Evolutionary Conservation of the Components in the TOR Signaling Pathways
Abstract
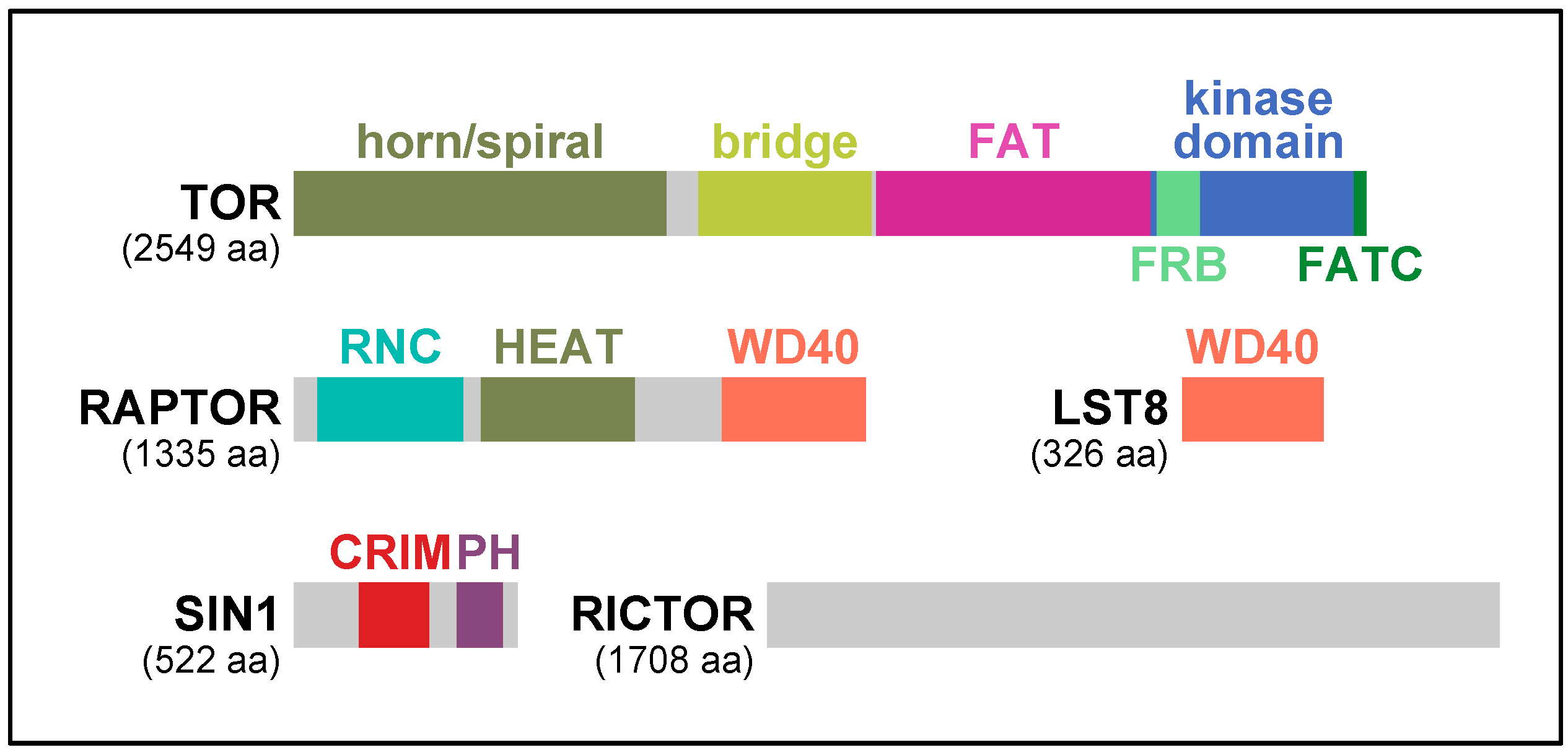
1. Introduction
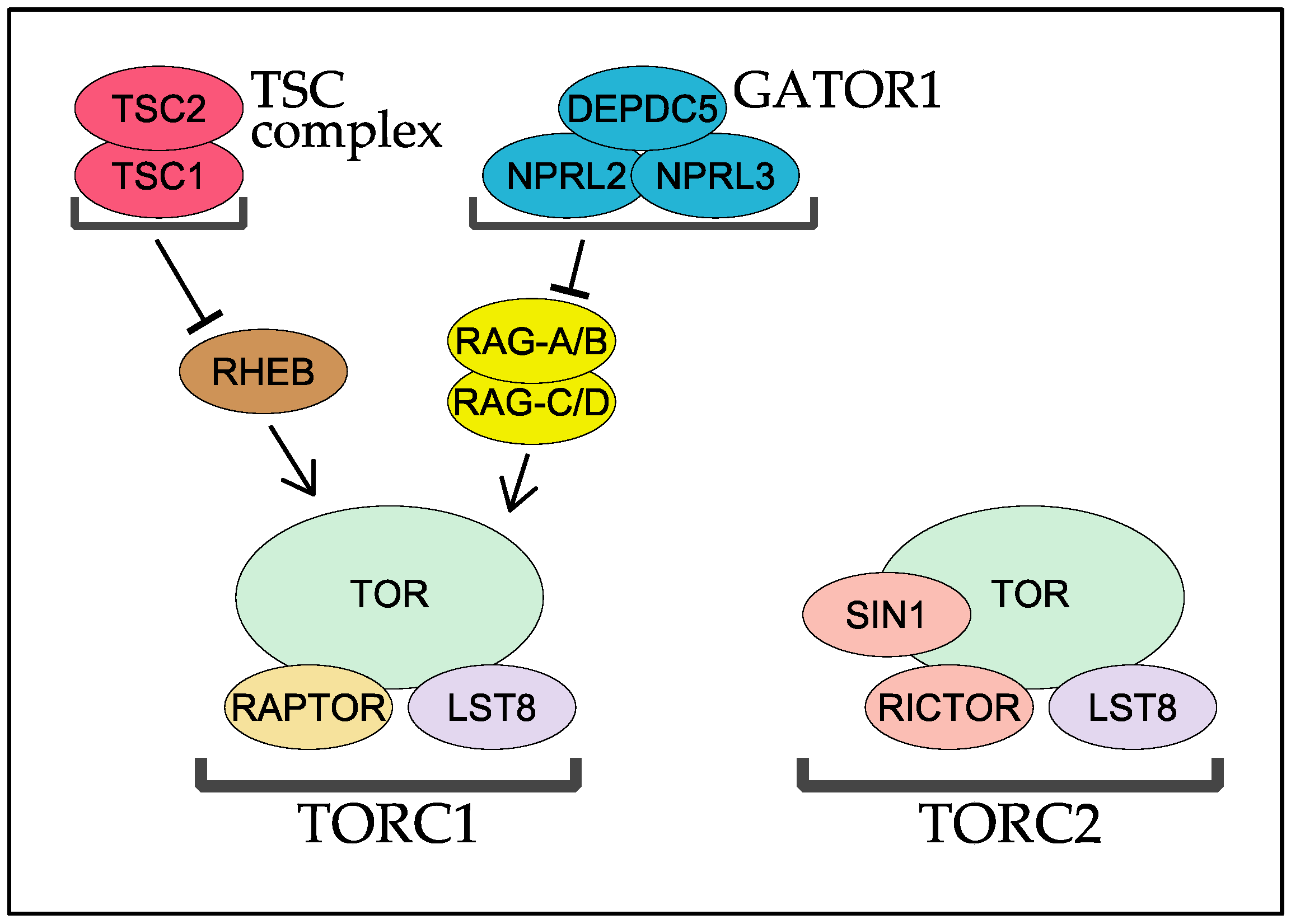
2. RAPTOR, the Signature Subunit of TORC1
3. TORC2 and Two Key Regulatory Subunits: RICTOR and SIN1
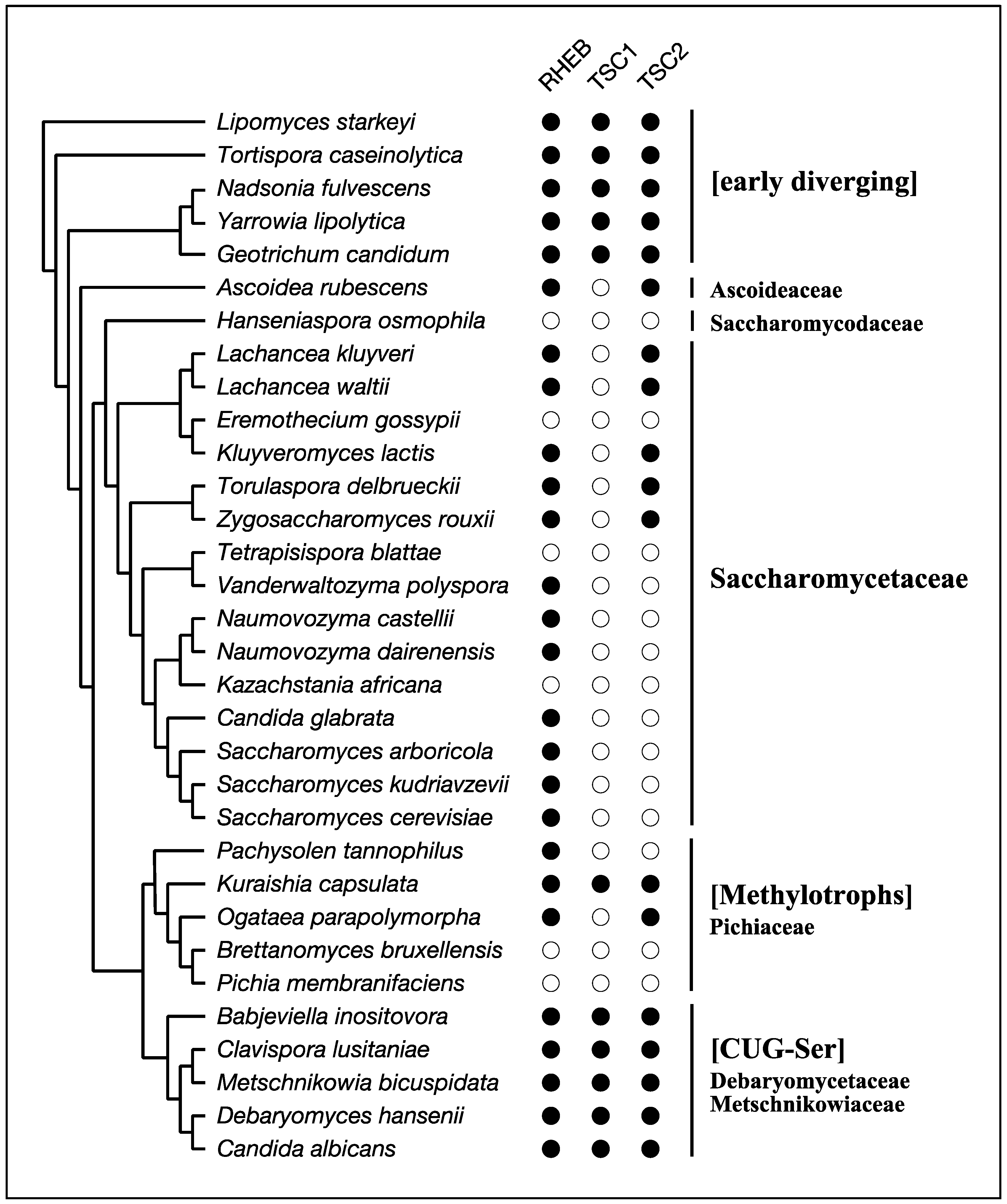
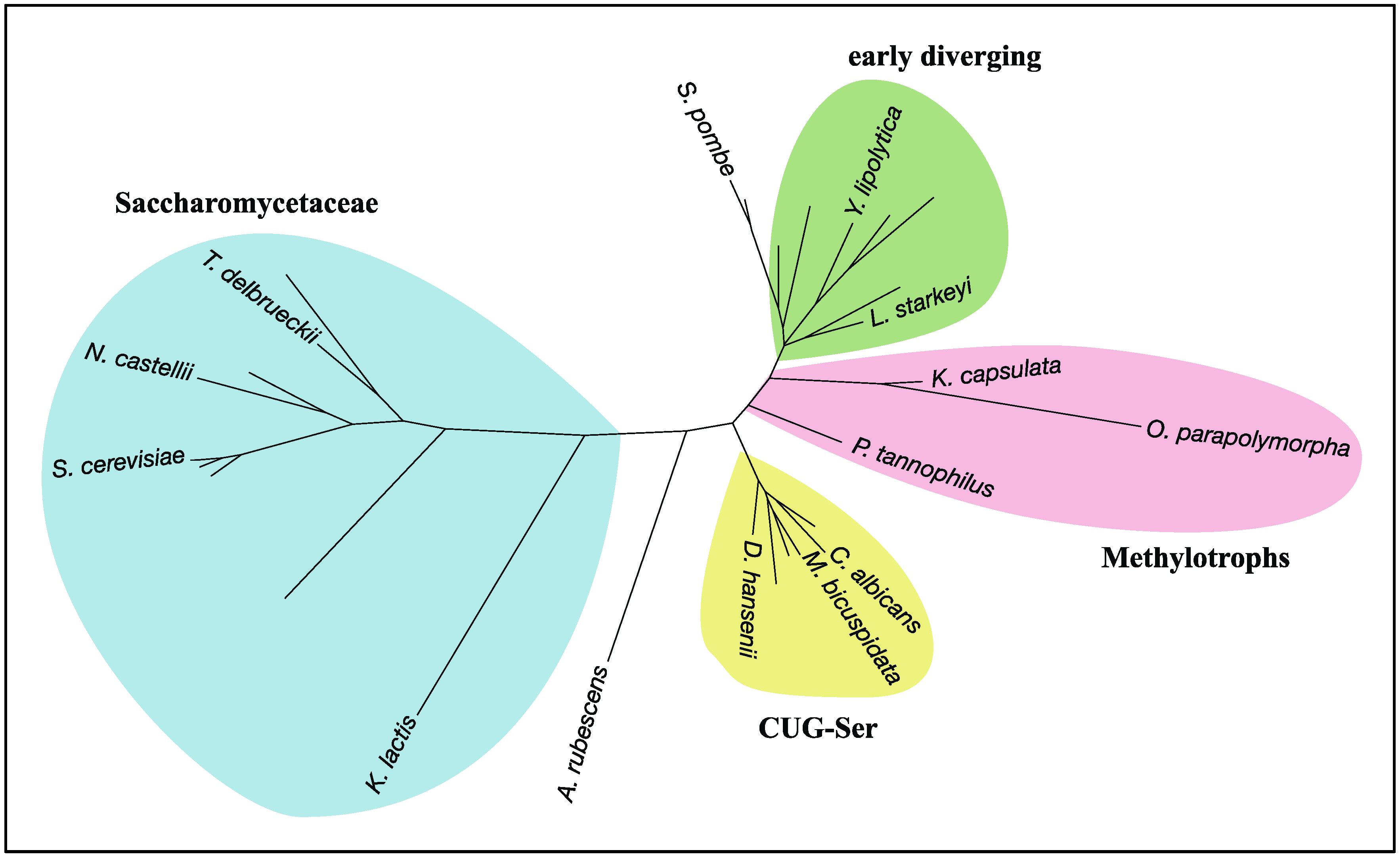
4. RHEB and TSC
5. The RAG GTPases and Their GAP Complex GATOR1
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shimobayashi, M.; Hall, M.N. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014, 15, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [PubMed]
- Soulard, A.; Cohen, A.; Hall, M.N. TOR signaling in invertebrates. Curr. Opin. Cell Biol. 2009, 21, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.A. Origin and diversification of eukaryotes. Annu. Rev. Microbiol. 2012, 66, 411–427. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, T.J.P.; Zwartkruis, F.J.T.; Bos, J.L.; Snel, B. Evolution of the TOR pathway. J. Mol. Evol. 2011, 73, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Barquilla, A.; Saldivia, M.; Diaz, R.; Bart, J.-M.; Vidal, I.; Calvo, E.; Hall, M.N.; Navarro, M. Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 2012, 109, 14399–14404. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Aylett, C.H.S.; Sauer, E.; Imseng, S.; Boehringer, D.; Hall, M.N.; Ban, N.; Maier, T. Architecture of human mTOR complex 1. Science 2016, 351, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Baretić, D.; Berndt, A.; Ohashi, Y.; Johnson, C.M.; Williams, R.L. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat. Commun. 2016, 7, 11016. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, J.; Liu, M.; Chen, X.; Huang, M.; Tan, D.; Dong, M.-Q.; Wong, C.C.L.; Wang, J.; Xu, Y.; et al. 4.4 Å Resolution Cryo-EM structure of human mTOR Complex 1. Protein Cell 2016, 7, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Cornu, M.; Albert, V.; Hall, M.N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013, 23, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Sabatini, D.M. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 2017, 26, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.N. Talks about TORCs: Recent advances in target of rapamycin signalling. On mTOR nomenclature. Biochem. Soc. Trans. 2013, 41, 887–888. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002, 110, 177–189. [Google Scholar] [CrossRef]
- Wedaman, K.P.; Reinke, A.; Anderson, S.; Yates, J.; McCaffery, J.M.; Powers, T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 1204–1220. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, E.A.; Dodd, K.M.; Seymour, L.A.; Tee, A.R. Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell Signal. 2009, 21, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Schalm, S.S.; Blenis, J. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 2002, 12, 632–639. [Google Scholar] [CrossRef]
- Schalm, S.S.; Fingar, D.C.; Sabatini, D.M.; Blenis, J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 2003, 13, 797–806. [Google Scholar] [CrossRef]
- Dunlop, E.A.; Tee, A.R. The kinase triad, AMPK, mTORC1 and ULK1, maintains energy and nutrient homoeostasis. Biochem. Soc. Trans. 2013, 41, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Morigasaki, S.; Tatebe, H.; Tamanoi, F.; Shiozaki, K. Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 2008, 7, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, E.; Facchinetti, V.; Liu, D.; Soto, N.; Wei, S.; Jung, S.Y.; Huang, Q.; Qin, J.; Su, B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006, 127, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.-H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Gaubitz, C.; Prouteau, M.; Kusmider, B.; Loewith, R. TORC2 Structure and Function. Trends Biochem. Sci. 2016, 41, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Oppliger, W.; Hall, M.N. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 2005, 280, 30697–30704. [Google Scholar] [CrossRef] [PubMed]
- Gaubitz, C.; Oliveira, T.M.; Prouteau, M.; Leitner, A.; Karuppasamy, M.; Konstantinidou, G.; Rispal, D.; Eltschinger, S.; Robinson, G.C.; Thore, S.; et al. Molecular Basis of the Rapamycin Insensitivity of Target of Rapamycin Complex 2. Mol. Cell 2015, 58, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.G.; Pino, T.S.; Tournier, S.; Buck, V.; Martin, H.; Christiansen, J.; Wilkinson, D.G.; Millar, J.B. Sin1: An evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 1999, 18, 4210–4221. [Google Scholar] [CrossRef] [PubMed]
- Schroder, W.A.; Buck, M.; Cloonan, N.; Hancock, J.F.; Suhrbier, A.; Sculley, T.; Bushell, G. Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell Signal. 2007, 19, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Schroder, W.; Cloonan, N.; Bushell, G.; Sculley, T. Alternative polyadenylation and splicing of mRNAs transcribed from the human Sin1 gene. Gene 2004, 339, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Z.; Roberts, R.M. The evolution of the Sin1 gene product, a little known protein implicated in stress responses and type I interferon signaling in vertebrates. BMC Evol. Biol. 2005, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Rebecchi, M.J.; Scarlata, S. Pleckstrin homology domains: A common fold with diverse functions. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 503–528. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Inoki, K.; Ikenoue, T.; Guan, K.-L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006, 20, 2820–2832. [Google Scholar] [CrossRef] [PubMed]
- Tatebe, H.; Murayama, S.; Yonekura, T.; Hatano, T.; Richter, D.; Furuya, T.; Kataoka, S.; Furuita, K.; Kojima, C.; Shiozaki, K. Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.J.M.; Linch, M.D.; Saurin, A.T.; Escribano, C.; Parker, P.J. mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 2011, 439, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-C.; Chen, M.-Y. Target of rapamycin complex 2 signals to downstream effector yeast protein kinase 2 (Ypk2) through adheres-voraciously-to-target-of-rapamycin-2 protein 1 (Avo1) in Saccharomyces cerevisiae. J. Biol. Chem. 2012, 287, 6089–6099. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, D.; Walther, T.C. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 2009, 20, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Tatebe, H.; Morigasaki, S.; Murayama, S.; Zeng, C.T.; Shiozaki, K. Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr. Biol. 2010, 20, 1975–1982. [Google Scholar] [CrossRef] [PubMed]
- Ebner, M.; Sinkovics, B.; Szczygieł, M.; Ribeiro, D.W.; Yudushkin, I. Localization of mTORC2 activity inside cells. J. Cell Biol. 2017, 216, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Betz, C.; Stracka, D.; Prescianotto-Baschong, C.; Frieden, M.; Demaurex, N.; Hall, M.N. Feature Article: MTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA 2013, 110, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013, 203, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Gan, W.; Chin, Y.R.; Ogura, K.; Guo, J.; Zhang, J.; Wang, B.; Blenis, J.; Cantley, L.C.; Toker, A.; et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov. 2015, 5, 1194–1209. [Google Scholar] [CrossRef] [PubMed]
- Heard, J.J.; Fong, V.; Bathaie, S.Z.; Tamanoi, F. Recent progress in the study of the Rheb family GTPases. Cell Signal. 2014, 26, 1950–1957. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 2007, 25, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Nakashima, A.; Guo, L.; Tamanoi, F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J. Biol. Chem. 2009, 284, 12783–12791. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Elis, W.; Menon, S.; Qin, W.; Klekota, J.; Asara, J.M.; Finan, P.M.; Kwiatkowski, D.J.; Murphy, L.O.; Manning, B.D. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 2012, 47, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Mazhab-Jafari, M.T.; Marshall, C.B.; Ishiyama, N.; Ho, J.; Di Palma, V.; Stambolic, V.; Ikura, M. An autoinhibited noncanonical mechanism of GTP hydrolysis by Rheb maintains mTORC1 homeostasis. Structure (Lond. Engl. 1993) 2012, 20, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.H.; Thapar, N.; Guo, L.; Martinez, M.; Maris, J.; Gau, C.-L.; Lengyel, J.A.; Tamanoi, F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 2003, 116, 3601–3610. [Google Scholar] [CrossRef] [PubMed]
- Stocker, H.; Radimerski, T.; Schindelholz, B.; Wittwer, F.; Belawat, P.; Daram, P.; Breuer, S.; Thomas, G.; Hafen, E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003, 5, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, X.; Saucedo, L.J.; Ru, B.; Edgar, B.A.; Pan, D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 2003, 5, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Honjoh, S.; Yamamoto, T.; Uno, M.; Nishida, E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 2009, 457, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, Y.; Yamamato, M. TOR signaling in fission yeast. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Urano, J.; Sato, T.; Matsuo, T.; Otsubo, Y.; Yamamoto, M.; Tamanoi, F. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. USA 2007, 104, 3514–3519. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Tabancay, A.P.; Urano, J.; Tamanoi, F. Failure to farnesylate Rheb protein contributes to the enrichment of G0/G1 phase cells in the Schizosaccharomyces pombe farnesyltransferase mutant. Mol. Microbiol. 2001, 41, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Panepinto, J.C.; Oliver, B.G.; Fortwendel, J.R.; Smith, D.L.H.; Askew, D.S.; Rhodes, J.C. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of Invasive pulmonary aspergillosis. Infect. Immun. 2003, 71, 2819–2826. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Nicastro, R.; Sardu, A.; Panchaud, N.; De Virgilio, C. The Architecture of the Rag GTPase Signaling Network. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Urano, J.; Tabancay, A.P.; Yang, W.; Tamanoi, F. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J. Biol. Chem. 2000, 275, 11198–11206. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Göker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H.; et al. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.-C.; Chen, Y.-T.; Lan, C.-Y. A small G protein Rhb1 and a GTPase-activating protein Tsc2 involved in nitrogen starvation-induced morphogenesis and cell wall integrity of Candida albicans. Fungal Genet. Biol. FGB 2009, 46, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-X.; Zhou, X.; Kominek, J.; Kurtzman, C.P.; Hittinger, C.T.; Rokas, A. Reconstructing the Backbone of the Saccharomycotina Yeast Phylogeny Using Genome-Scale Data. G3 (Bethesda Md.) 2016, 6, 3927–3939. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res. 2013, 13, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Jones, E.; Inoki, K. Lysosomal Regulation of mTORC1 by Amino Acids in Mammalian Cells. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Noda, T. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Kira, S.; Kumano, Y.; Ukai, H.; Takeda, E.; Matsuura, A.; Noda, T. Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol. Biol. Cell 2016, 27, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, T.; Kamada, Y.; Furuno, N.; Funakoshi, M.; Kobayashi, H. Amino acid residues required for Gtr1p-Gtr2p complex formation and its interactions with the Ego1p-Ego3p complex and TORC1 components in yeast. Genes Cells Devoted Mol. Cell. Mech. 2014, 19, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, N.; Péli-Gulli, M.-P.; De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 2013, 6, ra42. [Google Scholar] [CrossRef] [PubMed]
- Binda, M.; Péli-Gulli, M.-P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; De Virgilio, C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 2009, 35, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Morrison, H.G.; McArthur, A.G.; Gillin, F.D.; Aley, S.B.; Adam, R.D.; Olsen, G.J.; Best, A.A.; Cande, W.Z.; Chen, F.; Cipriano, M.J.; et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 2007, 317, 1921–1926. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, M.E.; Couso, I.; Crespo, J.L. The TOR Signaling Network in the Model Unicellular Green Alga Chlamydomonas reinhardtii. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]

| Major Clade | Kingdom | Species | RICTOR | SIN1 | TOR | LST8 | RAPTOR | RHEB | TSC1 | TSC2 | RAG-A/B | RAG-C/D | DEPDC5 | NPRL2 | NPRL3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Opisthokonta | Metazoa | Homo sapiens | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Metazoa | Ciona intestinalis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Metazoa | Drosophila melanogaster | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Metazoa | Schistosoma mansoni | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Opisthokonta | Metazoa | Caenorhabditis elegans | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||
| Opisthokonta | Metazoa | Nematostella vectensis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Metazoa | Trichoplax adhaerens | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Amphimedon queenslandica | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Capsaspora owczarzaki | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Mitosporidium daphniae | ✔ | ✔ | ✔ | ||||||||||
| Opisthokonta | Fungi | Encephalitozoon intestinalis | |||||||||||||
| Opisthokonta | Fungi | Rozella allomycis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Opisthokonta | Fungi | Allomyces macrogynus | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Opisthokonta | Fungi | Batrachochytrium dendrobatidis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Gonapodya prolifera | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Conidiobolus coronatus | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Smittium culicis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Rhizophagus irregularis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Lobosporangium transversale | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Mortierella elongata | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Mucor circinelloides | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Neocallimastix californiae | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Puccinia sorghi | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Schizophyllum commune | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Cutaneotrichosporon oleaginosus | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Tilletiaria anomala | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Ustilago maydis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Dothistroma septosporum | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Cladophialophora bantiana | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Aspergillus fumigatus | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Trichopyton equinum | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Botrytis cinerea | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Colletotrichum graminicola | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Fusarium fujikuroi | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Neurospora crassa | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Schizosaccharomyces pombe | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Saitoella complicata | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Yarrowia lipolytica | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Candida albicans | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Opisthokonta | Fungi | Ogataea parapolymorpha | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Eremothecium gossypii | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Opisthokonta | Fungi | Kluyveromyces lactis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Opisthokonta | Fungi | Candida glabrata | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Opisthokonta | Fungi | Saccharomyces cerevisiae | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||
| Amoebozoa | Acanthamoeba castellanii | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Amoebozoa | Dictyostelium discoideum | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||
| Amoebozoa | Entamoeba histolytica | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||||||
| Excavata | Naegleria gruberi | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||||
| Excavata | Bodo saltans | ✔ | ✔ | ✔ | ✔ 1 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||||
| Excavata | Angomonas deanei | ✔ | ✔ | ✔ | ✔ 1 | ✔ | ✔ | ✔ | ✔ | ||||||
| Excavata | Trypanosoma brucei | ✔ | ✔ | ✔ 1 | ✔ | ✔ | ✔ | ✔ | ✔ | ||||||
| Excavata | Leishmania major | ✔ | ✔ | ✔ | ✔ 1 | ✔ | ✔ | ✔ | ✔ | ||||||
| Excavata | Giardia intestinalis | ✔ | ✔ | ✔ | ✔ | ||||||||||
| Excavata | Spironucleus salmonicida | ✔ | ✔ | ✔ | ✔ | ||||||||||
| SAR | Rhizaria | Reticulomyxa filosa | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| SAR | Rhizaria | Plasmodiophora brassicae | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Rhizaria | Bigelowiella natans | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| SAR | Alveolata | Stylonychia lemnae | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||||
| SAR | Alveolata | Oxytricha trifallax | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||||
| SAR | Alveolata | Paramecium tetraurelia | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| SAR | Alveolata | Tetrahymena thermophila | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ||||
| SAR | Alveolata | Vitrella brassicaformis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| SAR | Alveolata | Plasmodium falciparum | |||||||||||||
| SAR | Alveolata | Hammondia hammondi | ✔ | ✔ | ✔ | ||||||||||
| SAR | Alveolata | Toxoplasma gondii | ✔ | ✔ | ✔ | ||||||||||
| SAR | Alveolata | Cyclospora cayetanensis | |||||||||||||
| SAR | Alveolata | Eimeria maxima | |||||||||||||
| SAR | Alveolata | Cryptosporidium pavrum | |||||||||||||
| SAR | Alveolata | Theileria annulata | |||||||||||||
| SAR | Stramenopiles | Aureococcus anophagefferens | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||||||
| SAR | Stramenopiles | Saprolegnia diclina | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Stramenopiles | Aphanomyces invadans | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| SAR | Stramenopiles | Pythium irregulare | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Stramenopiles | Phytophthora infestans | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Stramenopiles | Plasmopara halstedii | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Stramenopiles | Hyaloperonospora arabidopsis | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Stramenopiles | Albugo candida | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| SAR | Stramenopiles | Nannochloropsis gaditana | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| SAR | Stramenopiles | Blastocystis hominis | ✔ | ✔ | ✔ | ✔ | ✔ | ||||||||
| SAR | Stramenopiles | Phaeodactylum tricornutum | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||
| Plantae | Rhodophyta | Cyanidioschyzon melorae | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| Plantae | Rhodophyta | Galdieria sulphuraria | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |||||
| Plantae | Rhodophyta | Chondrus crispus | ✔ | ✔ | ✔ | ||||||||||
| Plantae | Chlorophyta | Ostreococcus lucimarinus | ✔ | ✔ | ✔ | ||||||||||
| Plantae | Streptophyta | Selaginella moellendorffii | ✔ | ✔ | ✔ | ||||||||||
| Plantae | Streptophyta | Amborella trichopoda | ✔ | ✔ | ✔ | ||||||||||
| Plantae | Streptophyta | Oryza sativa | ✔ | ✔ | ✔ | ||||||||||
| Plantae | Streptophyta | Arabidopsis thaliana | ✔ | ✔ | ✔ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatebe, H.; Shiozaki, K. Evolutionary Conservation of the Components in the TOR Signaling Pathways. Biomolecules 2017, 7, 77. https://doi.org/10.3390/biom7040077
Tatebe H, Shiozaki K. Evolutionary Conservation of the Components in the TOR Signaling Pathways. Biomolecules. 2017; 7(4):77. https://doi.org/10.3390/biom7040077
Chicago/Turabian StyleTatebe, Hisashi, and Kazuhiro Shiozaki. 2017. "Evolutionary Conservation of the Components in the TOR Signaling Pathways" Biomolecules 7, no. 4: 77. https://doi.org/10.3390/biom7040077
APA StyleTatebe, H., & Shiozaki, K. (2017). Evolutionary Conservation of the Components in the TOR Signaling Pathways. Biomolecules, 7(4), 77. https://doi.org/10.3390/biom7040077






