Preparation, Characterization, and Inhibition of Hyaluronic Acid Oligosaccharides in Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
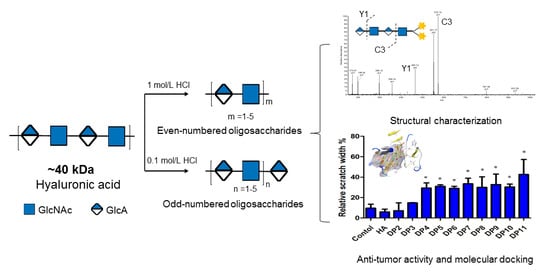
2.2. Preparation of Oligosaccharides from Hyaluronic Acid (HA)
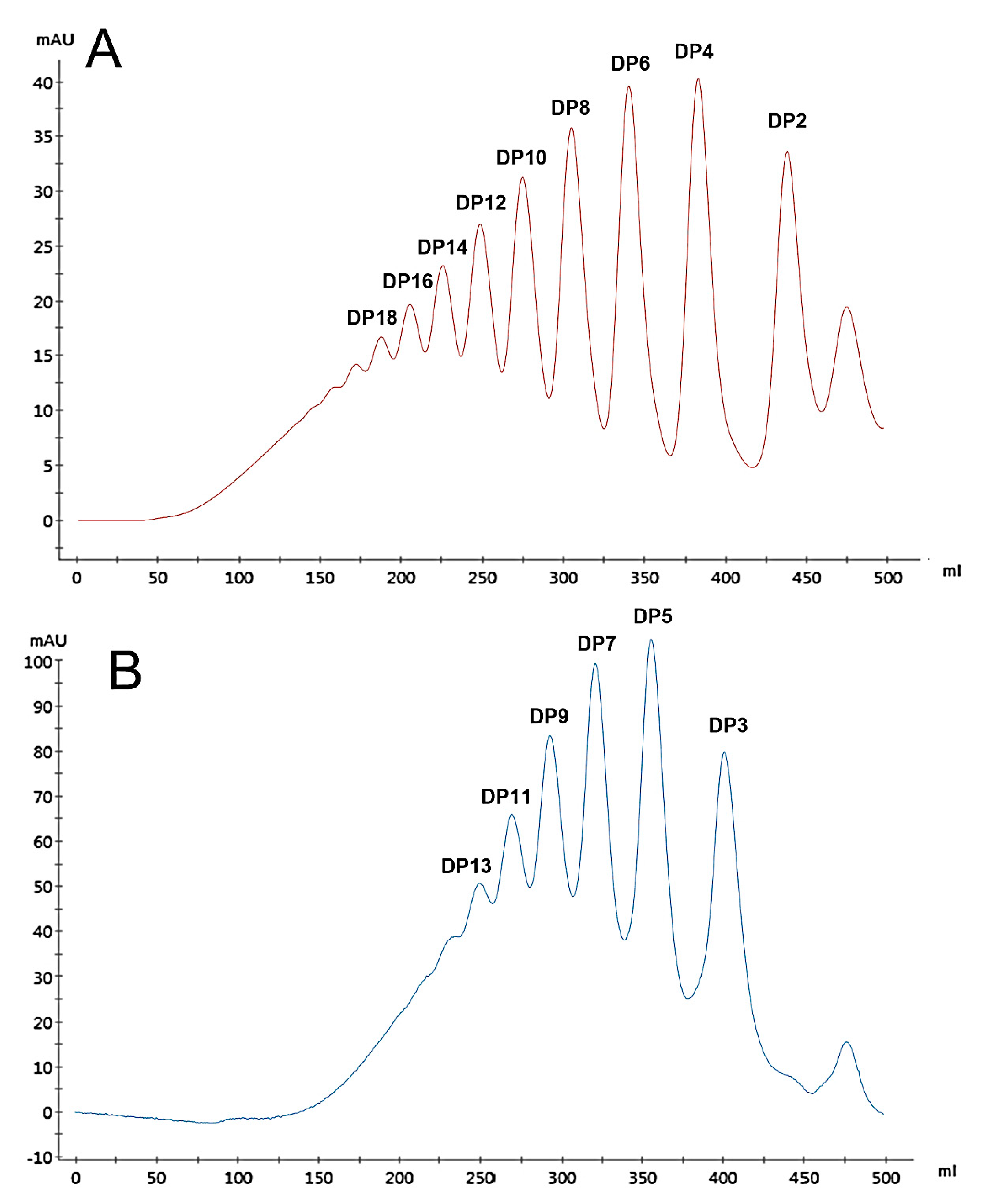
2.3. HPLC Analysis of HA Oligosaccharides
2.4. Thin Layer Chromatography of HA Oligosaccharides
2.5. Electrospray Mass Spectroscopy
2.6. NMR Spectroscopy Analysis
2.7. 1-Phenyl-3-methyl-5-pyrazolone (PMP) Labeling
2.8. Synthesis of Hyaluronic Acid Conjugated with Fluorescein Isothiocyanate (FITC) (HA-FITC)
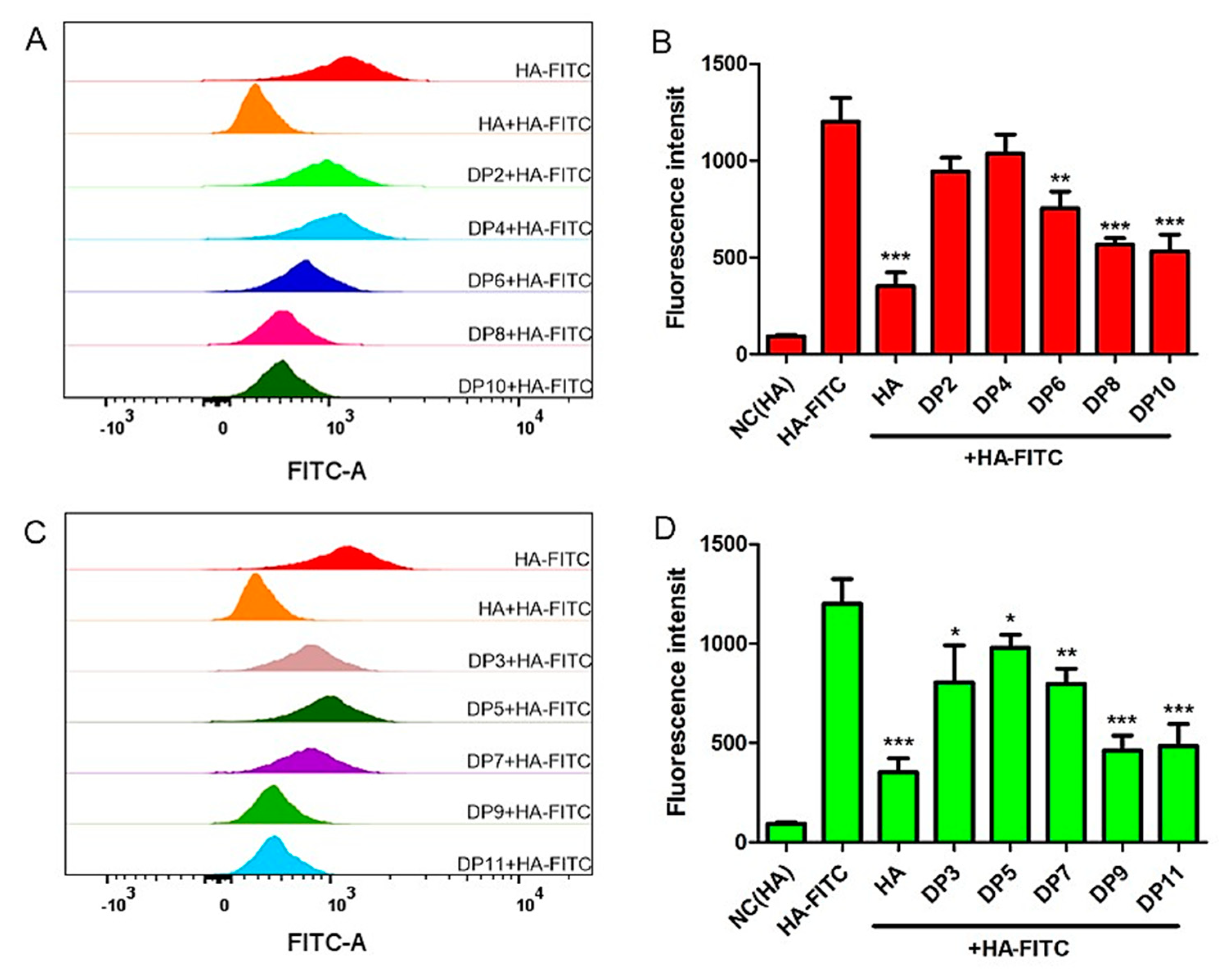
2.9. Binding Assay of HA Oligosaccharides (HAOs) to MDA-MB-231 Cells by Flow Cytometry Analysis
2.10. Molecular Docking of HAOs
2.11. Viability Assay of MDA-MB-231 Cells
2.12. Migration Assay of MDA-MB-231 Cells
2.13. Statistical Analysis
3. Results
3.1. Preparation of HA Oligosaccharides by Acid Hydrolysis
3.2. The Negative-Ion Negative-Ion Electrospray Ionization Mass Spectrometry (ESI-MS) of HA Oligosaccharide Fractions
3.3. NMR Spectroscopy of HA Oligosaccharide Fractions
3.4. Glycosidic Bond-Selective Depolymerization by Acid Hydrolysis
3.5. Binding Abilities of HAOs with MDA-MB-231 Cells
3.6. Molecular Docking of HAOs
3.7. Effect of HAOs on MDA-MB-231 Migration and Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liang, J.; Jiang, D.; Noble, P.W. Hyaluronan as a therapeutic target in human diseases. Adv. Drug. Deliv. Rev. 2016, 97, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Viola, M.; Vigetti, D.; Karousou, E.; Angelo, M.L.D.; Caon, I.; Moretto, P.; De Luca, G.; Passi, A. Biology and biotechnology of hyaluronan. Glycoconj. J. 2015, 32, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, C.; Hoare, K.; Owens, R.; Hohn, H.P.; Hook, M.; Moore, D.; Cripps, V.; Austen, L.; Nance, D.M.; Turley, E.A. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J. Cell Biol. 1992, 117, 1343–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calikoglu, E.; Augsburger, E.; Chavaz, P.; Saurat, J.; Kaya, G. CD44 and hyaluronate in the differential diagnosis of dermatofibroma and dermatofibrosarcoma protuberans. J. Cutan. Pathol. 2003, 30, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Palvai, S.; Kuman, M.M.; Basu, S. Hyaluronic acid cloaked oleic acid nanoparticles inhibit MAPK signaling with sub-cellular DNA damage in colon cancer cells. J. Mater. Chem. B 2017, 5, 3658–3666. [Google Scholar] [CrossRef]
- Yao, W.; Chen, M.; Dou, X.; Jin, H.; Zhang, X.; Zhu, Y.; Sha, M.; Liu, Z.; Meng, X.; Zhang, L.; et al. Unravel a neuroactive sHA sulfation pattern with neurogenesis activity by a library of defined oligosaccharides. Eur. J. Med. Chem. 2019, 163, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Ni, J.; Wang, S.; Clasper, S.; Su, J.; Tammi, R.; Jones, M.; Jackson, D.G. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J. Cell Biol. 1999, 144, 789–801. [Google Scholar] [CrossRef]
- Tesar, B.M.; Jiang, D.; Liang, J.; Palmer, S.M.; Noble, P.W.; Goldstein, D.R. The role of hyaluronan degradation products as innate alloimmune agonists. Am. J. Transplant. 2006, 6, 2622–2635. [Google Scholar] [CrossRef]
- Kothapalli, D.; Flowers, J.; Xu, T.; Pure, E.; Assoian, R.K. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. J. Biol. Chem. 2008, 283, 31823–31829. [Google Scholar] [CrossRef]
- Weigel, P.H. Planning, evaluating and vetting receptor signaling studies to assess hyaluronan size-dependence and specificity. Glycobiology 2017, 27, 796–799. [Google Scholar] [CrossRef]
- Bharadwaj, A.G.; Rector, K.; Simpson, M.A. Inducible hyaluronan production reveals differential effects on prostate tumor cell growth and tumor angiogenesis. J. Biol. Chem. 2007, 282, 20561–20572. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA-Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Heldin, P.; Basu, K.; Olofsson, B.; Porsch, H.; Kozlova, I.; Kahata, K. Deregulation of hyaluronan synthesis, degradation and binding promotes breast cancer. J. Biochem. 2013, 154, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Gabriel, W.; Christine, E.; Katherine, K.; Spevak, C.C. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 2010, 285, 36721–36735. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Mccarthy, J.B.; Yazdani, A.; Turley, E.A. Hyaluronan and RHAMM in wound repair and the “cancerization” of stromal tissues. Biomed Res. Int. 2014, 2014, 103923. [Google Scholar] [CrossRef]
- Wu, M.; Cao, M.L.; He, Y.Q.; Liu, Y.W.; Yang, C.X.; Du, Y.; Wang, W.J.; Gao, F. A novel role of low molecular weight hyaluronan in breast cancer metastasis. FASEB J. 2015, 29, 1290–1298. [Google Scholar] [CrossRef]
- Koyama, H.; Hibi, T.; Isogai, Z.; Yoneda, M.; Fujimori, M.; Amano, J.; Kawakubo, M.; Kannagi, R.; Kimata, K.; Taniguchi, S.; et al. Hyperproduction of hyaluronan in neu-induced mammary tumor accelerates angiogenesis through stromal cell recruitment: Possible involvement of versican/PG-M. Am. J. Pathol. 2007, 170, 1086–1099. [Google Scholar] [CrossRef]
- Bernert, B.; Porsch, H.; Heldin, P. Hyaluronan Synthase 2 (HAS2) promotes breast cancer cell invasion by suppression of tissue metalloproteinase inhibitor 1 (TIMP-1). J. Biol. Chem. 2011, 286, 42349–42359. [Google Scholar] [CrossRef]
- Zeng, C.X.; Toole, B.P.; Kinney, S.D.; Kuo, J.W.; Stamenkovic, I. Inhibition of tumor growth in vivo by hyaluronan oligomers. Int. J. Cancer 1998, 77, 396–401. [Google Scholar] [CrossRef]
- Egger, G.; Pfragner, R.; Siegl, V.; Zatloukal, K.; Glasner, A.; Bader, A.; Steindorfer, P. The affinity of MCF7 breast cancer cells to hyaluronan substrates of different molecular weight and concentrations in an in vitro model. Int. J. Oncol. 2000, 17, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Robyt, J.F.; Mukerjea, R. Separation and quantitative determination of nanogram quantities of maltodextrins and isomaltodextrins by thin-layer chromatography. Carbohydr. Res. 1994, 251, 187–202. [Google Scholar] [CrossRef]
- Pu, J.; Zhao, X.; Wang, Q.; Wang, Y.; Zhou, H. Development and validation of a HPLC method for determination of degree of polymerization of xylo-oligosaccharides. Food Chem. 2016, 213, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Zhao, X.; Wang, Q.; Xiao, L.; Zhao, H. Structural characterization of xylo-oligosaccharides from corncob residues. J. Carbohydr. Chem. 2016, 35, 344–354. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Svechkarev, D.; Souchek, J.J.; Hill, T.K.; Taylor, M.A.; Natarajan, A.; Mohs, A.M. Impact of structurally modifying hyaluronic acid on CD44 interaction. J. Mater. Chem. B 2017, 5, 8183–8192. [Google Scholar] [CrossRef]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the CD44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234–239. [Google Scholar] [CrossRef]
- Plazinski, W.; Knys-Dzieciuch, A. Interactions between CD44 protein and hyaluronan: Insights from the computational study. Mol. Biosyst. 2012, 8, 543–547. [Google Scholar] [CrossRef]
- Kim, H.M.; Park, B.S.; Kim, J.-I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef]
- Tian, Z.; Chu, Y.; Wang, H.; Zhong, L.; Deng, M.; Li, W. Biological activity and interaction mechanism of the diketopiperazine derivatives as tubulin polymerization inhibitors. RSC Adv. 2018, 8, 1055–1064. [Google Scholar] [CrossRef] [Green Version]
- D’Agostino, A.; Stellavato, A.; Corsuto, L.; Diana, P.; Filosa, R.; La Gatta, A.; De Rosa, M.; Schiraldi, C. Is molecular size a discriminating factor in hyaluronan interaction with human cells? Carbohydr. Polym. 2017, 157, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Tawada, A.; Masa, T.; Oonuki, Y.; Watanabe, A.; Matsuzaki, Y.; Asari, A. Large-scale preparation, purification, and characterization of hyaluronan oligosaccharides from 4-mers to 52-mers. Glycobiology 2002, 12, 421–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, S.; Yan, L.; Ding, T.; Linhardt, R.J.; Yu, Y.; Liu, X.; Liu, D.; Ye, X.; Chen, S. Fucosylated chondroitin sulfate oligosaccharides exert anticoagulant activity by targeting at intrinsic tenase complex with low FXII activation: Importance of sulfation pattern and molecular size. Eur. J. Med. Chem. 2017, 139, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Song, F.; Xu, N.; Li, D.; Linhardt, R.J.; Zhang, Z. New insights into the action of bacterial chondroitinase AC I and hyaluronidase on hyaluronic acid. Carbohydr. Polym. 2017, 158, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Tokita, Y.; Okamoto, A. Hydrolytic degradation of hyaluronic acid. Polym. Degrad. Stabil. 1995, 48, 269–273. [Google Scholar] [CrossRef]
- Teriete, P.; Banerji, S.; Noble, M.; Blundell, C.D.; Wright, A.J.; Pickford, A.R.; Lowe, E.; Mahoney, D.J.; Tammi, M.I.; Kahmann, J.D. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol. Cell 2004, 13, 483–496. [Google Scholar] [CrossRef]
- Campo, G.M.; Avenoso, A.; D’Ascola, A.; Scuruchi, M.; Prestipino, V.; Calatroni, A.; Campo, S. 6-Mer hyaluronan oligosaccharides increase IL-18 and IL-33 production in mouse synovial fibroblasts subjected to collagen-induced arthritis. Innate. Immun. 2012, 18, 675–684. [Google Scholar] [CrossRef]
- Campo, G.M.; Avenoso, A.; D’Ascola, A.; Scuruchi, M.; Calatroni, A.; Campo, S. Beta-arrestin-2 negatively modulates inflammation response in mouse chondrocytes induced by 4-mer hyaluronan oligosaccharide. Mol. Cell Biochem. 2015, 399, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; Mccarthy, J.B. Hyaluronan, inflammation, and breast cancer progression. Front. Immunol. 2015, 6, 236. [Google Scholar] [CrossRef]
- Gu, Z.M.; Cai, Q.Y.; He, Y.; Fu, T.M.; Li, F.S. Degradation of hyaluronan by an electrochemical process. Carbohydr. Polym. 2010, 82, 521–523. [Google Scholar] [CrossRef]
- Yue, W. Preparation of low-molecular-weight hyaluronic acid by ozone treatment. Carbohydr. Polym. 2012, 89, 709–712. [Google Scholar]
- Lenormand, H.; Amar-Bacoup, F.; Vincent, J.-C. pH effects on the hyaluronan hydrolysis catalysed by hyaluronidase in the presence of proteins. Part III. The electrostatic non-specific hyaluronan-hyaluronidase complex. Carbohydr. Polym. 2011, 86, 1491–1500. [Google Scholar] [CrossRef]
- Guglieri, S.; Hricovini, M.; Raman, R.; Polito, L.; Torri, G.; Casu, B.; Sasisekharan, R.; Guerrini, M. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochemistry 2008, 47, 13862–13869. [Google Scholar] [CrossRef] [PubMed]
- Solera, C.; Macchione, G.; Maza, S.; Kayser, M.M.; Corzana, F.; de Paz, J.L.; Nieto, P.M. Chondroitin sulfate tetrasaccharides: Synthesis, three-dimensional structure and interaction with midkine. Chemistry 2016, 22, 2356–2369. [Google Scholar] [CrossRef] [PubMed]
- Ooki, T.; Murata-Kamiya, N.; Takahashi-Kanemitsu, A.; Wu, W.; Hatakeyama, M. High-nolecular-weight hyaluronan is a hippo pathway ligand directing cell density-dependent growth inhibition via PAR1b. Dev. Cell. 2019, 49, 590. [Google Scholar] [CrossRef] [PubMed]
- Kolar, S.L.; Kyme, P.; Tseng, C.W.; Soliman, A.; Kaplan, A.; Liang, J.; Nizet, V.; Jiang, D.; Murali, R.; Arditi, M.; et al. Group B Streptococcus evades host immunity by degrading hyaluronan. Cell Host Microbe 2015, 18, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Garcia, J.C.; Chabrol, E.; Vives, R.R.; Thomas, A.; de Paz, J.L.; Rojo, J.; Imberty, A.; Fieschi, F.; Nieto, P.M.; Angulo, J. Langerin-Heparin Interaction: Two binding sites for small and large ligands as revealed by a combination of NMR spectroscopy and cross-linking mapping experiments. J. Am. Chem. Soc. 2015, 137, 4100–4110. [Google Scholar] [CrossRef] [PubMed]





| Fraction | Rf a | [M −H]− | [M −2H]2− | [M −3H]3− | [M −4H]4− | Molecular Mass | Purity (%) b |
|---|---|---|---|---|---|---|---|
| DP2 | 0.79 | 396.20 | 397.33 | 99.5 | |||
| DP3 | 0.76 | 572.17 | 285.58 | 573.46 | 98.4 | ||
| DP4 | 0.54 | 775.22 | 387.11 | 776.65 | 99.3 | ||
| DP5 | 0.53 | 475.15 | 316.43 | 952.78 | 98.6 | ||
| DP6 | 0.36 | 576.66 | 384.11 | 1155.79 | 99.5 | ||
| DP7 | 0.32 | 664.71 | 442.81 | 1332.09 | 98.0 | ||
| DP8 | 0.23 | 766.60 | 1535.29 | 99.0 | |||
| DP9 | 0.18 | 854.28 | 569.18 | 426.64 | 1711.41 | 99.4 | |
| DP10 | 0.15 | 956.20 | 637.20 | 1914.61 | 98.8 | ||
| DP11 | 0.11 | 695.56 | 521.42 | 2090.73 | 98.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, W.; Song, L.; Wang, Y.; Lv, Y.; Chen, X.; Zhao, X. Preparation, Characterization, and Inhibition of Hyaluronic Acid Oligosaccharides in Triple-Negative Breast Cancer. Biomolecules 2019, 9, 436. https://doi.org/10.3390/biom9090436
Han W, Song L, Wang Y, Lv Y, Chen X, Zhao X. Preparation, Characterization, and Inhibition of Hyaluronic Acid Oligosaccharides in Triple-Negative Breast Cancer. Biomolecules. 2019; 9(9):436. https://doi.org/10.3390/biom9090436
Chicago/Turabian StyleHan, Wenwei, Lili Song, Yingdi Wang, Youjing Lv, Xiangyan Chen, and Xia Zhao. 2019. "Preparation, Characterization, and Inhibition of Hyaluronic Acid Oligosaccharides in Triple-Negative Breast Cancer" Biomolecules 9, no. 9: 436. https://doi.org/10.3390/biom9090436