Small, Smaller, Smallest: Minimal Structural Requirements for a Fully Functional Box C/D Modification Guide RNA
Abstract
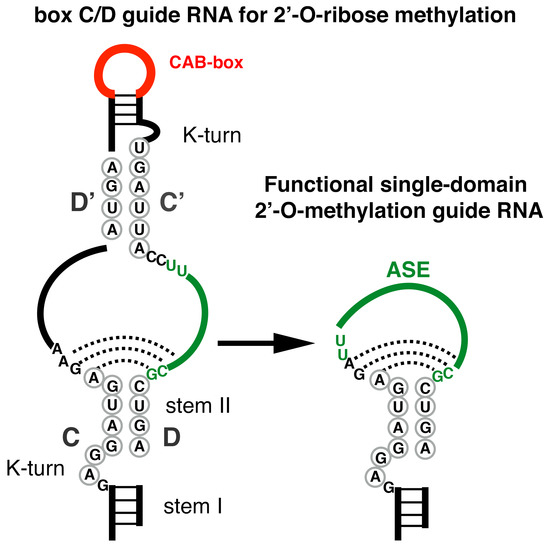
:1. Introduction
2. Materials and Methods
2.1. Animals and Oocytes
2.2. In Vitro Transcription
2.3. In Vitro Modification Assays
2.4. 2’-O-methylation Mapping
2.5. Expression of Drosophila snoRNAs in HeLa Cells
2.6. Northern Blot
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bachellerie, J.-P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Yu, Y.-T.; Terns, R.M.; Terns, M.P. Mechanisms and functions of RNA-guided RNA modification. In Fine-Tuning of RNA Functions by Modification and Editing. Topics in Current Genetics; Grosjean, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 12, pp. 223–262. [Google Scholar]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Smith, C.M.; Shu, M.-D.; Steitz, J.A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. USA 1996, 93, 14480–14485. [Google Scholar] [CrossRef] [PubMed]
- Kiss-László, Z.; Henry, Y.; Bachellerie, J.-P.; Caizergues-Ferrer, M.; Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef]
- Nicoloso, M.; Qu, L.H.; Michot, B.; Bachellerie, J.-P. Intron-encoded, antisense small nucleolar RNAs: The characterization of nine novel species points to their direct role as guides for the 2’-O-ribose methylation of rRNAs. J. Mol. Biol. 1996, 260, 178–195. [Google Scholar] [CrossRef]
- Cavaillé, J.; Bachellerie, J.P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: An exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie 1996, 78, 443–456. [Google Scholar] [CrossRef]
- Watkins, N.J.; Ségault, V.; Charpentier, B.; Nottrott, S.; Fabrizio, P.; Bachi, A.; Wilm, M.; Rosbash, M.; Branlant, C.; Lührmann, R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 2000, 103, 457–466. [Google Scholar] [CrossRef]
- Kiss-László, Z.; Henry, Y.; Kiss, T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998, 17, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Watkins, N.J.; Dickmanns, A.; Luhrmann, R. Conserved Stem II of the Box C/D Motif Is Essential for Nucleolar Localization and Is Required, Along with the 15.5K Protein, for the Hierarchical Assembly of the Box C/D snoRNP. Mol. Cell. Biol. 2002, 22, 8342–8352. [Google Scholar] [CrossRef] [Green Version]
- Qu, G.; van Nues, R.W.; Watkins, N.J.; Maxwell, E.S. The Spatial-Functional Coupling of Box C/D and C’/D’ RNPs Is an Evolutionarily Conserved Feature of the Eukaryotic Box C/D snoRNP Nucleotide Modification Complex. Mol. Cell. Biol. 2011, 31, 365–374. [Google Scholar] [CrossRef]
- Van Nues, R.W.; Watkins, N.J. Unusual C’/D’ motifs enable box C/D snoRNPs to modify multiple sites in the same rRNA target region. Nucleic Acids Res. 2017, 45, 2016–2028. [Google Scholar] [CrossRef] [PubMed]
- Deryusheva, S.; Gall, J.G. Small Cajal Body-specific RNAs of Drosophila Function in the Absence of Cajal Bodies. Mol. Biol. Cell 2009, 20, 5250–5259. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, Z.-H.; Terns, R.M.; Terns, M.P.; Yu, Y.-T. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA 2002, 8, 1515–1525. [Google Scholar] [PubMed]
- Deryusheva, S.; Gall, J.G. Novel small Cajal-body-specific RNAs identified in Drosophila: Probing guide RNA function. RNA 2013, 19, 1802–1814. [Google Scholar] [CrossRef] [PubMed]
- Deryusheva, S.; Choleza, M.; Barbarossa, A.; Gall, J.G.; Bordonne, R. Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells. RNA 2012, 18, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yu, Y.T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Krogh, N.; Kongsbak-Wismann, M.; Geisler, C.; Nielsen, H. Substoichiometric ribose methylations in spliceosomal snRNAs. Org. Biomol. Chem. 2017, 15, 8872–8876. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.A.; Jared, D.W.; Dumont, J.N.; Sega, M.W. Protein incorporation by isolated amphibian oocytes III. Optimum incubation conditions. J. Exp. Zool. 1973, 184, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Gall, J.G.; Murphy, C.; Callan, H.G.; Wu, Z. Lampbrush Chromosomes. Methods Cell Biol. 1991, 36, 149–166. [Google Scholar]
- Cahill, N.M.; Friend, K.; Speckmann, W.; Li, Z.H.; Terns, R.M.; Terns, M.P.; Steitz, J.A. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002, 21, 3816–3828. [Google Scholar] [CrossRef] [Green Version]
- Szewczak, L.B.W.; Gabrielsen, J.S.; Degregorio, S.J.; Strobel, S.A.; Steitz, J.A. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA 2005, 11, 1407–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tycowski, K.T.; Aab, A.; Steitz, J.A. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in Metazoa. Curr. Biol. 2004, 14, 1985–1995. [Google Scholar] [CrossRef] [PubMed]
- Deryusheva, S.; Gall, J.G. Orchestrated positioning of post-transcriptional modifications at the branch point recognition region of U2 snRNA. RNA 2018, 24, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.J.; Zhang, X.; Maxwell, E.S. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 2003, 22, 3930–3940. [Google Scholar] [CrossRef] [PubMed]
- Bleichert, F.; Baserga, S.J. Dissecting the role of conserved box C/D sRNA sequences in di-sRNP assembly and function. Nucleic Acids Res. 2010, 38, 8295–8305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghalei, H.; Hsiao, H.H.; Urlaub, H.; Wahl, M.C.; Watkins, N.J. A novel Nop5-sRNA interaction that is required for efficient archaeal box C/D sRNP formation. RNA 2010, 16, 2341–2348. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Wang, R.; Yang, F.; Terns, R.M.; Terns, M.P.; Zhang, X.; Maxwell, E.S.; Li, H. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol. Cell 2010, 39, 939–949. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deryusheva, S.; Gall, J.G. Small, Smaller, Smallest: Minimal Structural Requirements for a Fully Functional Box C/D Modification Guide RNA. Biomolecules 2019, 9, 457. https://doi.org/10.3390/biom9090457
Deryusheva S, Gall JG. Small, Smaller, Smallest: Minimal Structural Requirements for a Fully Functional Box C/D Modification Guide RNA. Biomolecules. 2019; 9(9):457. https://doi.org/10.3390/biom9090457
Chicago/Turabian StyleDeryusheva, Svetlana, and Joseph G. Gall. 2019. "Small, Smaller, Smallest: Minimal Structural Requirements for a Fully Functional Box C/D Modification Guide RNA" Biomolecules 9, no. 9: 457. https://doi.org/10.3390/biom9090457