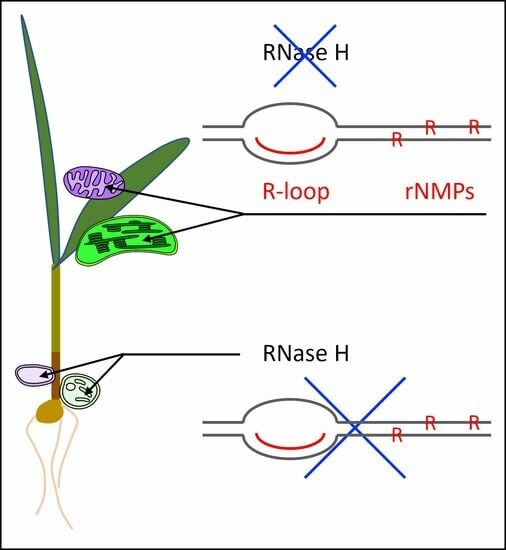
Ribonucleotide and R-Loop Damage in Plastid DNA and Mitochondrial DNA during Maize Development
Abstract
:1. Introduction
- (1)
- During DNA replication, DNA polymerases must distinguish between deoxyribose and ribose in the sugar moiety, as well as choose the correct complimentary base [6]. Misincorporation of ribonucleotides may occur, however, because of the inadequate ability of DNA polymerases to distinguish between these two sugars. The outcome of this misincorporation is typically a single ribonucleotide. Misincorporation rates can be fairly high, even in healthy cells [7,8]. Ribonucleoside monophosphate has been reported as the most common aberrant nucleotide found in DNA: one per 10,000 to 100,000 nucleotides [7,8,9,10,11]. Genome instability may be caused by the reactivity of the 2′-OH in the sugar portion of a ribonucleotide, which increases its susceptibility to strand cleavage [7].
- (2)
- For lagging-strand DNA synthesis, a short stretch of ribonucleotides (an RNA primer) anneals to the template strand and primes replication [12]. As DNA synthesis proceeds, the RNA primer is removed, the resulting gap is filled with deoxynucleotides, and the nascent strand is sealed by DNA ligase. If the RNA primer is not removed, however, it may be deleterious to the organism.
- (3)
- Another type of RNA-DNA structure may be generated during DNA replication, repair, and transcription: an R-loop. R-loops are three-stranded nucleic acid structures containing a displaced DNA strand and an RNA-DNA hybrid (RNA base-paired with its complementary DNA) that influence many biological processes [13,14,15]. In yeast, mammals, and plants, R-loops can occupy approximately 5 to 10% of the nuclear genome [16,17,18], suggesting that they are typically tolerated. Nevertheless, persistent R-loops may lead to DNA replication fork stalling, genome instability [19], DNA damage [20], chromosomal DNA rearrangements, recombination [21], and transcription–replication conflict [22]. The effects of persistent R-loops may result in human diseases, including cancer, neurodegeneration, and inflammatory diseases [15,23,24]. In summary, R-loops can affect genome stability.
2. Materials and Methods
2.1. Plant Tissue and Growth Conditions
2.2. Isolation of Maize Organelles (Plastids and Mitochondria)
2.3. Isolation of Organelle Proteins
2.4. Isolation of Organelle DNA
2.5. Measurement of RNase H Protein Levels Using Slot-Blot Assays
2.6. R-Loop Detection in Organelle DNA
2.7. Ribonucleotide Detection in Organelle DNA
2.8. Statistical Analysis
3. Results
3.1. Changes in the Level of R-Loops in orgDNA during Leaf Development and for Light-Grown Compared to Dark-Grown Plants
3.2. Changes in the Level of rNMPs in orgDNA during Leaf Development and for Light-Grown Compared to Dark-Grown Plants
3.3. Changes in the Level of Organellar RNase H during Leaf Development and for Light-Grown Compared to Dark-Grown Plants
4. Discussion
5. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Oldenburg, D.J.; Bendich, A.J. DNA maintenance in plastids and mitochondria of plants. Front. Plant Sci. 2015, 6, 883. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Oldenburg, D.J.; Bendich, A.J. Oxidative and glycation damage to mitochondrial DNA and plastid DNA during plant development. Antioxidants 2023, 12, 891. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Williams, R.S. Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA Biol. 2014, 11, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Cerritelli, S.M.; Crouch, R.J. The balancing act of ribonucleotides in DNA. Trends Biochem. Sci. 2016, 41, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Nava, G.M.; Grasso, L.; Sertic, S.; Pellicioli, A.; Muzi Falconi, M.; Lazzaro, F. One, no one, and one hundred thousand: The many forms of ribonucleotides in DNA. Int. J. Mol. Sci. 2020, 21, 1706. [Google Scholar] [CrossRef]
- Joyce, C.M. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. USA 1997, 94, 1619–1622. [Google Scholar] [CrossRef] [PubMed]
- Nick McElhinny, S.A.; Kumar, D.; Clark, A.B.; Watt, D.L.; Watts, B.E.; Lundström, E.B.; Johansson, E.; Chabes, A.; Kunkel, T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010, 6, 774–781. [Google Scholar] [CrossRef]
- Clausen, A.R.; Zhang, S.; Burgers, P.M.; Lee, M.Y.; Kunkel, T.A. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair 2013, 12, 121–127. [Google Scholar] [CrossRef]
- Nick McElhinny, S.A.; Watts, B.E.; Kumar, D.; Watt, D.L.; Lundström, E.B.; Burgers, P.M.; Johansson, E.; Chabes, A.; Kunkel, T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA 2010, 107, 4949–4954. [Google Scholar] [CrossRef]
- Reijns, M.A.; Rabe, B.; Rigby, R.E.; Mill, P.; Astell, K.R.; Lettice, L.A.; Boyle, S.; Leitch, A.; Keighren, M.; Kilanowski, F.; et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 2012, 149, 1008–1022. [Google Scholar] [CrossRef]
- Kalhorzadeh, P.; Hu, Z.; Cools, T.; Amiard, S.; Willing, E.M.; De Winne, N.; Gevaert, K.; De Jaeger, G.; Schneeberger, K.; White, C.I.; et al. Arabidopsis thaliana RNase H2 deficiency counteracts the needs for the WEE1 checkpoint kinase but triggers genome instability. Plant Cell 2014, 26, 3680–3692. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, A.; Baker, T.A. DNA Replication, 2nd ed.; Freeman, W.H., Ed.; Nuclear Acid Science Books: New York, NY, USA, 2005. [Google Scholar]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, P.; Li, X.; Wu, W.; Wei, H.; Zhang, W. Toward an understanding of the detection and function of R-loops in plants. J. Exp. Bot. 2021, 72, 6110–6122. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Lan, L.; Zou, L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, dynamic, and conserved R-Loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef]
- Xu, W.; Xu, H.; Li, K.; Fan, Y.; Liu, Y.; Yang, X.; Sun, Q. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 2017, 3, 704–714. [Google Scholar] [CrossRef]
- Fang, Y.; Chen, L.; Lin, K.; Feng, Y.; Zhang, P.; Pan, X.; Sanders, J.; Wu, Y.; Wang, X.E.; Su, Z.; et al. Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice. Genome Res. 2019, 29, 1287–1297. [Google Scholar] [CrossRef]
- Tuduri, S.; Crabbé, L.; Conti, C.; Tourrière, H.; Holtgreve-Grez, H.; Jauch, A.; Pantesco, V.; De Vos, J.; Thomas, A.; Theillet, C.; et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009, 11, 1315–1324. [Google Scholar] [CrossRef]
- Brambati, A.; Zardoni, L.; Nardini, E.; Pellicioli, A.; Liberi, G. The dark side of RNA:DNA hybrids. Mutat. Res. Mol. Mech. Mutagen. 2020, 784, 108300. [Google Scholar] [CrossRef]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef] [PubMed]
- Mannini, L.; Musio, A. The dark side of cohesin: The carcinogenic point of view. Mutat. Res. 2011, 728, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Cimprich, K.A. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015, 25, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Tannous, E.; Kanaya, E.; Kanaya, S. Role of RNase H1 in DNA repair: Removal of single ribonucleotide misincorporated into DNA in collaboration with RNase H2. Sci. Rep. 2015, 5, 9969. [Google Scholar] [CrossRef] [PubMed]
- Holt, I.J. The Jekyll and Hyde character of RNase H1 and its multiple roles in mitochondrial DNA metabolism. DNA Repair 2019, 84, 102630. [Google Scholar] [CrossRef]
- Posse, V.; Al-Behadili, A.; Uhler, J.P.; Clausen, A.R.; Reyes, A.; Zeviani, M.; Falkenberg, M.; Gustafsson, C.M. RNase H1 directs origin-specific initiation of DNA replication in human mitochondria. PLoS Genet. 2019, 15, e1007781. [Google Scholar] [CrossRef]
- Yang, Z.; Hou, Q.; Cheng, L.; Xu, W.; Hong, Y.; Li, S.; Sun, Q. RNase H1 cooperates with DNA Gyrases to restrict R-Loops and maintain genome integrity in Arabidopsis chloroplasts. Plant Cell 2017, 29, 2478–2497. [Google Scholar] [CrossRef]
- Bubeck, D.; Reijns, M.A.; Graham, S.C.; Astell, K.R.; Jones, E.Y.; Jackson, A.P. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011, 39, 3652–3666. [Google Scholar] [CrossRef]
- Kojima, K.; Baba, M.; Tsukiashi, M.; Nishimura, T.; Yasukawa, K. RNA/DNA structures recognized by RNase H2. Brief. Funct. Genom. 2018, 18, 169–173. [Google Scholar] [CrossRef]
- Kuciński, J.; Chamera, S.; Kmera, A.; Rowley, M.J.; Fujii, S.; Khurana, P.; Nowotny, M.; Wierzbicki, A.T. Evolutionary history and activity of RNase H1-like proteins in Arabidopsis thaliana. Plant Cell Physiol. 2020, 61, 1107–1119. [Google Scholar] [CrossRef]
- Yang, Z.; Li, M.; Sun, Q. RHON1 Co-transcriptionally resolves R-loops for Arabidopsis chloroplast genome maintenance. Cell Rep. 2020, 30, 243–256.e245. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, K.; Yang, Z.; Hou, Q.; Zhao, W.W.; Sun, Q. RNase H1C collaborates with ssDNA binding proteins WHY1/3 and recombinase RecA1 to fulfill the DNA damage repair in Arabidopsis chloroplasts. Nucleic Acids Res. 2021, 49, 6771–6787. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Nam, A.; Oldenburg, D.J.; Bendich, A.J. Reactive oxygen species, antioxidant agents, and DNA damage in developing maize mitochondria and plastids. Front. Plant Sci. 2020, 11, 596. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Oldenburg, D.J.; Bendich, A.J. Glycation damage to organelles and their DNA increases during maize seedling development. Sci. Rep. 2022, 12, 2688. [Google Scholar] [CrossRef]
- Witzel, K.; Shahzad, M.; Matros, A.; Mock, H.-P.; Mühling, K.H. Comparative evaluation of extraction methods for apoplastic proteins from maize leaves. Plant Methods 2011, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef]
- Ramirez, P.; Crouch, R.J.; Cheung, V.G.; Grunseich, C. R-Loop analysis by dot-blot. J. Vis. Exp. 2021, 167, e62069. [Google Scholar] [CrossRef]
- Meroni, A.; Nava, G.M.; Sertic, S.; Plevani, P.; Muzi-Falconi, M.; Lazzaro, F. measuring the levels of ribonucleotides embedded in genomic DNA. Methods Mol. Biol. 2018, 1672, 319–327. [Google Scholar] [CrossRef]
- Pham, P.; Shao, Y.; Cox, M.M.; Goodman, M.F. Genomic landscape of single-stranded DNA gapped intermediates in Escherichia coli. Nucleic Acids Res. 2022, 50, 937–951. [Google Scholar] [CrossRef]
- Sylvester, A.W.; Cande, W.Z.; Freeling, M. Division and differentiation during normal and liguleless-1 maize leaf development. Development 1990, 110, 985–1000. [Google Scholar] [CrossRef]
- Stern, D.B.; Hanson, M.R.; Barkan, A. Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci. 2004, 9, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, D.J.; Bendich, A.J. Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J. Mol. Biol. 2004, 344, 1311–1330. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, D.J.; Rowan, B.A.; Zhao, L.; Walcher, C.L.; Schleh, M.; Bendich, A.J. Loss or retention of chloroplast DNA in maize seedlings is affected by both light and genotype. Planta 2006, 225, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, D.J.; Kumar, R.A.; Bendich, A.J. The amount and integrity of mtDNA in maize decline with development. Planta 2013, 237, 603–617. [Google Scholar] [CrossRef]
- Zheng, Q.; Oldenburg, D.J.; Bendich, A.J. Independent effects of leaf growth and light on the development of the plastid and its DNA content in Zea species. J. Exp. Bot. 2011, 62, 2715–2730. [Google Scholar] [CrossRef]
- Smolka, J.A.; Sanz, L.A.; Hartono, S.R.; Chédin, F. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J. Cell Biol. 2021, 220, e202004079. [Google Scholar] [CrossRef]
- Kumar, R.A.; Oldenburg, D.J.; Bendich, A.J. Changes in DNA damage, molecular integrity, and copy number for plastid DNA and mitochondrial DNA during maize development. J. Exp. Bot. 2014, 65, 6425–6439. [Google Scholar] [CrossRef]
- Qiu, Z.; Zhu, L.; He, L.; Chen, D.; Zeng, D.; Chen, G.; Hu, J.; Zhang, G.; Ren, D.; Dong, G.; et al. DNA damage and reactive oxygen species cause cell death in the rice local lesions 1 mutant under high light and high temperature. New Phytol. 2019, 222, 349–365. [Google Scholar] [CrossRef]
- Xu, W.; Li, K.; Li, S.; Hou, Q.; Zhang, Y.; Liu, K.; Sun, Q. The R-Loop atlas of Arabidopsis development and responses to environmental stimuli. Plant Cell 2020, 32, 888–903. [Google Scholar] [CrossRef]
- Rinaldi, C.; Pizzul, P.; Longhese, M.P.; Bonetti, D. Sensing R-loop-associated DNA damage to safeguard genome stability. Front Cell Dev. Biol. 2020, 8, 618157. [Google Scholar] [CrossRef]
- Teng, Y.; Yadav, T.; Duan, M.; Tan, J.; Xiang, Y.; Gao, B.; Xu, J.; Liang, Z.; Liu, Y.; Nakajima, S.; et al. ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat. Commun. 2018, 9, 4115. [Google Scholar] [CrossRef] [PubMed]
- Renaudin, X.; Lee, M.; Shehata, M.; Surmann, E.M.; Venkitaraman, A.R. BRCA2 deficiency reveals that oxidative stress impairs RNaseH1 function to cripple mitochondrial DNA maintenance. Cell Rep. 2021, 36, 109478. [Google Scholar] [CrossRef] [PubMed]
- Renaudin, X.; Venkitaraman, A.R. A mitochondrial response to oxidative stress mediated by unscheduled RNA-DNA hybrids (R-loops). Mol. Cell. Oncol. 2021, 8, 2007028. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A. DNA replication fidelity. J. Biol. Chem. 2004, 279, 16895–16898. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Gao, Y. Translesion and repair DNA polymerases: Diverse structure and mechanism. Annu. Rev. Biochem. 2018, 87, 239–261. [Google Scholar] [CrossRef]
- Göksenin, A.Y.; Zahurancik, W.; LeCompte, K.G.; Taggart, D.J.; Suo, Z.; Pursell, Z.F. Human DNA polymerase ε is able to efficiently extend from multiple consecutive ribonucleotides. J. Biol. Chem. 2012, 287, 42675–42684. [Google Scholar] [CrossRef]
- Lazzaro, F.; Novarina, D.; Amara, F.; Watt, D.L.; Stone, J.E.; Costanzo, V.; Burgers, P.M.; Kunkel, T.A.; Plevani, P.; Muzi-Falconi, M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell 2012, 45, 99–110. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, W.; Yao, Y.; Sun, Q. Mitochondrial RNase H1 activity regulates R-loop homeostasis to maintain genome integrity and enable early embryogenesis in Arabidopsis. PLoS Biol. 2021, 19, e3001357. [Google Scholar] [CrossRef]
- Hao, Z.; Gowder, M.; Proshkin, S.; Bharati, B.K.; Epshtein, V.; Svetlov, V.; Shamovsky, I.; Nudler, E. RNA polymerase drives ribonucleotide excision DNA repair in E. coli. Cell 2023, 186, 2425–2437.e2421. [Google Scholar] [CrossRef]
- Klein, H.L. Genome instabilities arising from ribonucleotides in DNA. DNA Repair 2017, 56, 26–32. [Google Scholar] [CrossRef]
- Misra, S.; Bennett, J.; Friew, Y.N.; Abdulghani, J.; Irvin-Wilson, C.V.; Tripathi, M.K.; Williams, S.; Chaudhuri, M.; Chaudhuri, G. A type II ribonuclease H from Leishmania mitochondria: An enzyme essential for the growth of the parasite. Mol. Biochem. Parasitol. 2005, 143, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E. Transcription-coupled global genomic repair in E. coli. Trends Biochem. Sci. 2023, in press. [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripathi, D.; Oldenburg, D.J.; Bendich, A.J. Ribonucleotide and R-Loop Damage in Plastid DNA and Mitochondrial DNA during Maize Development. Plants 2023, 12, 3161. https://doi.org/10.3390/plants12173161
Tripathi D, Oldenburg DJ, Bendich AJ. Ribonucleotide and R-Loop Damage in Plastid DNA and Mitochondrial DNA during Maize Development. Plants. 2023; 12(17):3161. https://doi.org/10.3390/plants12173161
Chicago/Turabian StyleTripathi, Diwaker, Delene J. Oldenburg, and Arnold J. Bendich. 2023. "Ribonucleotide and R-Loop Damage in Plastid DNA and Mitochondrial DNA during Maize Development" Plants 12, no. 17: 3161. https://doi.org/10.3390/plants12173161