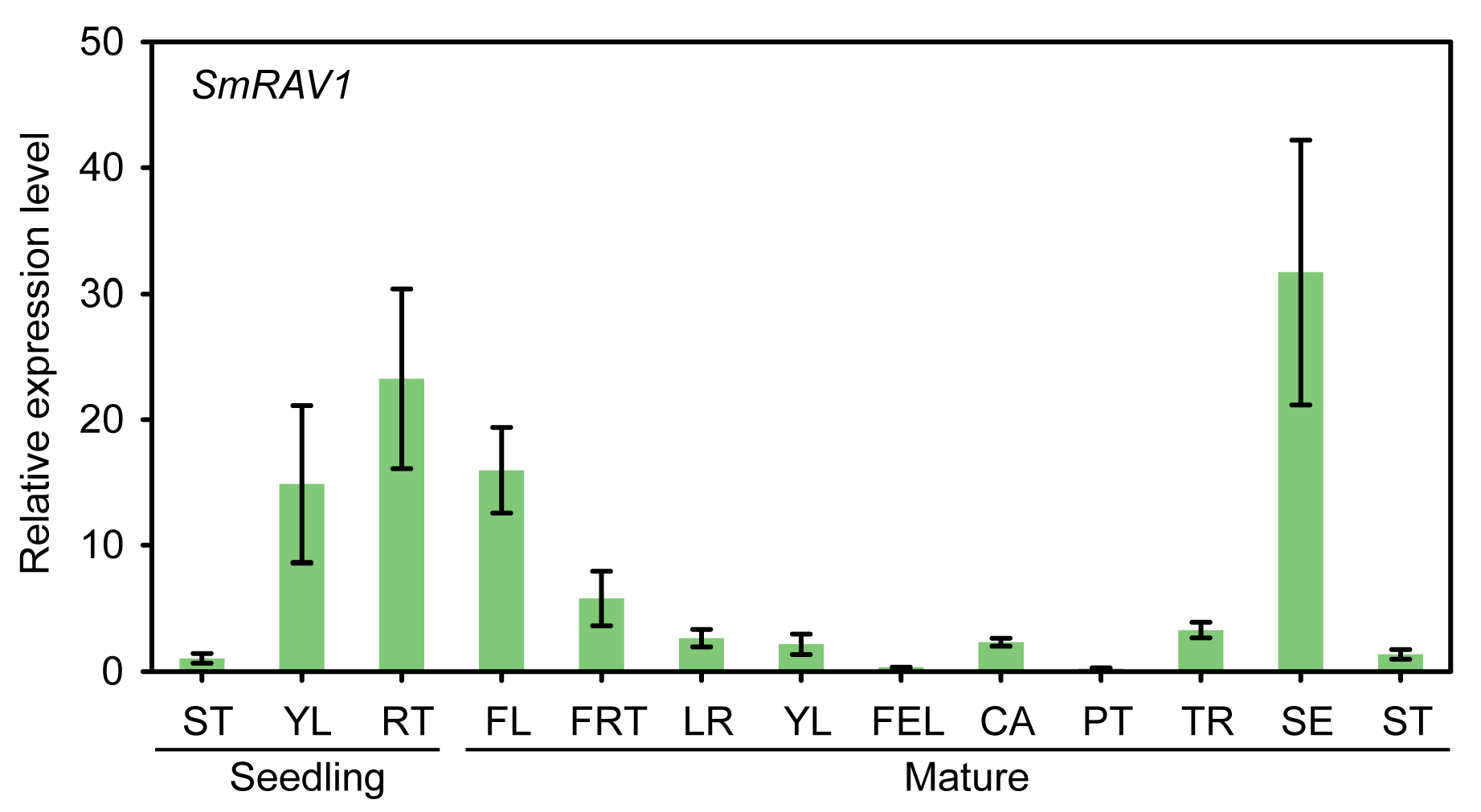
2.3. Analysis of Tissue Specific Expression of SmRAV1
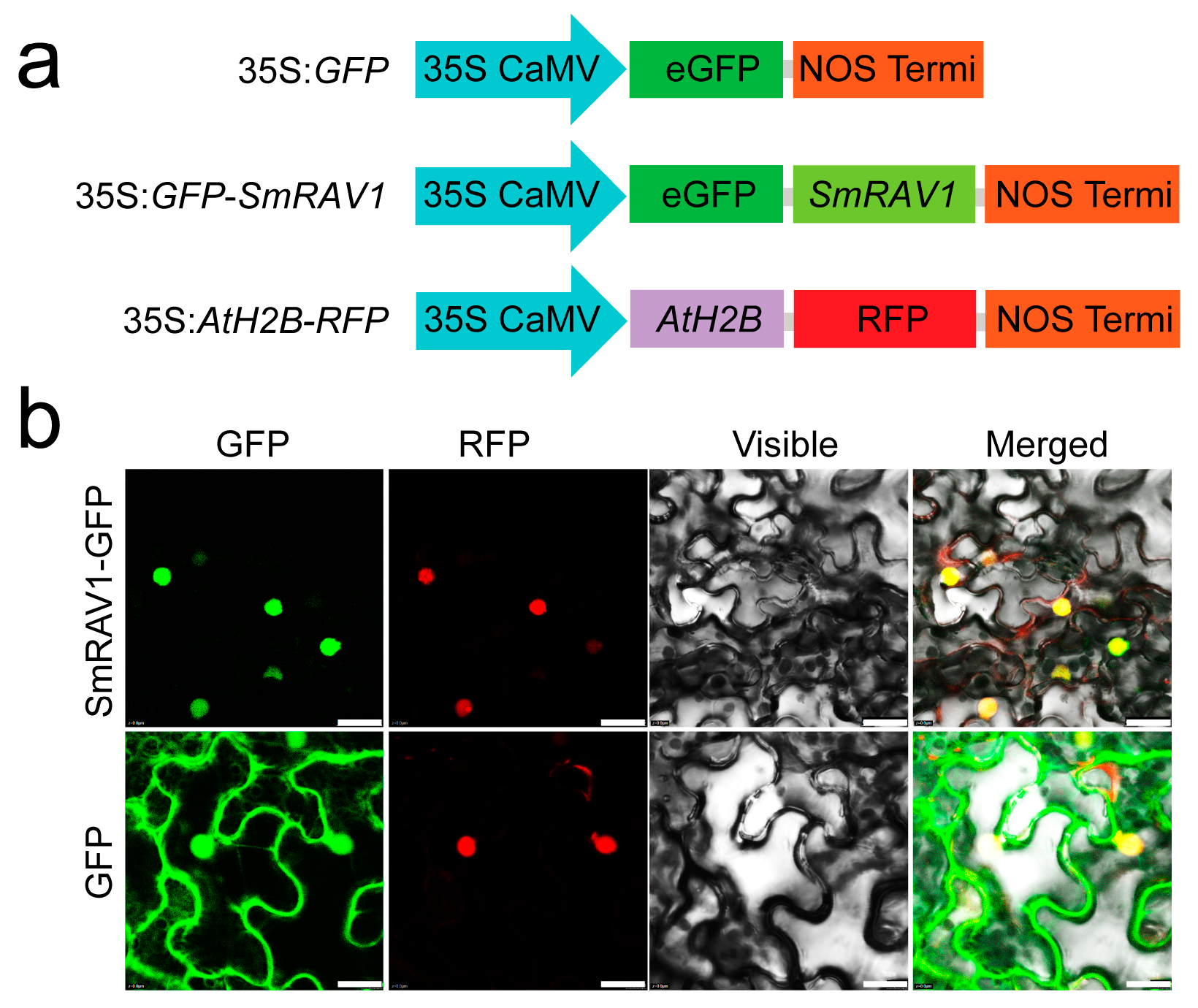
2.4. Subcellular Localization of SmRAV1
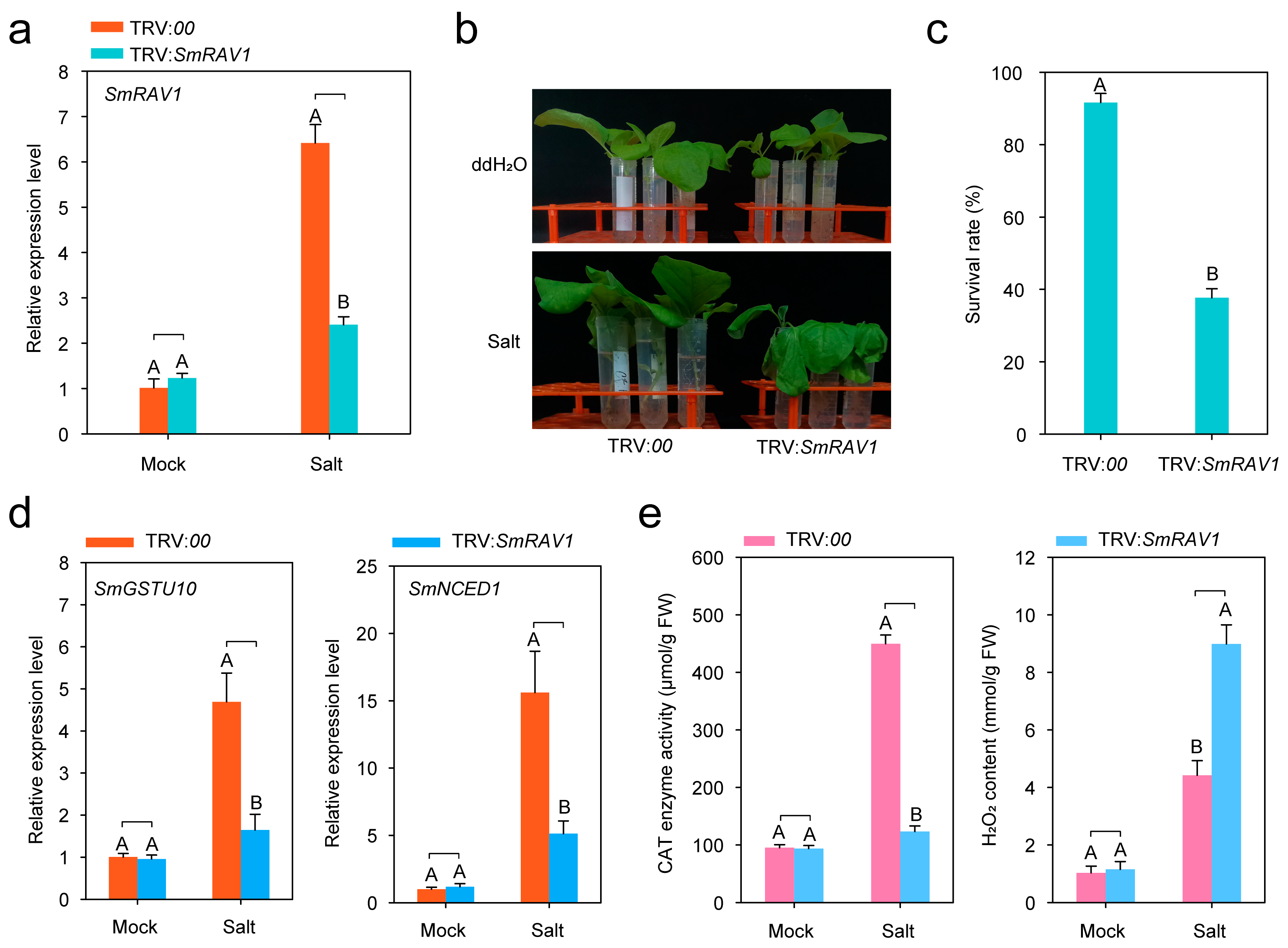
2.5. Silencing of SmRAV1 Enhances Susceptibility of Eggplant against Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material Growth and Conditions
4.2. Abiotic Stresses Treatment
4.3. Plant Total RNA Extraction, cDNA Synthesis, and RT-qPCR Analysis
4.4. Agrobacterium tumefaciens Cultivation and Infiltration
4.5. Subcellular Localization Assay
4.6. VIGS Assay
4.7. Measurement of Physiological Indices
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, Y.; Fricke, W.; Schmidhalter, U. Salinity and the growth of non-halophytic grass leaves: The role of mineral nutrient distribution. Funct. Plant Biol. 2005, 32, 973–985. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrao, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Negrao, S.; Schmockel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Involvement of auxin in growth and stress response of cucumber. Veg. Res. 2022, 2, 13. [Google Scholar] [CrossRef]
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic. Res. 2022, 9, uhab024. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhou, H.; Fan, Y.; Guo, Y.; Zhang, M.; Shabala, S.; Zhao, C.; Lv, C.; Guo, B.; Wang, F.; et al. HvNCX, a prime candidate gene for the novel qualitative locus qS7.1 associated with salinity tolerance in barley. Theor. Appl. Genet. 2023, 136, 9. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Jeridi, M.; Siddiqui, S.; Shah, A.Z.; Ali, S. Genome-wide identification, characterization, and expression analysis of the Chalcone Synthase gene family in Oryza sativa under Abiotic Stresses. Plant Stress 2023, 9, 100201. [Google Scholar] [CrossRef]
- Ahmad, S.; Ali, S.; Shah, A.Z.; Khan, A.; Faria, S. Chalcone synthase (CHS) family genes regulate the growth and response of cucumber (Cucumis sativus L.) to Botrytis cinerea and abiotic stresses. Plant Stress 2023, 8, 100159. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, Q.; Zhang, Z.; Cheng, P.; Song, A.; Zhou, L.; Wang, L.; Chen, S.; Chen, F.; Jiang, J. The RAV transcription factor TEMPRANILLO1 involved in ethylene-mediated delay of chrysanthemum flowering. Plant J. 2023. [Google Scholar] [CrossRef]
- Swaminathan, K.; Peterson, K.; Jack, T. The plant B3 superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef]
- Matias-Hernandez, L.; Aguilar-Jaramillo, A.E.; Marin-Gonzalez, E.; Suarez-Lopez, P.; Pelaz, S. RAV genes: Regulation of floral induction and beyond. Ann. Bot. 2014, 114, 1459–1470. [Google Scholar] [CrossRef]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef]
- Tavares, E.Q.P.; De Souza, A.P.; Romim, G.H.; Grandis, A.; Plasencia, A.; Gaiarsa, J.W.; Grima-Pettenati, J.; de Setta, N.; Van Sluys, M.A.; Buckeridge, M.S. The control of endopolygalacturonase expression by the sugarcane RAV transcription factor during aerenchyma formation. J. Exp. Bot. 2019, 70, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Hao, D.; Chen, L.; Lu, Q.; Zhang, Y.; Li, Y.; Duan, Y.; Li, W. Roles for a soybean RAV-like orthologue in shoot regeneration and photoperiodicity inferred from transgenic plants. J. Exp. Bot. 2012, 63, 3257–3270. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Zhao, L.; Li, D.; Hao, D.; Zhan, Y.; Li, W. A GmRAV ortholog is involved in photoperiod and sucrose control of flowering time in soybean. PLoS ONE 2014, 9, e89145. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, M.; Zhang, L.; Jiang, Y.; Zhao, C.; Shahid, M.Q.; Bai, Y.; Hao, J.; Peng, J.; Gao, Y.; et al. EjRAV1/2 Delay Flowering through Transcriptional Repression of EjFTs and EjSOC1s in Loquat. Front. Plant Sci. 2021, 12, 816086. [Google Scholar] [CrossRef]
- Zhang, K.; Zhao, L.; Yang, X.; Li, M.; Sun, J.; Wang, K.; Li, Y.; Zheng, Y.; Yao, Y.; Li, W. GmRAV1 regulates regeneration of roots and adventitious buds by the cytokinin signaling pathway in Arabidopsis and soybean. Physiol. Plant. 2019, 165, 814–829. [Google Scholar] [CrossRef]
- Fu, M.; Kang, H.K.; Son, S.H.; Kim, S.K.; Nam, K.H. A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 2014, 55, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yan, Y.; Lu, Y.; Liu, G.; Liu, J.; Shi, H. The co-modulation of RAV transcription factors in ROS burst and extensive transcriptional reprogramming underlies disease resistance in cassava. Plant Cell Rep. 2022, 41, 1261–1272. [Google Scholar] [CrossRef]
- Wei, Y.; Chang, Y.; Zeng, H.; Liu, G.; He, C.; Shi, H. RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. J. Pineal Res. 2018, 64, e12454. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; You, S.J.; Hsieh, T.H.; Chao, T.C.; Chan, M.T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011, 156, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.H.; Lee, S.C.; Jung, H.W.; Hong, J.K.; Hwang, B.K. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006, 61, 897–915. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhang, K.; Yang, C. Functional roles of the birch BpRAV1 transcription factor in salt and osmotic stress response. Plant Sci. 2022, 315, 111131. [Google Scholar] [CrossRef]
- Karami, M.; Fatahi, N.; Lohrasebi, T.; Razavi, K. RAV transcription factor regulatory function in response to salt stress in two Iranian wheat landraces. J. Plant Res. 2022, 135, 121–136. [Google Scholar] [CrossRef]
- Osnato, M.; Cereijo, U.; Sala, J.; Matias-Hernandez, L.; Aguilar-Jaramillo, A.E.; Rodriguez-Goberna, M.R.; Riechmann, J.L.; Rodriguez-Concepcion, M.; Pelaz, S. The floral repressors TEMPRANILLO1 and 2 modulate salt tolerance by regulating hormonal components and photo-protection in Arabidopsis. Plant J. 2021, 105, 7–21. [Google Scholar] [CrossRef]
- Gao, Y.; Han, D.; Jia, W.; Ma, X.; Yang, Y.; Xu, Z. Molecular characterization and systematic analysis of NtAP2/ERF in tobacco and functional determination of NtRAV-4 under drought stress. Plant Physiol. Biochem. 2020, 156, 420–435. [Google Scholar] [CrossRef]
- Luo, Y.X.; Chen, S.K.; Wang, P.D.; Peng, D.; Zhang, X.; Li, H.F.; Feng, C.Z. Genome-Wide Analysis of the RAV Gene Family in Wheat and Functional Identification of TaRAV1 in Salt Stress. Int. J. Mol. Sci. 2022, 23, 8834. [Google Scholar] [CrossRef]
- Kabir, N.; Lin, H.; Kong, X.; Liu, L.; Qanmber, G.; Wang, Y.; Zhang, L.; Sun, Z.; Yang, Z.; Yu, Y.; et al. Identification, evolutionary analysis and functional diversification of RAV gene family in cotton (G. hirsutum L.). Planta 2021, 255, 14. [Google Scholar] [CrossRef]
- Liu, J.; Deng, Z.; Liang, C.; Sun, H.; Li, D.; Song, J.; Zhang, S.; Wang, R. Genome-Wide Analysis of RAV Transcription Factors and Functional Characterization of Anthocyanin-Biosynthesis-Related RAV Genes in Pear. Int. J. Mol. Sci. 2021, 22, 5567. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, Y.; Zhang, H.; Ma, Q.; Wei, Z.; Chen, J.; Sun, Z. Genome-Wide Analysis of the RAV Transcription Factor Genes in Rice Reveals Their Response Patterns to Hormones and Virus Infection. Viruses 2021, 13, 752. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.P.; Xu, Z.S.; Zheng, W.J.; Zhao, W.; Wang, Y.X.; Yu, T.F.; Chen, M.; Zhou, Y.B.; Min, D.H.; Ma, Y.Z.; et al. Genome-Wide Analysis of the RAV Family in Soybean and Functional Identification of GmRAV-03 Involvement in Salt and Drought Stresses and Exogenous ABA Treatment. Front. Plant Sci. 2017, 8, 905. [Google Scholar] [CrossRef]
- Zhang, C.H.; Shangguan, L.F.; Ma, R.J.; Sun, X.; Tao, R.; Guo, L.; Korir, N.K.; Yu, M.L. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet. Mol. Res. 2012, 11, 4789–4809. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.; Li, H.; Wang, J.; Zeng, Q.; Huang, W.; Huang, H.; Xie, Y.; Yu, S.; Kan, Q.; et al. GLRaV-2 protein p24 suppresses host defenses by interaction with a RAV transcription factor from grapevine. Plant Physiol. 2022, 189, 1848–1865. [Google Scholar] [CrossRef] [PubMed]
- Yarra, R.; Kirti, P.B. Expressing class I wheat NHX (TaNHX2) gene in eggplant (Solanum melongena L.) improves plant performance under saline condition. Funct. Integr. Genom. 2019, 19, 541–554. [Google Scholar] [CrossRef]
- Shen, L.; Zhao, E.; Liu, R.; Yang, X. Transcriptome Analysis of Eggplant under Salt Stress: AP2/ERF Transcription Factor SmERF1 Acts as a Positive Regulator of Salt Stress. Plants 2022, 11, 2205. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Guo, Y.; Yang, Y.; Huang, Y.; Shen, H.B. Discovering nuclear targeting signal sequence through protein language learning and multivariate analysis. Anal. Biochem. 2020, 591, 113565. [Google Scholar] [CrossRef] [PubMed]
- Launholt, D.; Merkle, T.; Houben, A.; Schulz, A.; Grasser, K.D. Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 2006, 18, 2904–2918. [Google Scholar] [CrossRef]
- Boisnard-Lorig, C.; Colon-Carmona, A.; Bauch, M.; Hodge, S.; Doerner, P.; Bancharel, E.; Dumas, C.; Haseloff, J.; Berger, F. Dynamic analyses of the expression of the HISTONE::YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 2001, 13, 495–509. [Google Scholar] [CrossRef]
- Li, J.; Song, C.; Li, H.; Wang, S.; Hu, L.; Yin, Y.; Wang, Z.; He, W. Comprehensive analysis of cucumber RAV family genes and functional characterization of CsRAV1 in salt and ABA tolerance in cucumber. Front. Plant Sci. 2023, 14, 1115874. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Doll, J. Arabidopsis WRKY53, a Node of Multi-Layer Regulation in the Network of Senescence. Plants 2019, 8, 578. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef]
- Wang, X.; Chen, K.; Zhou, M.; Gao, Y.; Huang, H.; Liu, C.; Fan, Y.; Fan, Z.; Wang, Y.; Li, X. GmNAC181 promotes symbiotic nodulation and salt tolerance of nodulation by directly regulating GmNINa expression in soybean. New Phytol. 2022, 236, 656–670. [Google Scholar] [CrossRef]
- Segarra, G.; Van der Ent, S.; Trillas, I.; Pieterse, C.M. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef]
- Son, G.H.; Wan, J.; Kim, H.J.; Nguyen, X.C.; Chung, W.S.; Hong, J.C.; Stacey, G. Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 2012, 25, 48–60. [Google Scholar] [CrossRef]
- Wang, S.; Guo, T.; Shen, Y.; Wang, Z.; Kang, J.; Zhang, J.; Yi, F.; Yang, Q.; Long, R. Overexpression of MtRAV3 enhances osmotic and salt tolerance and inhibits growth of Medicago truncatula. Plant Physiol. Biochem. 2021, 163, 154–165. [Google Scholar] [CrossRef]
- Li, X.J.; Li, M.; Zhou, Y.; Hu, S.; Hu, R.; Chen, Y.; Li, X.B. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS ONE 2015, 10, e0118056. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fu, J.; Shen, Q.; Wang, Q. OsWRKY10 extensively activates multiple rice diterpenoid phytoalexin biosynthesis genes to enhance rice blast resistance. Plant J. 2023, 115, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Sun, B.; Cai, W.; Zhou, X.; Mao, Y.; Chen, C.; Wei, J.; Cao, B.; Chen, C.; Chen, G.; et al. Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. New Phytol. 2019, 223, 922–938. [Google Scholar] [CrossRef]
- Guo, Q.; Major, I.T.; Kapali, G.; Howe, G.A. MYC transcription factors coordinate tryptophan-dependent defence responses and compromise seed yield in Arabidopsis. New Phytol. 2022, 236, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, J.; Duan, W.; Ma, X.; Qu, L.; Xu, Z.; Yang, Y.; Xu, J. NtRAV4 negatively regulates drought tolerance in Nicotiana tabacum by enhancing antioxidant capacity and defence system. Plant Cell Rep. 2022, 41, 1775–1788. [Google Scholar] [CrossRef]
- Yang, S.; Luo, C.; Song, Y.; Wang, J. Two Groups of Thellungiella salsuginea RAVs Exhibit Distinct Responses and Sensitivity to Salt and ABA in Transgenic Arabidopsis. PLoS ONE 2016, 11, e0153517. [Google Scholar] [CrossRef]
- Zhao, L.; Luo, Q.; Yang, C.; Han, Y.; Li, W. A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 2008, 227, 1389–1399. [Google Scholar] [CrossRef]
- Shen, L.; He, J.; Yang, X. Genome-wide identification of calmodulin-binding protein 60 gene family and function of SmCBP60A1 in eggplant response to salt stress. Sci. Hortic. 2023, 322, 112448. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, J.; Yao, B.; Yang, X.; Shen, L. SmRAV1, an AP2 and B3 Transcription Factor, Positively Regulates Eggplant’s Response to Salt Stress. Plants 2023, 12, 4174. https://doi.org/10.3390/plants12244174
Ding J, Yao B, Yang X, Shen L. SmRAV1, an AP2 and B3 Transcription Factor, Positively Regulates Eggplant’s Response to Salt Stress. Plants. 2023; 12(24):4174. https://doi.org/10.3390/plants12244174
Chicago/Turabian StyleDing, Junjie, Bowen Yao, Xu Yang, and Lei Shen. 2023. "SmRAV1, an AP2 and B3 Transcription Factor, Positively Regulates Eggplant’s Response to Salt Stress" Plants 12, no. 24: 4174. https://doi.org/10.3390/plants12244174




