Insights into Ergosterol Peroxide’s Trypanocidal Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Structure Preparation
2.2. Inverse Docking
2.3. Preliminary Surface Plasmon Resonance Experimental Procedures
2.4. CYP51 Protein Expression Trypanosoma cruzi and Candida albicans
2.5. Mushroom Strain
2.6. General Procedures
2.7. Isolation and Purification of Ergosterol Peroxide
2.8. Parasite Cultures
2.9. Carbonylation Activity in Extracellular Epimastigotes
2.10. Electron Microscopy Assay
2.11. Confocal Imaging
2.12. Transmission Electron Microscopy
2.13. Candida albicans Viability Assay
2.14. Analysis of Intermolecular SPR Interactions
3. Results
3.1. Ergosterol Peroxide Interacts with Tc Sterols and Does Not Target Tc Glucose Metabolism
3.2. CYP51 Interacts with Ergosterol Peroxide in Tc but Not in Candida albicans
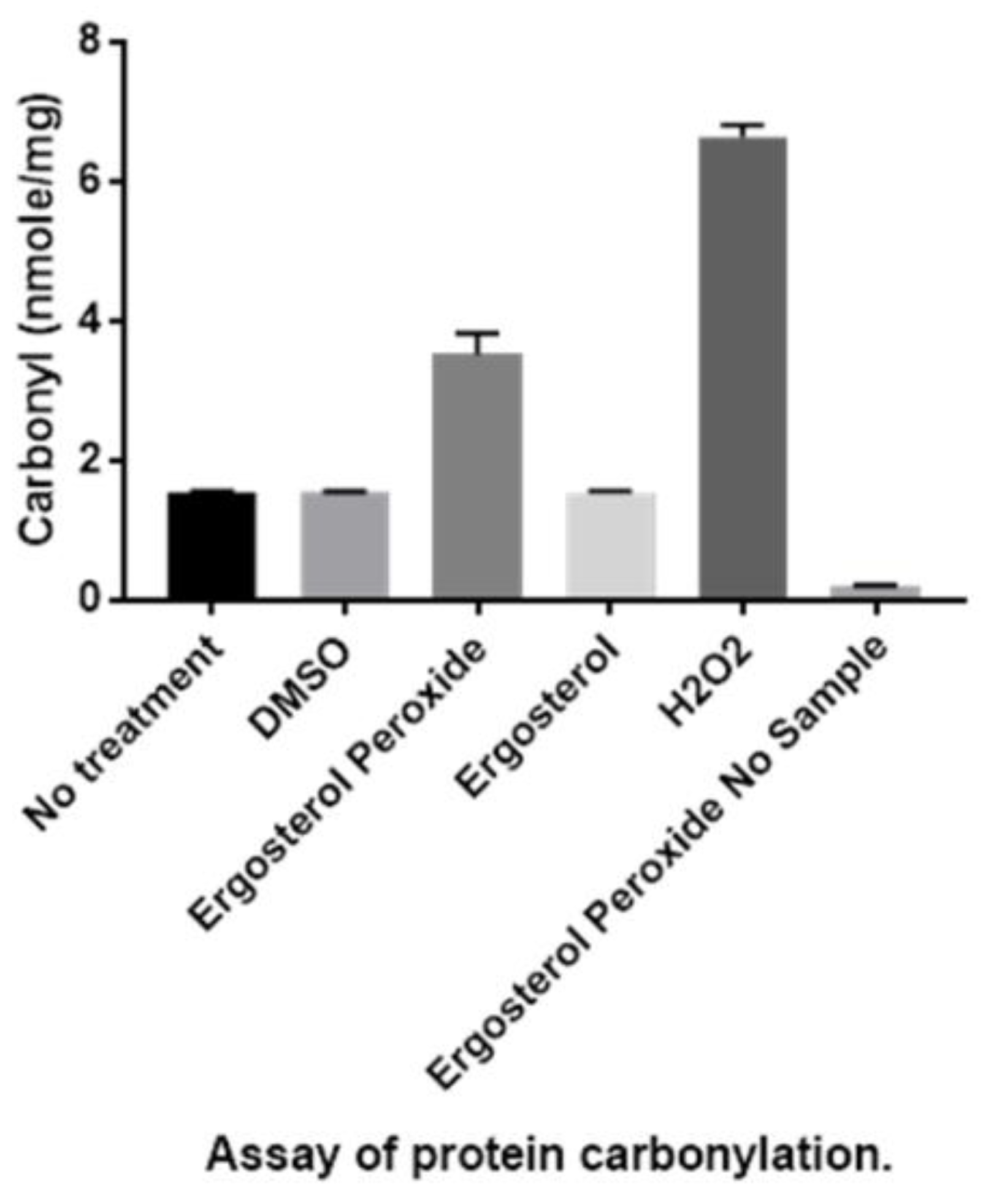
3.3. Detection of Protein Carbonylation Assay
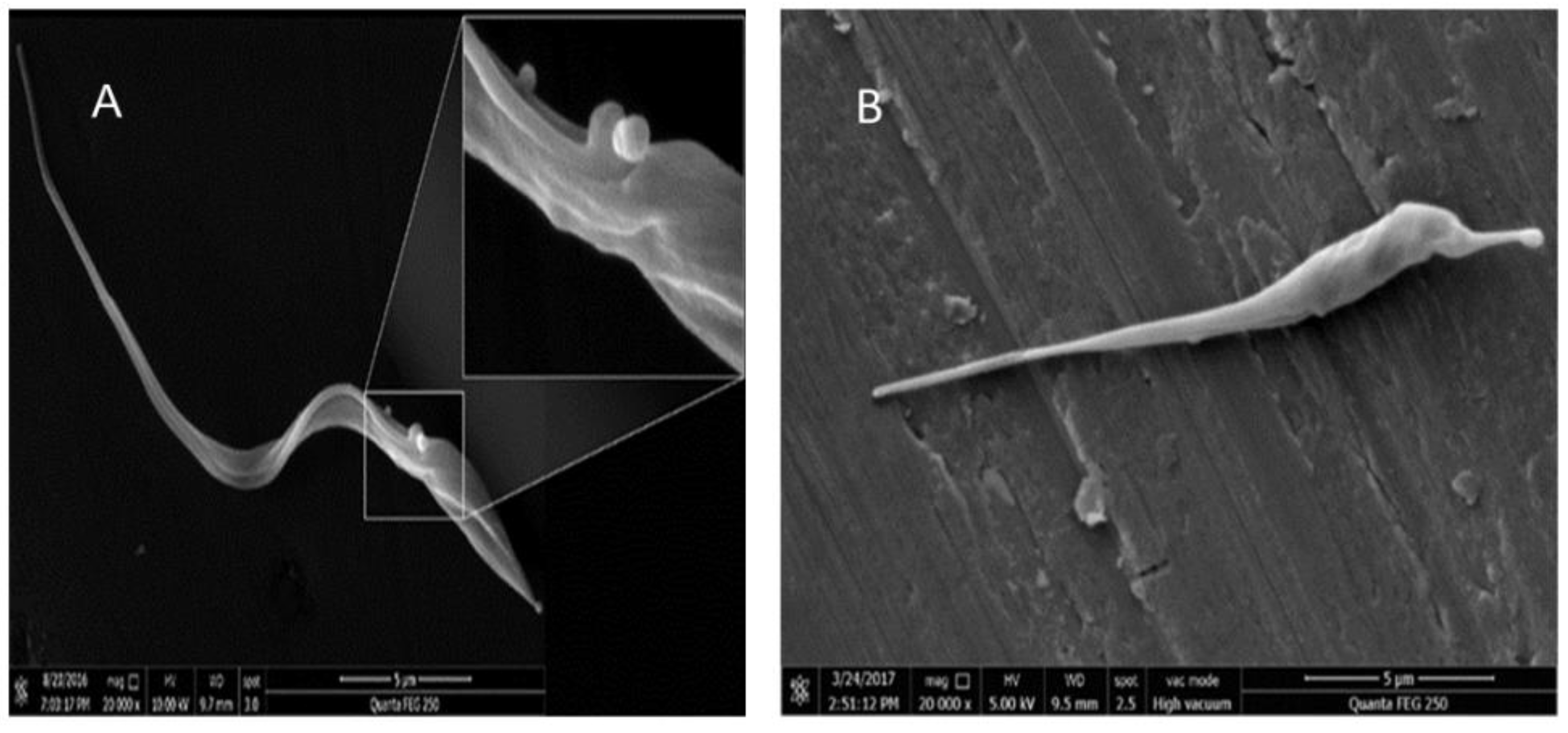
3.4. Detection of Enlarged Extracellular Vesicle Structures after Ergosterol Peroxide Treatment
3.5. Ergosterol Peroxide Disruption of the Cytoplasmic and Nuclear Membranes
3.6. Ergosterol Peroxide Disruption of the Golgi Apparatus and the Cytoplasmic and Nuclear Membranes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rassi, A., Jr.; Rassi, A.; Marín, J.A. Chagas Disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Aufderheidea, A.C.; Saloa, W.; Maddena, M.; Streitzc, J.; Buikstrad, J.; Guhle, F.; Arriaza, B.; Renier, C.; Wittmers, L.E., Jr.; Fornaciari, G.; et al. 9000-year record of Chagas’ disease. Proc. Natl. Acad. Sci. USA 2004, 101, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- Guilbert, J.J. The world health report 2002 reducing risks, promoting healthy life. Educ. Health 2003, 16, 230. [Google Scholar]
- Urbina, J.A.; Docampo, R. Specific chemotherapy of Chagas disease: Controversies and advances. Trends Parasitol. 2003, 19, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Vinhaes, M.C.; Schofield, C.J. Trypanosomiasis control: Surmounting diminishing returns. Trends Parasitol. 2003, 19, 112–113. [Google Scholar] [CrossRef]
- PAHO. 2006. Available online: https://www.paho.org/hq/index.php?option=com_topics&view=rdmore&cid=5507&item=neglected (accessed on 12 March 2018).
- Schmunis, G.A.; Yadon, Z.E. Chagas’ disease: A Latin American health problem becoming a world health problem. Acta Trop. 2010, 115, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Sustainable Development Department. The Millennium Development Goals in Latin America and the Caribbean: Progress, Priorities and IDB Support for Their Implementation; Inter-American Development Bank: Washington, DC, USA, 2005. [Google Scholar]
- Teixeira, A.R.; Nitz, N.; Guimaro, M.C.; Gomes, C.; Santos-Buch, C.A. Chagas disease. Postgrad. Med. J. 2006, 82, 788–798. [Google Scholar] [CrossRef]
- Howard, E.J.; Xiong, X.; Carlier, Y.; Sosa, S.; Buekens, P. Frequency of the congenital transmission of Trypanosoma Cruzi: A systematic review and meta-analysis. BJOG 2014, 121, 22–33. [Google Scholar] [CrossRef]
- Coura, J.; De Castro, S.L. A critical review in Chagas disease chemotherapy. Memórias do Instituto Oswaldo Cruz 2002, 97, 3–24. [Google Scholar] [CrossRef]
- Nagel, R.; Nepomnaschy, I. Mutagenicity of 2 antichagasic drugs and their metabolic deactivation. Mut. Res. 1983, 117, 237–242. [Google Scholar] [CrossRef]
- Maya, J.D.; Cassels, B.K.; Iturriaga, P.; Ferreira, J.; Faúndez, M.; Galanti, N.; Ferreira, A.; Morello, A. Mode of action of natural and synthetic drugs against Trypanosoma cruzi and their interaction with the mammalian host. Comp. Bichem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 601–620. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, C.A.; Malani, A.N.; Easley, C.; Kirkpatrick, P. Posaconazole. Nat. Rev. Drug Discov. 2007, 6, 183–184. [Google Scholar] [CrossRef] [PubMed]
- Boglione-Kerrien, C.; Picard, S.; Tron, C.; Nimubona, S.; Gangneux, J.P.; Lalanne, S.; Lemaitre, F.; Bellissant, E.; Verdier, M.C.; Petitcollin, A. Safety study and therapeutic drug monitoring of the oral tablet formulation of posaconazole in patients with haematological malignancies. J. Cancer Res. Clin. Oncol. 2007, 144, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.B.; Tiwari, V.K. Chemistry Natural products: An evolving role in future drug discovery. Eur. J. Med. 2011, 46, 4769–4807. [Google Scholar] [CrossRef] [PubMed]
- Youyou, T. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar]
- Yasukawa, K.; Akihisa, T.; Kanno, H.; Kaminaga, T.; Izumida, M.; Sakoh, T.; Tamura, T.; Takido, M. Inhibitory effects of sterols isolated from Chlorella vulgaris on 12-Otetradecanoylphorbol-13-acetate-induced inflammation and tumor promotion in mouse skin. Bio. Pharm. Bull. 1996, 19, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Murata, H.; Inatomi, Y.; Inada, A.; Murata, J.; Lang, F.A.; Yamasaki, K.; Nakano, M.; Kawahata, T.; Mori, H.; et al. Screening of anti-HIV-1 activity of North American plants. Anti-HIV-1 activities of plant extracts, and active components of Lethalia vulpine (L.). Hue. J. Nat. Med. 1988, 52, 521–526. [Google Scholar]
- Takahashi, R.; Tanaka, O.; Shibata, S. Ergosterol peroxide from Peltigera aphthosa and Peltigera dolichorrhiza. Phytochemistry 1972, 11, 1850. [Google Scholar] [CrossRef]
- Jimenez, C.; Quinoa, E.; Rignera, R.; Vilalta, R.; Quintella, J.M. The dietary origin of epidioxy steroids in Actinia equina. A carbon-14 incorporation experiment. J. Nat. Prod. 1989, 52, 619–622. [Google Scholar] [CrossRef]
- Deghrigue, M.C.; Festa, C.; Ghribi, L.; D’auria, M.V.; de Marino, S.; Jannet, H.; Ben Said, R.; Bouraoui, A. Pharmacological evaluation of the semipurified fractions from the soft coral Eunicella singularis and isolation of pure compounds. J. Pharma. Sci. 2014, 22, 64. [Google Scholar]
- Tchouankeu, J.C.; Nyasse, B.; Tsamo, E.; Sondengam, B.; Morin, C. An ergostane derivative from the bark of Entandrophragma Utile. Phytochem. 1992, 31, 704–705. [Google Scholar] [CrossRef]
- Wu, Q.P.; Xie, Y.Z.; Deng, Z.; Li, X.M.; Yang, W.; Jiao, C.W.; Fang, L.; Li, S.Z.; Pan, H.H.; Yee, A.J.; et al. Ergosterol peroxide isolated from Ganoderma lucidum abolishes microRNA miR-378-mediated tumor cells on chemoresistance. PLoS ONE 2012, 7, e44579. [Google Scholar] [CrossRef] [PubMed]
- Sgarbi, D.B.; Da Silva, A.J.; Carlos, I.Z.; Silva, C.L.; Angluster, J.; Alviano, C.S. Isolation of ergosterol peroxide and its reversion to ergosterol in the pathogenic fungus Sporothrix Schenckii. Mycopathologia 1997, 139, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Ligonio, A.; López, A.; Trigos, Á. Trypanocidal activity of ergosterol peroxide from Pleurotus ostreatus. Phytother. Res. 2012, 26, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.S.; Jo, Y.S.; Kim, Y.H.; Hyun, J.W.; Kim, H.W. Cytotoxic activities of acetoxyscirpenediol and ergosterol peroxide from Paecilomyces Tenuipes. Life Sci. 2001, 69, 229–237. [Google Scholar] [CrossRef]
- Gao, J.; Wang, M.; Liu, L.; Wei, G.H.; Zhang, A.L.; Draghici, C.; Konishi, Y. Ergosterol peroxides as phospholipase A2 inhibitors from the fungus Lactarius Hatsudake. Phytomedicine 2007, 14, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Trigos, Á.; Suárez, J. Biologically active metabolites of the genus Ganoderma: Three decades of myco-chemistry research. Rev. Mex. Micol. 2011, 34, 63–83. [Google Scholar]
- Ma, L.; Chen, H.; Dong, P.; Lu, X. Anti-inflammatory and anticancer activities of extracts and compounds from the mushroom Inonotus Obliquus. Food Chem. 2013, 139, 503–508. [Google Scholar] [CrossRef]
- Kim, D.H.; Jung, S.J.; Chung, I.S.; Lee, Y.H.; Kim, D.K.; Kim, S.H.; Kwon, B.M.; Jeong, T.S.; Park, M.H.; Seoung, N.S.; et al. Ergosterol peroxide from flowers of Erigeron annuus L. as an anti-atherosclerosis agent. Arch. Pharm. Res. 2005, 28, 541–545. [Google Scholar] [CrossRef]
- Kobori, M.; Yoshida, M.; Ohnishi-Kameyama, M.; Shinmoto, H. Ergosterol peroxide from an edible mushroom suppresses inflammatory responses in RAW264.7 macrophages and growth of HT29 colon adenocarcinoma cells. Br. J. Pharmacol. 2007, 150, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Lindequist, U.; Lesnau, A.; Teuscher, E.; Pilgrim, H. Antiviral activity of ergosterol peroxide. Pharm 1989, 44, 579–580. [Google Scholar]
- Duarte, N.; Ferreira, M.J.; Martins, M.; Viveiros, M.; Amaral, L. Antibacterial activity of ergosterol peroxide against Mycobacterium tuberculosis: Dependence upon system and medium employed. Phytother. Res. 2007, 21, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Jang, J.E.; Mishra, S.K.; Lee, H.J.; Nho, C.W.; Shin, D.; Jin, M.; Kim, M.K.; Choi, C.; Oh, S.H. Ergosterol peroxide from Chaga mushroom (Inonotus obliquus) exhibits anti-cancer activity by down-regulation of the β-catenin pathway in colorectal cancer. J. Ethnopharmacol. 2015, 173, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Meza-Menchaca, T.; Suárez-Medellín, J.; Del Ángel-Piña, C.; Trigos, Á. The Amoebicidal Effect of Ergosterol Peroxide Isolated from Pleurotus Ostreatus. Phyto. Res. 2015, 19, 1982–1986. [Google Scholar] [CrossRef] [PubMed]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameter, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theo. Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Morris, G.; Huey, R.; Lindstrom, W.; Sanner, M.; Belew, R.; Goodsell, D.; Olson, A. Autodock4 and AutodockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Barnes, H.J.; Arlotto, M.P.; Waterman, M.R. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. USA 1991, 88, 5597–5601. [Google Scholar] [CrossRef]
- Chen, C.K.; Leung, S.S.; Guilbert, C.; Jacobson, M.P.; McKerrow, J.H.; Podust, L.M. Structural Characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei Bound to the Antifungal Drugs Posaconazole and Fluconazole. Pharm. Bull. 1998, 5, 444–448. [Google Scholar] [CrossRef]
- Shkel, T.V.; Vasilevskaya, A.V.; Gilep, A.A.; Usanov, S.A.; Chernovetsky, M.A.; Lukyanea’ko, I.G. Molecular cloning, expression, purification and ligand-binding properties of CYP51 from Candida Albicans. Doklady Nat. Akad. Nauk. Belaru. 2017, 56, 64–71. [Google Scholar]
- Martinez-Carrera, D.; Morales, P.; Sobal, M. Cultivation of Several Mexican Strains of Pleurotus ostreatus on coffe pulp and Barley Straw. Rev. Mex. Micol. 1988, 4, 153–160. [Google Scholar]
- Camargo, E.P. Growth and differentiation in Trypasoma cruzi I. Origin in metacyclic tripanosomes in liquid media. Rev. Inst. Med. Trop. Sao Paulo 1964, 6, 93–100. [Google Scholar]
- Hartley, D.; Jroll, D.; Petersen, D. Prooxidant-initiated lipid peroxidation in isolated rat hepatocytes: Detection of 4-hydroxynonenal and malondialdehyde-protein adducts. Chem. Res. Toxicol. 1997, 10, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-García, L.F.; Segura-Valdez, M.L. Visualizing nuclear structure in situ by atomic force microscopy. Atomic Force Microscopy: Methods and Protocols in Biomedical Applications Methods IN Molecular Medicine; Braga, P.C., Ricci, D., Eds.; Humana Press: Totowa, NJ, USA, 2008; pp. 191–199. [Google Scholar]
- Noble, S.M.; Johnson, A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Euk. Cell. 2005, 4, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Bu, M.; Tingting, C.; Hongxia, L.; Mingzhou, G.; Burton, B.Y.; Chengchu, Z.; Yue, Z.; Na, Z.; Liming, H. Synthesis and biological evaluation of novel steroidal 5α,8α-epidioxyandrost-6-ene-3β-ol-17-(O-phenylacetamide) oxime derivatives as potential anticancer agents. Bioorg. Med. Chem. Lett. 2017, 27, 3856–3861. [Google Scholar] [CrossRef]
- Rosa, L.H.; Machado, K.M.; Rabello, A.L.; Souza-Fagundes, E.M.; Correa-Oliveira, R.; Rosa, C.A.; Zani, C.L. Cytotoxic, immunosuppressive, trypanocidal and antileishmanial activities of Basidiomycota fungi present in Atlantic rainforest in Brazil. Antonie Van Leeuwenhoek 2009, 95, 227–237. [Google Scholar] [CrossRef]
- Tatiana, R.A.; Marta, L.L.; Mariana, K.G.; Juliana, T.M.; Matilia, A.D.; Augusto, L.D.; Patricia, S.; Daniel, C.P.; Andre, G.T. Ergosterol isolated from the basidiomycete Pleurotus salmoneostramineus affects Trypanosoma cruzi plasma membrane and mitochondria. J. Venom. Anim. Toxi. Trop. Dis. 2017, 23, 30. [Google Scholar]
- Frédéric-Antoine, D.; Mélanie, B.; Nicolas, L.; Denis, D.; Pierrette, C.; Florian, L.; Sylvie, D.; Philippe, V.; Derrick, R.R. Trypanosoma brucei CYP51: Essentiality and Targeting Therapy in an Experimental Model. PLoS Negl. Trop. Dis. 2016, 11, e0005328. [Google Scholar]
- Abe, F.; Usui, K.; Hiraki, T. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces Cerevisiae. Biochemistry 2009, 48, 8494–8504. [Google Scholar] [CrossRef]
- Daum, G.; Lees, N.D.; Bard, M.; Dickson, R. Biochemistry, Cell Biology and Molecular Biology of Lipids of Saccharomyces Cervisiae. Yeast 1998, 14, 1471–1510. [Google Scholar] [CrossRef]
- Hawkins, C.L.; Morgan, P.E.; Davies, M.J. Quantification of protein modification by oxidants. Free Radic. Biol. Med. 2009, 46, 965–988. [Google Scholar] [CrossRef] [PubMed]
- Rahmanto, A.S.; Morgan, P.E.; Hawkins, C.L.; Davies, M.J. Cellular effects of peptide and protein hydroperoxides. Free Radic. Biol. Med. 2010, 48, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Correa, J. Trypanosoma cruzi Dihydrolipoamide Dehydrogenase as Target for Phenothiazine Cationic Radicals. Effect of Antioxidants. Curr. Drug Targ. 2006, 7, 1155–1179. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.L.; Sahai, E.A.; Yeo, M.; Bosch, M.; Dewart, A.; Olson, M.F. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 2001, 3, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Pyrshev, K.A.; Yesylevskyy, S.O.; Mély, Y.; Demchenko, A.P.; Klymchenko, A.S. Caspase-3 activation decreases lipid order in the outer plasma membrane leaflet during apoptosis: A fluorescent probe study. Biochim. Biophys. Acta Biomem. 2017, 1859, 2123–2132. [Google Scholar] [CrossRef]
- Seliskar, M.; Rozman, D. Mammalian cytochromes P450-importance of tissue specificity. Biochim. Biophys. Acta 2007, 1770, 458–466. [Google Scholar] [CrossRef]
- Lepesheva, G.; Waterman, M. Sterol 14alpha-Demethylase (CYP51) as a Therapeutic Target for Human Trypanosomiasis and Leishmaniasis. Curr. Top. Med. Chem. 2011, 11, 2060–2071. [Google Scholar] [CrossRef]
- Hong, J.; Dai, J.; Jin, G. Mechanism of Tachyplesin I Injury to Bacterial Membranes and Intracellular Enzymes, Determined by Laser Confocal Scanning Microscopy and Flow Citometry. Microbiol. Res. 2014, 170, 69–77. [Google Scholar] [CrossRef]
- Marcilla, A.; Martín-Jaular, L.; Trelis, M.; de Menezes-Neto, A.; Osuna, A.; Bernal, D.; Fernandez-Becerra, C.; Almeida, I.C.; del Portillo, H.A. Extracellular Vesicles in Parasitic Diseases. J. Extracell. Ves. 2014, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.H.; Soares, R.P.; Ribeiro, K.; Andrade, A.C.; Batista, W.L.; Torrecilhas, A.C. Extracellular vesicles: Role in inflammatory responses and potential uses in vaccination in cancer and infectious diseases. J. Immunol. Res. 2015, 2015, 832057. [Google Scholar] [CrossRef] [PubMed]
- Mizushina, Y.; Watanabe, I.; Togashi, H.; Hanashima, L.; Takemura, M.; Ohta, K. An Ergosterol Peroxide, a Natural Product that Selectively Enhances the Inhibitory Effect of Linoleic Acid on DNA Polymerase β. Biol. Pharm. Bull. 1998, 5, 444–448. [Google Scholar] [CrossRef] [PubMed]

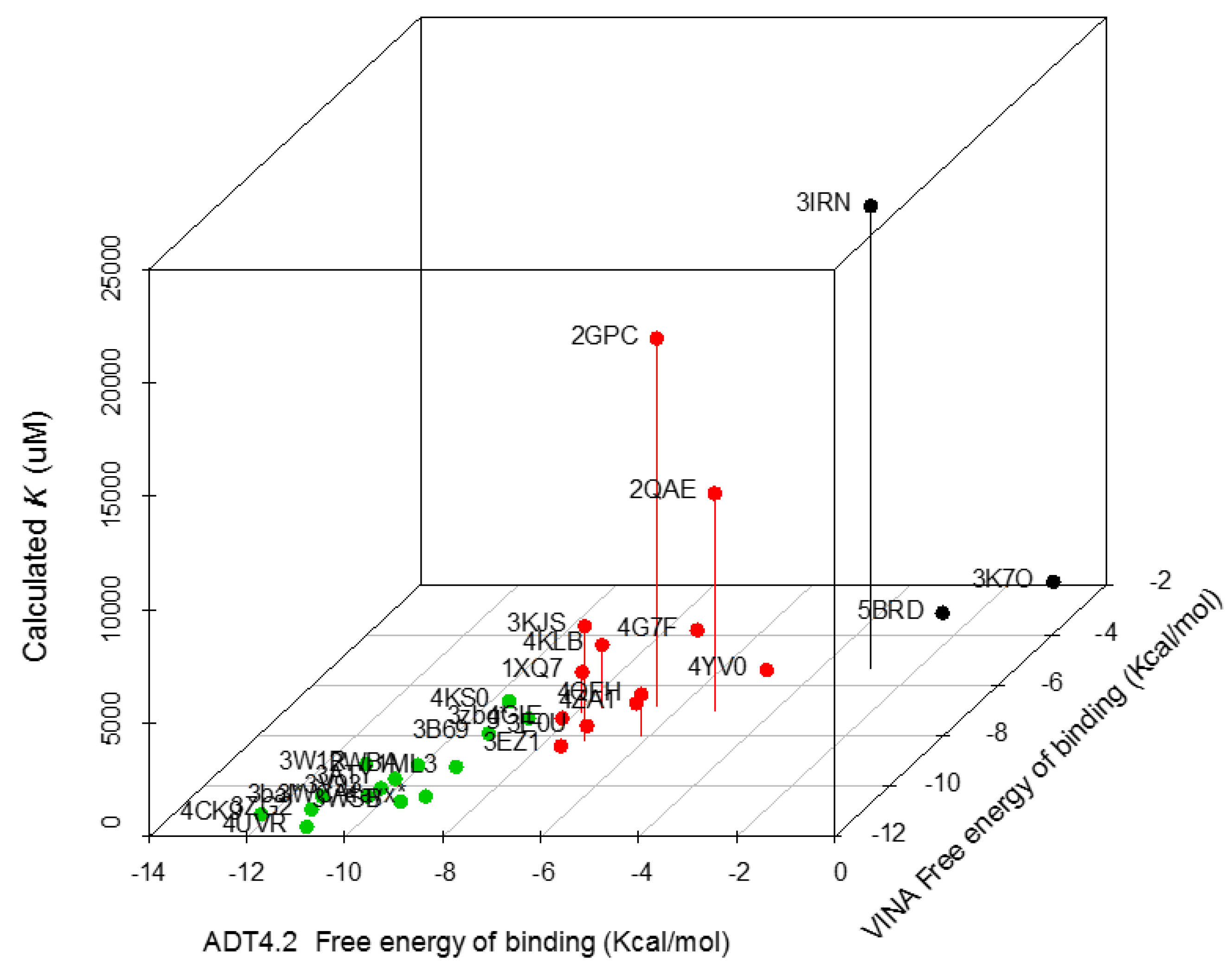




| Trypanosoma spp. Protein Set | PDB Code | VINA Free energy of Binding (Kcal/moL) | ADT4.2 Free energy of Binding (Kcal/moL) | Calculated Ki (µM) |
|---|---|---|---|---|
| Sterol 14-alpha demethylase (CYP51) | 4CK9 | −11.1 | −12.21 | 0.002 |
| Lanosterol 14 alpha demethylase | 3ZG2 | −10.9 | −11.29 | 0.013 |
| Farnesyl transferase | 3WSB | −10.6 | −9.64 | 0.087 |
| C-8 sterol demethylase model | 3bal * | −10.4 | −11.35 | 4.537 |
| Squalene synthase (TcSQS) | 3WCA | −10.4 | −10.42 | 0.021 |
| ABC b model | 4ayx * | −10.4 | −9.24 | 0.171 |
| Phosphodiesterase | 3V93 | −10.1 | −10.32 | 28.66 |
| Old Yellow enzyme (FMN oxidereductase) | 3ATY | −9.7 | −10.25 | 0.029 |
| Glyceraldehyde 3-phosphate dehydrogenase | 1ML3 | −9.4 | −9.17 | 191.147 |
| Trypanothione reductase | 2WBA | −9.2 | −10.06 | 44.723 |
| Dihydroorotate dehydrogenase | 3W1R | −9.1 | −11.17 | 6.407 |
| Chagasin | 3EZ1 | −8.4 | −7.59 | 2.9 |
| Dihydrofolate reductase | 3KJS | −8.2 | −7.21 | 5060 |
| Gluc-6-Pho Isomerase | 4QFH | −8 | −6.17 | 1830 |
| Trans-sialidase | 3B69 | −7.9 | −9.34 | 0.145 |
| Glutathione peroxidase | 3E0U | −7.6 | −7.5 | 3.24 |
| Prostaglandin F synthase | 4GIE | −7.3 | −8.17 | 1.507 |
| Sterol carrier protein 2 model | 3zbg * | −7.3 | −8.86 | 0.321 |
| Cyclophilin | 1XQ7 | −7.1 | −7.88 | 1810 |
| Lipoamide dehydrogenase | 2QAE | −7 | −5.22 | 9576.667 |
| Cruzain | 4KLB | −6.9 | −7.58 | 2783.333 |
| Fe-Superoxide dismutase | 2GPC | −6.8 | −6.52 | 16,176.667 |
| Pyruvate kinase | 4KS0 | −6.7 | −9.59 | 91.953 |
| Spermidine synthase | 4YV0 | −5.6 | −4.94 | 241.627 |
| Dihydrofolate Reductase | 3IRN | −5.3 | −2.98 | 2,0330 |
| Enolase | 4G7F | −3.9 | −7.3 | 102.843 |
| Glucokinase | 5BRD | −3.4 | −2.57 | 310.043 |
| Ribose 5-phosphate isomerase | 3K7O | −2.1 | −1.03 | 238.85 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meza-Menchaca, T.; Ramos-Ligonio, A.; López-Monteon, A.; Vidal Limón, A.; Kaluzhskiy, L.A.; V. Shkel, T.; V. Strushkevich, N.; Jiménez-García, L.F.; Agredano Moreno, L.T.; Gallegos-García, V.; et al. Insights into Ergosterol Peroxide’s Trypanocidal Activity. Biomolecules 2019, 9, 484. https://doi.org/10.3390/biom9090484
Meza-Menchaca T, Ramos-Ligonio A, López-Monteon A, Vidal Limón A, Kaluzhskiy LA, V. Shkel T, V. Strushkevich N, Jiménez-García LF, Agredano Moreno LT, Gallegos-García V, et al. Insights into Ergosterol Peroxide’s Trypanocidal Activity. Biomolecules. 2019; 9(9):484. https://doi.org/10.3390/biom9090484
Chicago/Turabian StyleMeza-Menchaca, Thuluz, Angel Ramos-Ligonio, Aracely López-Monteon, Abraham Vidal Limón, Leonid A. Kaluzhskiy, Tatjana V. Shkel, Natallia V. Strushkevich, Luis Felipe Jiménez-García, Lourdes Teresa Agredano Moreno, Verónica Gallegos-García, and et al. 2019. "Insights into Ergosterol Peroxide’s Trypanocidal Activity" Biomolecules 9, no. 9: 484. https://doi.org/10.3390/biom9090484





