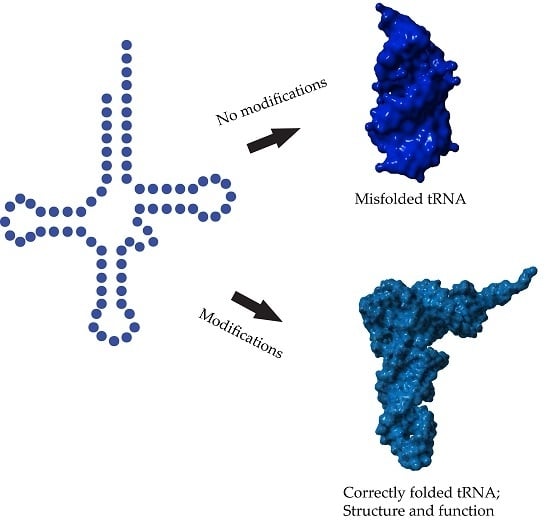
Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function
Abstract
:1. Introduction
2. Modifications Responsible for tRNA Structure
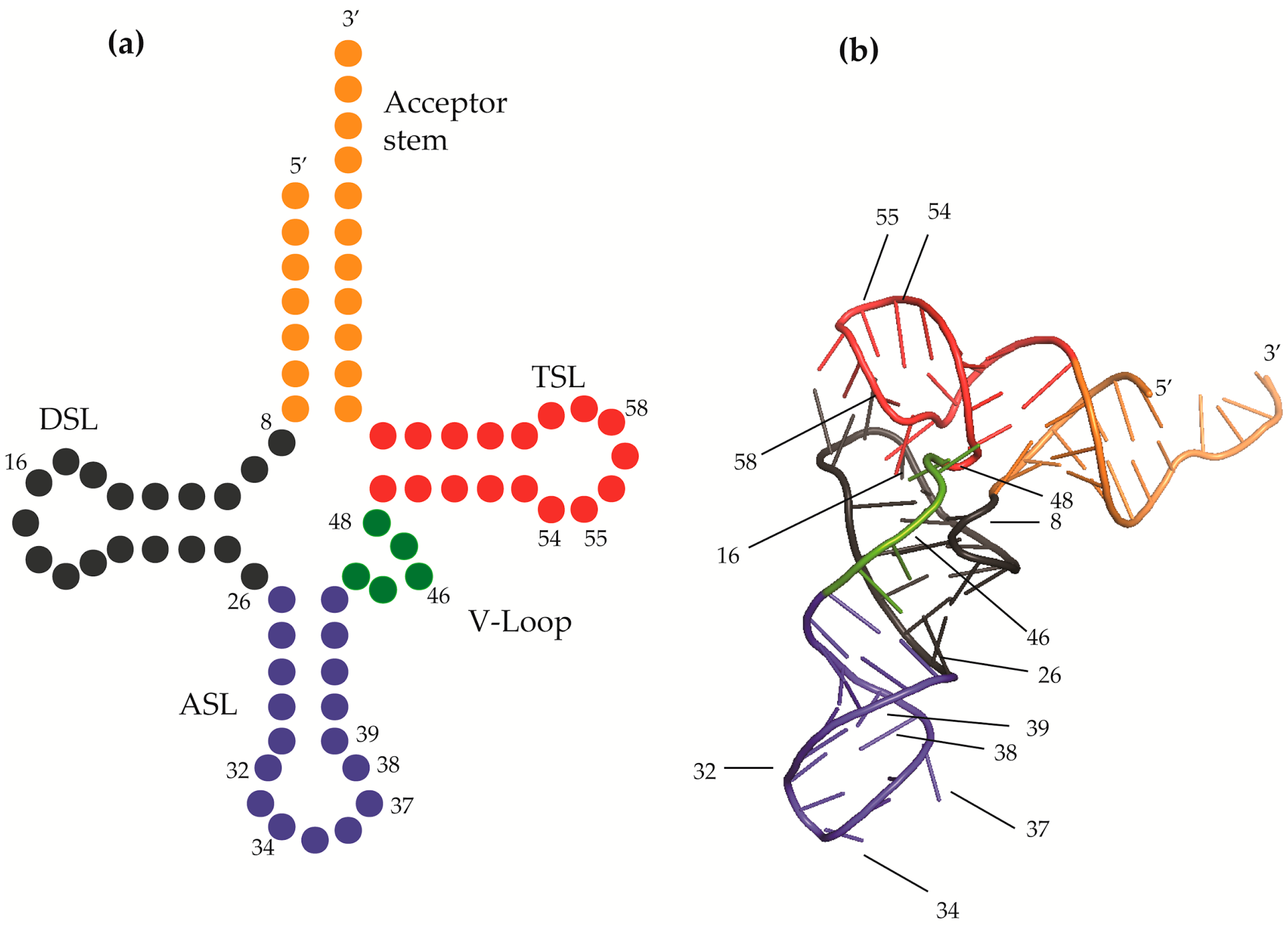
2.1. tRNA Junction
2.1.1. Position 9 and 10 (A9, G9 and G10)
2.1.2. G10: 2-Methylguanosines
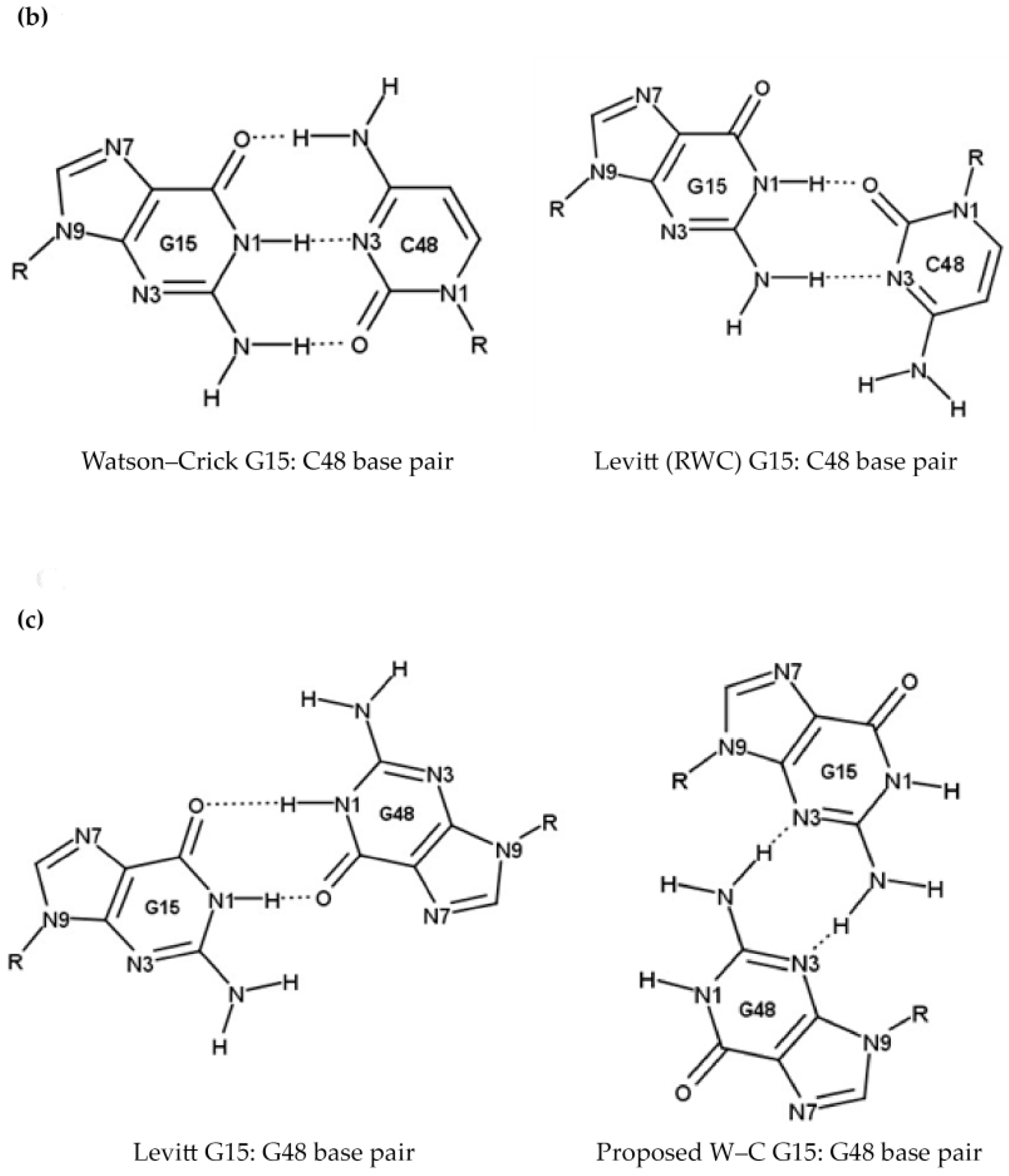
2.1.3. Position 26 (G26)
2.1.4. Position 48 and 49 (m5C48 and m5C49)
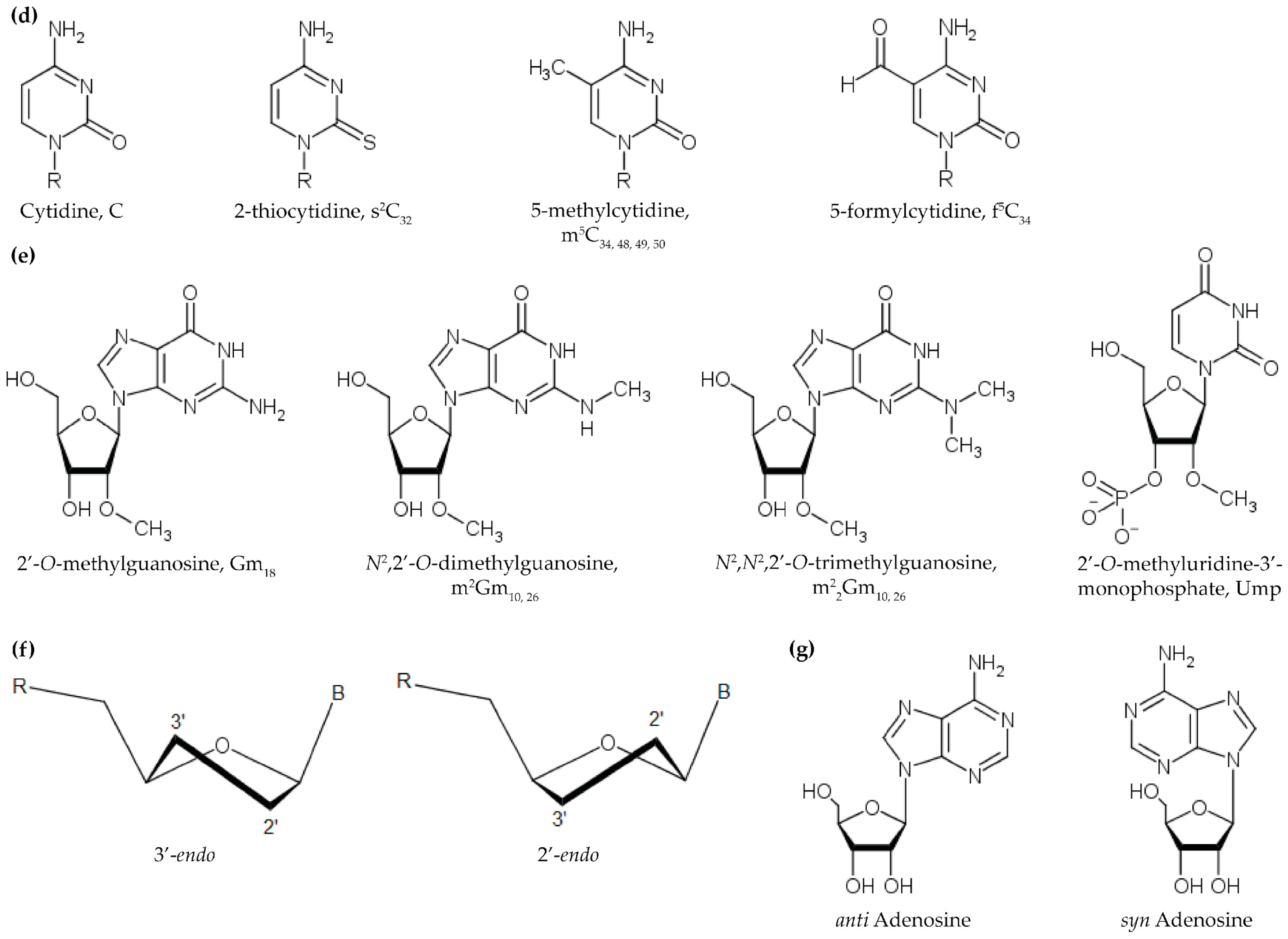
2.2. Methylations of the Ribose 2’-OH and Nucleobase
2.3. Uridine Modifications Affecting Core Structure
2.3.1. Dihydrouridine
2.3.2. Pseudouridine
3. Modifications Impacting Decoding
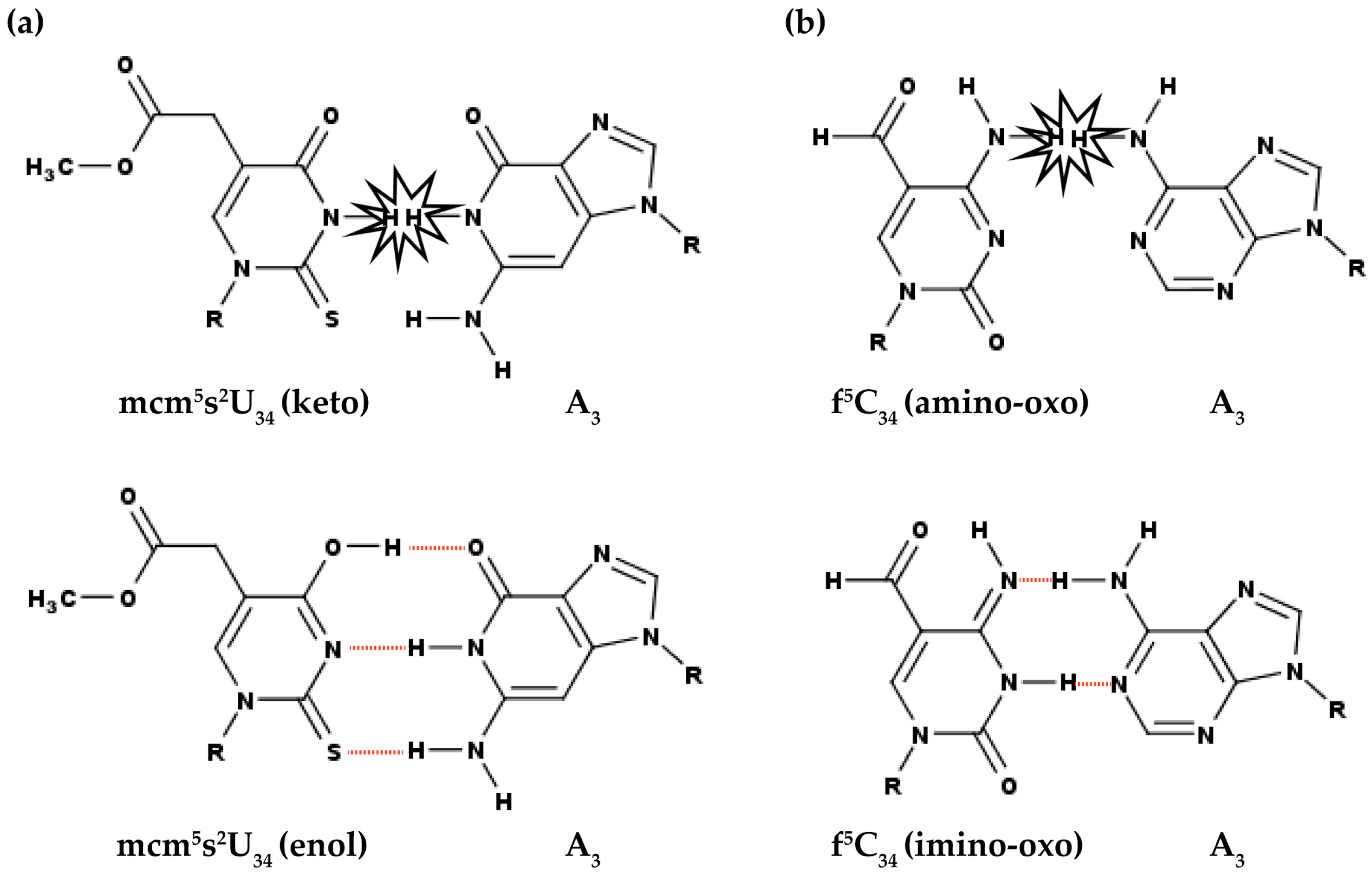
3.1. Modifications at Position 34
3.2. Modifications at Position 37
3.3. Metal Ions and the Modification of the tRNA Anticodon Domain
3.4. Modifications at Position 32
4. Discussion
Supplementary Materials
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. The importance of being modified: An unrealized code to RNA structure and function. RNA 2015, 21, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Duechler, M.; Leszczyńska, G.; Sochacka, E.; Nawrot, B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life Sci. 2016, 73, 3075–3095. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1996, 53, 79–129. [Google Scholar] [PubMed]
- Schmidt, P.G.; Sierzputowska-Gracz, H.; Agris, P.F. Internal motions in yeast phenylalanine transfer RNA from 13C NMR relaxation rates of modified base methyl groups: A model-free approach. Biochemistry 1987, 26, 8529–8534. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: A modified-wobble hypothesis. Biochimie 1991, 73, 1345–1349. [Google Scholar] [CrossRef]
- Björk, G.R. Chapter 11: Biosynthesis and Function of Modified Nucleosides. In tRNA: Structure, Biosynthesis, and Function; Söll, D., RajBhandary, U., Eds.; American Society for Microbiology: Washington, DC, USA, 1995; pp. 165–205. [Google Scholar]
- Suzuki, T.; Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014, 42, 7346–7357. [Google Scholar] [PubMed]
- Björk, G.; Hagervall, T. Transfer RNA modification: Presence, synthesis, and function. EcoSal Plus 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA modifications: Playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Carell, T.; Brandmayr, C.; Hienzsch, A.; Muller, M.; Pearson, D.; Reiter, V.; Thoma, I.; Thumbs, P.; Wagner, M. Structure and function of noncanonical nucleobases. Angew. Chem. Int. Ed. Engl. 2012, 51, 7110–7131. [Google Scholar] [CrossRef] [PubMed]
- Basavappa, R.; Sigler, P.B. The 3 Å crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J. 1991, 10, 3105–3111. [Google Scholar] [PubMed]
- Lewis, C.J.; Pan, T.; Kalsotra, A. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ferre-D’Amare, A.R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 2013, 500, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Vendeix, F.A.; Graham, W.D. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hamann, C.S.; Hou, Y.-M. Probing a tRNA core that contributes to aminoacylation1. J. Mol. Biol. 2000, 295, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Edqvist, J.; Stråby, K.B.; Giegé, R. Enzymatic formation of modified nucleosides in tRNA: Dependence on tRNA Architecture. J. Mol. Biol. 1996, 255, 67–85. [Google Scholar] [CrossRef] [PubMed]
- Dirheimer, G.; Keith, G.; Dumas, P.; Westhof, E. tRNA: Structure, Biosynthesis, and Function; Söll, D., RajBhandary, U.L., Eds.; ASM Press: Washington, DC, USA, 1995; pp. 93–126. [Google Scholar]
- Giegé, R.; Puglisi, J.D.; Florentz, C. tRNA Structure and Aminoacylation Efficiency. In Progress in Nucleic Acid Research and Molecular Biology; Waldo, E.C., Kivie, M., Eds.; Academic Press: New York, NY, USA, 1993; Volume 45, pp. 129–206. [Google Scholar]
- Christian, T.; Lipman, R.S.; Evilia, C.; Hou, Y.M. Alternative design of a tRNA core for aminoacylation. J. Mol. Biol. 2000, 303, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Klug, A.; Ladner, J.; Robertus, J.D. The structural geometry of co-ordinated base changes in transfer RNA. J. Mol. Biol. 1974, 89, 511–516. [Google Scholar] [CrossRef]
- Sprinzl, M.; Horn, C.; Brown, M.; Ioudovitch, A.; Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998, 26, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.R.; DiRenzo, A.B.; Behlen, L.S.; Uhlenbeck, O.C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science 1989, 243, 1363–1366. [Google Scholar] [CrossRef] [PubMed]
- McClain, W.H.; Foss, K. Nucleotides that contribute to the identity of Escherichia coli tRNAPhe. J. Mol. Biol. 1988, 202, 697–709. [Google Scholar] [CrossRef]
- Helm, M.; Giegé, R.; Florentz, C. A Watson–Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 1999, 38, 13338–13346. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Ohtsuki, T.; Watanabe, K. Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res. 2005, 33, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Vilardo, E.; Nachbagauer, C.; Buzet, A.; Taschner, A.; Holzmann, J.; Rossmanith, W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012, 40, 11583–11593. [Google Scholar] [CrossRef] [PubMed]
- Oerum, S.; Dégut, C.; Barraud, P.; Tisné, C. m1A post-transcriptional modification in tRNAs. Biomolecules 2017, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Gillis, D.; Krishnamohan, A.; Yaacov, B.; Shaag, A.; Jackman, J.E.; Elpeleg, O. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J. Med. Genet. 2014, 51, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Voigts-Hoffmann, F.; Hengesbach, M.; Kobitski, A.Y.; van Aerschot, A.; Herdewijn, P.; Nienhaus, G.U.; Helm, M. A methyl group controls conformational equilibrium in human mitochondrial tRNALys. J. Am. Chem. Soc. 2007, 129, 13382–13383. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Attardi, G. Nuclear control of cloverleaf structure of human mitochondrial tRNALys. J. Mol. Biol. 2004, 337, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Sissler, M.; Helm, M.; Frugier, M.; Giege, R.; Florentz, C. Aminoacylation properties of pathology-related human mitochondrial tRNALys variants. RNA 2004, 10, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, J.; Frank, P.; Löffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Kobitski, A.Y.; Hengesbach, M.; Helm, M.; Nienhaus, G.U. Sculpting an RNA conformational energy landscape by a methyl group modification—A single-molecule FRET study. Angew. Chem. Int. Ed. 2008, 47, 4326–4330. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef] [PubMed]
- Nobles, K.N.; Yarian, C.S.; Liu, G.; Guenther, R.H.; Agris, P.F. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002, 30, 4751–4760. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Montange, R.K.; Malik, H.S.; Phizicky, E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 2003, 9, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Saenger, W. Intercalation. In Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1984; pp. 350–367. [Google Scholar]
- Noon, K.R.; Guymon, R.; Crain, P.F.; McCloskey, J.A.; Thomm, M.; Lim, J.; Cavicchioli, R. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23 °C) and Stetteria hydrogenophila (Topt, 95 °C). J. Bacteriol. 2003, 185, 5483–5490. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Moore, P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: A classic structure revisited. RNA 2000, 6, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Hayase, Y.; Jahn, M.; Rogers, M.J.; Sylvers, L.A.; Koizumi, M.; Inoue, H.; Ohtsuka, E.; Söll, D. Recognition of bases in Escherichia coli tRNAGln by glutaminyl-tRNA synthetase: A complete identity set. EMBO J. 1992, 11, 4159–4165. [Google Scholar] [PubMed]
- Sprinzl, M.; Hartmann, T.; Weber, J.; Blank, J.; Zeidler, R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989, 17, r1–r172. [Google Scholar] [CrossRef] [PubMed]
- Edqvist, J.; Grosjean, H.; Straby, K.B. Identity elements for N2-dimethylation of guanosine-26 in yeast tRNAs. Nucleic Acids Res. 1992, 20, 6575–6581. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Blewett, N.H.; Iben, J.R.; Lamichhane, T.N.; Cherkasova, V.; Hafner, M.; Maraia, R.J. RNA polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet. 2015, 11, e1005671. [Google Scholar] [CrossRef] [PubMed]
- Hori, H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Burgess, A.L.; David, R.; Searle, I.R. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 2015, 15, 199. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Grosjean, H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 1999, 5, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Kurowski, A.; Nichols, J.; Watt, F.M.; Benitah, S.A.; Frye, M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011, 7, e1002403. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Gamper, H.; Yang, W. Post-transcriptional modifications to tRNA—A response to the genetic code degeneracy. RNA 2015, 21, 642–644. [Google Scholar] [CrossRef] [PubMed]
- Auxilien, S.; Guerineau, V.; Szweykowska-Kulinska, Z.; Golinelli-Pimpaneau, B. The human tRNA m5C methyltransferase Misu is multisite-specific. RNA Biol. 2012, 9, 1331–1338. [Google Scholar] [CrossRef] [PubMed]
- Militello, K.T.; Chen, L.M.; Ackerman, S.E.; Mandarano, A.H.; Valentine, E.L. A map of 5-methylcytosine residues in Trypanosoma brucei tRNA revealed by sodium bisulfite sequencing. Mol. Biochem. Parasitol. 2014, 193, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M. Detailed molecular model for transfer ribonucleic acid. Nature 1969, 224, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, M.E.; Turner, D.H.; Tinoco, I., Jr. The interactions that shape RNA structure. In The RNA World, 2nd ed.; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1999; pp. 233–264. [Google Scholar]
- Oliva, R.; Tramontano, A.; Cavallo, L. Mg2+ binding and archaeosine modification stabilize the G15–C48 Levitt base pair in tRNAs. RNA 2007, 13, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Gregson, J.M.; Crain, P.F.; Edmonds, C.G.; Gupta, R.; Hashizume, T.; Phillipson, D.W.; McCloskey, J.A. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-β-d-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)). J. Biol. Chem. 1993, 268, 10076–10086. [Google Scholar] [PubMed]
- Quigley, G.J.; Rich, A. Structural domains of transfer RNA molecules. Science 1976, 194, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Sherlin, L.D.; Bullock, T.L.; Newberry, K.J.; Lipman, R.S.; Hou, Y.M.; Beijer, B.; Sproat, B.S.; Perona, J.J. Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases. J. Mol. Biol. 2000, 299, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Nissen, P.; Thirup, S.; Kjeldgaard, M.; Nyborg, J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 1999, 7, 143–156. [Google Scholar] [CrossRef]
- Hou, Y.M.; Westhof, E.; Giege, R. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6776–6780. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Sterner, T.; Jansen, M. Permutation of a pair of tertiary nucleotides in a transfer RNA. Biochemistry 1995, 34, 2978–2984. [Google Scholar] [CrossRef] [PubMed]
- Cusack, S.; Yaremchuk, A.; Tukalo, M. The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNASer and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 1996, 15, 2834–2842. [Google Scholar] [PubMed]
- Kawai, G.; Ue, H.; Yasuda, M.; Sakamoto, K.; Hashizume, T.; McCloskey, J.A.; Miyazawa, T.; Yokoyama, S. Relation between functions and conformational characteristics of modified nucleosides found in tRNAs. Nucleic Acids Symp. Ser. 1991, 25, 49–50. [Google Scholar]
- Kawai, G.; Yamamoto, Y.; Kamimura, T.; Masegi, T.; Sekine, M.; Hata, T.; Iimori, T.; Watanabe, T.; Miyazawa, T.; Yokoyama, S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2’-hydroxyl group. Biochemistry 1992, 31, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Koh, H.; Soll, D. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch. Biochem. Biophys. 1973, 154, 277–282. [Google Scholar] [CrossRef]
- Kumagai, I.; Watanabe, K.; Oshima, T. Thermally induced biosynthesis of 2’-O-methylguanosine in tRNA from an extreme thermophile, Thermus thermophilus HB27. Proc. Natl. Acad. Sci. USA 1980, 77, 1922–1926. [Google Scholar] [CrossRef] [PubMed]
- Droogmans, L.; Roovers, M.; Bujnicki, J.M.; Tricot, C.; Hartsch, T.; Stalon, V.; Grosjean, H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003, 31, 2148–2156. [Google Scholar] [CrossRef] [PubMed]
- Tomikawa, C.; Yokogawa, T.; Kanai, T.; Hori, H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 2010, 38, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Abdel-Salam, G.M.H.; Guy, M.P.; Alomar, R.; Abdel-Hamid, M.S.; Afifi, H.H.; Ismail, S.I.; Emam, B.A.; Phizicky, E.M.; Alkuraya, F.S. Mutation in WDR4 impairs tRNA m7G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015, 16, 210. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Phan, L.; Cuesta, R.; Carlson, B.A.; Pak, M.; Asano, K.; Bjork, G.R.; Tamame, M.; Hinnebusch, A.G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998, 12, 3650–3662. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Fu, Y.; Pavon-Eternod, M.; He, C.; Pan, T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 2010, 16, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Cavallo, L.; Tramontano, A. Accurate energies of hydrogen bonded nucleic acid base pairs and triplets in tRNA tertiary interactions. Nucleic Acids Res. 2006, 34, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Morla-Folch, J.; Xie, H.N.; Alvarez-Puebla, R.A.; Guerrini, L. Fast optical chemical and structural classification of RNA. ACS Nano 2016, 10, 2834–2842. [Google Scholar] [CrossRef] [PubMed]
- Davanloo, P.; Sprinzl, M.; Watanabe, K.; Albani, M.; Kersten, H. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979, 6, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Dalluge, J.J.; Hashizume, T.; Sopchik, A.E.; McCloskey, J.A.; Davis, D.R. Conformational flexibility in RNA: The role of dihydrouridine. Nucleic Acids Res. 1996, 24, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Dyubankova, N.; Sochacka, E.; Kraszewska, K.; Nawrot, B.; Herdewijn, P.; Lescrinier, E. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem. 2015, 13, 4960–4966. [Google Scholar] [CrossRef] [PubMed]
- Nawrot, B.; Malkiewicz, A.; Smith, W.S.; Sierzputowska-Gracz, H.; Agris, P.F. RNA modified uridines VII: Chemical synthesis and initial analysis of tRNA D-Loop oligomers with tandem modified uridines. Nucleosides Nucleotides 1995, 14, 143–165. [Google Scholar] [CrossRef]
- Stuart, J.W.; Basti, M.M.; Smith, W.S.; Forrest, B.; Guenther, R.; Sierzputowska-Gracz, H.; Nawrot, B.; Malkiewicz, A.; Agris, P.F. Structure of the trinucleotide D-acp3U-A with coordinated Mg2+ demonstrates that modified nucleosides contribute to regional conformations of RNA. Nucleosides Nucleotides 1996, 15, 1009–1028. [Google Scholar] [CrossRef]
- Suck, D.; Saenger, W.; Zechmeister, K. Molecular and crystal structure of the tRNA minor constituent dihydrouridine. Acta Crystallogr. Sect. B: Struct. Sci. 1972, 28, 596–605. [Google Scholar] [CrossRef]
- Egert, E.; Lindner, H.J.; Hillen, W.; Boehm, M.C. Influence of substituents at the 5 position on the structure of uridine. J. Am. Chem. Soc. 1980, 102, 3707–3713. [Google Scholar] [CrossRef]
- Uhl, W.; Reiner, J.; Gassen, H.G. On the conformation of 5-substituted uridines as studied by proton magnetic resonance. Nucleic Acids Res. 1983, 11, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Sundaralingam, M.; Rao, S.T.; Abola, J. Stereochemistry of nucleic acids and their constituents. XXIII. Crystal and molecular structure of dihydrouridine “hemihydrate”, a rare nucleoside with a saturated base occurring in the dihydrouridine loop of transfer ribonucleic acids. J. Am. Chem. Soc. 1971, 93, 7055–7062. [Google Scholar] [CrossRef] [PubMed]
- Deslauriers, R.; Smith, I.C.P. A comparison of the conformations of uridine, β-pseudouridine, and dihydrouridine in dimethyl sulfoxide and water. A 1H nuclear magnetic resonance study. Can. J. Chem. 1973, 51, 833–838. [Google Scholar] [CrossRef]
- Sipa, K.; Sochacka, E.; Kazmierczak-Baranska, J.; Maszewska, M.; Janicka, M.; Nowak, G.; Nawrot, B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA 2007, 13, 1301–1316. [Google Scholar] [CrossRef] [PubMed]
- Westhof, E.; Sundaralingam, M. Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles. Biochemistry 1986, 25, 4868–4878. [Google Scholar] [CrossRef] [PubMed]
- Westhof, E.; Dumas, P.; Moras, D. Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol. 1985, 184, 119–145. [Google Scholar] [CrossRef]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [PubMed]
- Dalluge, J.J.; Hamamoto, T.; Horikoshi, K.; Morita, R.Y.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional modification of tRNA in psychrophilic bacteria. J. Bacteriol. 1997, 179, 1918–1923. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Oshima, T.; Iijima, K.; Yamaizumi, Z.; Nishimura, S. Purification and thermal stability of several amino acid-specific tRNAs from an extreme thermophile, Thermus thermophilus HB8. J. Biochem. 1980, 87, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Yokoyama, S.; Miyazawa, T.; Watanabe, K.; Higuchi, S. NMR analyses on the molecular mechanism of the conformational rigidity of 2-thioribothymidine, a modified nucleoside in extreme thermophile tRNAs. FEBS Lett. 1983, 157, 95–99. [Google Scholar] [CrossRef]
- Horie, N.; Hara-Yokoyama, M.; Yokoyama, S.; Watanabe, K.; Kuchino, Y.; Nishimura, S.; Miyazawa, T. Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: Effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 1985, 24, 5711–5715. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.A.; Bloomingdale, R.J.; Znosko, B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA 2013, 19, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [PubMed]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Hamma, T.; Ferre-D’Amare, A.R. Pseudouridine synthases. Chem. Biol. 2006, 13, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Veerareddygari, G.R.; Singh, S.K.; Mueller, E.G. The pseudouridine synthases proceed through a glycal intermediate. J. Am. Chem. Soc. 2016, 138, 7852–7855. [Google Scholar] [CrossRef] [PubMed]
- Spenkuch, F.; Motorin, Y.; Helm, M. Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biol. 2014, 11, 1540–1554. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995, 23, 5020–5026. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Vanommeslaeghe, K.; Aleksandrov, A.; MacKerell, A.D., Jr.; Nilsson, L. Additive CHARMM force field for naturally occurring modified ribonucleotides. J. Comput. Chem. 2016, 37, 896–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanda, R.; Tewari, R.; Govil, G.; Smith, I.C. The conformation of β-pseudouridine about the glycosidic bond as studied by 1H homonuclear overhauser measurements and molecular orbital calculations. Can. J. Chem. 1974, 52, 371–375. [Google Scholar] [CrossRef]
- Neumann, J.M.; Bernassau, J.M.; Gueron, M.; Tran-Dinh, S. Comparative conformations of uridine and pseudouridine and their derivatives. Eur. J. Biochem. 1980, 108, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Yarian, C.S.; Basti, M.M.; Cain, R.J.; Ansari, G.; Guenther, R.H.; Sochacka, E.; Czerwinska, G.; Malkiewicz, A.; Agris, P.F. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucleic Acids Res. 1999, 27, 3543–3549. [Google Scholar] [CrossRef] [PubMed]
- Arluison, V.; Buckle, M.; Grosjean, H. Pseudouridine synthetase pus1 of Saccharomyces cerevisiae: Kinetic characterisation, tRNA structural requirement and real-time analysis of its complex with tRNA1. J. Mol. Biol. 1999, 289, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Juhling, F.; Morl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Putz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–162. [Google Scholar] [CrossRef] [PubMed]
- Lusic, H.; Gustilo, E.M.; Vendeix, F.A.; Kaiser, R.; Delaney, M.O.; Graham, W.D.; Moye, V.A.; Cantara, W.A.; Agris, P.F.; Deiters, A. Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res. 2008, 36, 6548–6557. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.W.; Koshlap, K.M.; Guenther, R.; Agris, P.F. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J. Mol. Biol. 2003, 334, 901–918. [Google Scholar] [CrossRef] [PubMed]
- Yarian, C.; Marszalek, M.; Sochacka, E.; Malkiewicz, A.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry 2000, 39, 13390–13395. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.R.; Durant, P.C. Nucleoside modifications affect the structure and stability of the anticodon of tRNALys,3. Nucleosides Nucleotides 1999, 18, 1579–1581. [Google Scholar] [CrossRef] [PubMed]
- Denmon, A.P.; Wang, J.; Nikonowicz, E.P. Conformation effects of base modification on the anticodon stem-loop of Bacillus subtilis tRNATyr. J. Mol. Biol. 2011, 412, 285–303. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Villegas, J.; Winkler, M.E.; Nikonowicz, E.P. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J. Mol. Biol. 2002, 319, 1015–1034. [Google Scholar] [CrossRef]
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002, 277, 16391–16395. [Google Scholar] [CrossRef] [PubMed]
- Kurata, S.; Weixlbaumer, A.; Ohtsuki, T.; Shimazaki, T.; Wada, T.; Kirino, Y.; Takai, K.; Watanabe, K.; Ramakrishnan, V.; Suzuki, T. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008, 283, 18801–18811. [Google Scholar] [CrossRef] [PubMed]
- Phelps, S.S.; Malkiewicz, A.; Agris, P.F.; Joseph, S. Modified nucleotides in tRNALys and tRNAVal are important for translocation. J. Mol. Biol. 2004, 338, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.S.; Sochacka, E.; Cain, R.; Guenther, R.; Malkiewicz, A.; Agris, P.F. Single atom modification (O→S) of tRNA confers ribosome binding. RNA 1999, 5, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Esberg, B.; Curran, J.F.; Bjork, G.R. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 1997, 271, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Chiari, Y.; Dion, K.; Colborn, J.; Parmakelis, A.; Powell, J.R. On the possible role of tRNA base modifications in the evolution of codon usage: Queuosine and Drosophila. J. Mol. Evol. 2010, 70, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Ninio, J. Multiple stages in codon-anticodon recognition: Double-trigger mechanisms and geometric constraints. Biochimie 2006, 88, 963–992. [Google Scholar] [CrossRef] [PubMed]
- Gustilo, E.M.; Vendeix, F.A.; Agris, P.F. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008, 11, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Urbonavicius, J.; Qian, Q.; Durand, J.M.; Hagervall, T.G.; Bjork, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001, 20, 4863–4873. [Google Scholar] [CrossRef] [PubMed]
- Bjork, G.R.; Durand, J.M.; Hagervall, T.G.; Leipuviene, R.; Lundgren, H.K.; Nilsson, K.; Chen, P.; Qian, Q.; Urbonavicius, J. Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett. 1999, 452, 47–51. [Google Scholar] [CrossRef]
- Brierley, I.; Meredith, M.R.; Bloys, A.J.; Hagervall, T.G. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: Influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 1997, 270, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F. Bringing order to translation: The contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008, 9, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Lagerkvist, U. “Two out of three”: An alternative method for codon reading. Proc. Natl. Acad. Sci. USA 1978, 75, 1759–1762. [Google Scholar] [CrossRef] [PubMed]
- Lagerkvist, U. Unorthodox codon reading and the evolution of the genetic code. Cell 1981, 23, 305–306. [Google Scholar] [CrossRef]
- Wakasugi, T.; Ohme, M.; Shinozaki, K.; Sugiura, M. Structures of tobacco chloroplast genes for tRNAIle (CAU), tRNALeu (CAA), tRNACys (GCA), tRNASer (UGA) and tRNAThr (GGU): A compilation of tRNA genes from tobacco chloroplasts. Plant Mol. Biol. 1986, 7, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, M.; Karcher, D.; Bock, R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008, 15, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Weixlbaumer, A.; Murphy, F.V.t.; Dziergowska, A.; Malkiewicz, A.; Vendeix, F.A.; Agris, P.F.; Ramakrishnan, V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007, 14, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Ishikura, H.; Yamada, Y.; Nishimura, S. Structure of serine tRNA from Escherichia coli. I. Purification of serine tRNA’s with different codon responses. Biochim. Biophys. Acta 1971, 228, 471–481. [Google Scholar] [CrossRef]
- Mitra, S.K.; Lustig, F.; Akesson, B.; Axberg, T.; Elias, P.; Lagerkvist, U. Relative efficiency of anticodons in reading the valine codons during protein synthesis in vitro. J. Biol. Chem. 1979, 254, 6397–6401. [Google Scholar] [PubMed]
- Samuelsson, T.; Elias, P.; Lustig, F.; Axberg, T.; Folsch, G.; Akesson, B.; Lagerkvist, U. Aberrations of the classic codon reading scheme during protein synthesis in vitro. J. Biol. Chem. 1980, 255, 4583–4588. [Google Scholar] [PubMed]
- Sakamoto, K.; Kawai, G.; Niimi, T.; Satoh, T.; Sekine, M.; Yamaizumi, Z.; Nishimura, S.; Miyazawa, T.; Yokoyama, S. A modified uridine in the first position of the anticodon of a minor species of arginine tRNA, the argU gene product, from Escherichia coli. Eur. J. Biochem. 1993, 216, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Spanjaard, R.A.; Chen, K.; Walker, J.R.; van Duin, J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: Assigning a codon to argU tRNA and T4 tRNAArg. Nucleic Acids Res. 1990, 18, 5031–5036. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.; Esberg, A.; Huang, B.; Bjork, G.R.; Bystrom, A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef] [PubMed]
- Takai, K.; Yokoyama, S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003, 31, 6383–6391. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Guenther, R.; Ingram, P.C.; Basti, M.M.; Stuart, J.W.; Sochacka, E.; Malkiewicz, A. Unconventional structure of tRNALysSUU anticodon explains tRNA’s role in bacterial and mammalian ribosomal frameshifting and primer selection by HIV-1. RNA 1997, 3, 420–428. [Google Scholar] [PubMed]
- Agris, P.F.; Soll, D.; Seno, T. Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry 1973, 12, 4331–4337. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Watanabe, T.; Murao, K.; Ishikura, H.; Yamaizumi, Z.; Nishimura, S.; Miyazawa, T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. USA 1985, 82, 4905–4909. [Google Scholar] [CrossRef] [PubMed]
- Lustig, F.; Elias, P.; Axberg, T.; Samuelsson, T.; Tittawella, I.; Lagerkvist, U. Codon reading and translational error. Reading of the glutamine and lysine codons during protein synthesis in vitro. J. Biol. Chem. 1981, 256, 2635–2643. [Google Scholar] [PubMed]
- Bilbille, Y.; Vendeix, F.A.; Guenther, R.; Malkiewicz, A.; Ariza, X.; Vilarrasa, J.; Agris, P.F. The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Res. 2009, 37, 3342–3353. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Nishimura, S. Modified nucleosides and codon recognition. In tRNA: Structure, Biosynthesis and Function; Schimmel, P.R., Söll, D., RajBhandary, U.L., Eds.; ASM Press: Washington, DC, USA, 1995; pp. 207–233. [Google Scholar]
- Yasukawa, T.; Suzuki, T.; Ishii, N.; Ueda, T.; Ohta, S.; Watanabe, K. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000, 467, 175–178. [Google Scholar] [CrossRef]
- Kirino, Y.; Suzuki, T. Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol. 2005, 2, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Kirino, Y.; Ishii, N.; Holt, I.J.; Jacobs, H.T.; Makifuchi, T.; Fukuhara, N.; Ohta, S.; Suzuki, T.; Watanabe, K. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 2005, 579, 2948–2952. [Google Scholar] [CrossRef] [PubMed]
- Kamble, A.S.; Kumbhar, B.V.; Sambhare, S.B.; Bavi, R.S.; Sonawane, K.D. Conformational preferences of modified nucleoside 5-taurinomethyluridine, τm5U occur at ‘wobble’ 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys. 2015, 71, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Vangaveti, S.; Ranganathan, S.V.; Basanta-Sanchez, M.; Haruehanroengra, P.; Chen, A.; Sheng, J. Synthesis, base pairing and structure studies of geranylated RNA. Nucleic Acids Res. 2016, 44, 6036–6045. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ranganathan, S.V.; Basanta-Sanchez, M.; Shen, F.; Chen, A.; Sheng, J. Synthesis and base pairing studies of geranylated 2-thiothymidine, a natural variant of thymidine. Chem. Commun. 2015, 51, 16369–16372. [Google Scholar] [CrossRef] [PubMed]
- Sierant, M.; Leszczynska, G.; Sadowska, K.; Dziergowska, A.; Rozanski, M.; Sochacka, E.; Nawrot, B. S-Geranyl-2-thiouridine wobble nucleosides of bacterial tRNAs; chemical and enzymatic synthesis of S-geranylated-RNAs and their physicochemical characterization. Nucleic Acids Res. 2016, 44, 10986–10998. [Google Scholar] [CrossRef] [PubMed]
- Andachi, Y.; Yamao, F.; Iwami, M.; Muto, A.; Osawa, S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc. Natl. Acad. Sci. USA 1987, 84, 7398–7402. [Google Scholar] [CrossRef] [PubMed]
- Andachi, Y.; Yamao, F.; Muto, A.; Osawa, S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 1989, 209, 37–54. [Google Scholar] [CrossRef]
- Guindy, Y.S.; Samuelsson, T.; Johansen, T.I. Unconventional codon reading by Mycoplasma mycoides tRNAs as revealed by partial sequence analysis. Biochem. J. 1989, 258, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Samuelsson, T.; Guindy, Y.S.; Lustig, F.; Boren, T.; Lagerkvist, U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc. Natl. Acad. Sci. USA 1987, 84, 3166–3170. [Google Scholar] [CrossRef] [PubMed]
- Sibler, A.P.; Dirheimer, G.; Martin, R.P. Codon reading patterns in Saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett. 1986, 194, 131–138. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tsurui, H.; Ueda, T.; Furusihima-Shimogawara, R.; Takamiya, S.; Kita, K.; Nishikawa, K.; Watanabe, K. Primary sequence of mitochondrial tRNAArg of a nematode Ascaris suum: occurrence of unmodified adenosine at the first position of the anticodon. Biochim. Biophys. Acta 1997, 1350, 119–122. [Google Scholar] [CrossRef]
- Murphy, F.V.t.; Ramakrishnan, V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004, 11, 1251–1252. [Google Scholar] [CrossRef] [PubMed]
- Deffit, S.N.; Hundley, H.A. To edit or not to edit: Regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 2016, 7, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Zinshteyn, B.; Nishikura, K. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Mannion, N.; Arieti, F.; Gallo, A.; Keegan, L.P.; O’Connell, M.A. New insights into the biological role of mammalian ADARs; the RNA editing proteins. Biomolecules 2015, 5, 2338–2362. [Google Scholar] [CrossRef] [PubMed]
- Vendeix, F.A.; Murphy, F.V.t.; Cantara, W.A.; Leszczynska, G.; Gustilo, E.M.; Sproat, B.; Malkiewicz, A.; Agris, P.F. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol. 2012, 416, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Rozov, A.; Demeshkina, N.; Khusainov, I.; Westhof, E.; Yusupov, M.; Yusupova, G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016, 7, 10457. [Google Scholar] [CrossRef] [PubMed]
- Rozov, A.; Westhof, E.; Yusupov, M.; Yusupova, G. The ribosome prohibits the G*U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res. 2016, 44, 6434–6441. [Google Scholar] [CrossRef] [PubMed]
- Kimsey, I.J.; Petzold, K.; Sathyamoorthy, B.; Stein, Z.W.; Al-Hashimi, H.M. Visualizing transient Watson–Crick-like mispairs in DNA and RNA duplexes. Nature. 2015, 519, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Murphy, F.V.t.; Demirci, H.; Agris, P.F. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. USA 2013, 110, 10964–10969. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016, 44, 8020–8040. [Google Scholar] [CrossRef] [PubMed]
- Manickam, N.; Joshi, K.; Bhatt, M.J.; Farabaugh, P.J. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res. 2016, 44, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Vendeix, F.A.; Dziergowska, A.; Gustilo, E.M.; Graham, W.D.; Sproat, B.; Malkiewicz, A.; Agris, P.F. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry 2008, 47, 6117–6129. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.W.; Gdaniec, Z.; Guenther, R.; Marszalek, M.; Sochacka, E.; Malkiewicz, A.; Agris, P.F. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 2000, 39, 13396–13404. [Google Scholar] [CrossRef] [PubMed]
- Durant, P.C.; Bajji, A.C.; Sundaram, M.; Kumar, R.K.; Davis, D.R. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: The effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 2005, 44, 8078–8089. [Google Scholar] [CrossRef] [PubMed]
- Witts, R.N.; Hopson, E.C.; Koballa, D.E.; Van Boening, T.A.; Hopkins, N.H.; Patterson, E.V.; Nagan, M.C. Backbone-base interactions critical to quantum stabilization of transfer RNA anticodon structure. J. Phys. Chem. B 2013, 117, 7489–7497. [Google Scholar] [CrossRef] [PubMed]
- McCrate, N.E.; Varner, M.E.; Kim, K.I.; Nagan, M.C. Molecular dynamics simulations of human tRNALys,3UUU: The role of modified bases in mRNA recognition. Nucleic Acids Res. 2006, 34, 5361–5368. [Google Scholar] [CrossRef] [PubMed]
- Murphy, F.V.; Ramakrishnan, V.; Malkiewicz, A.; Agris, P.F. The role of modifications in codon discrimination by tRNALysUUU. Nat. Struct. Mol. Biol. 2004, 11, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Blewett, N.H.; Crawford, A.K.; Cherkasova, V.A.; Iben, J.R.; Begley, T.J.; Farabaugh, P.J.; Maraia, R.J. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol. Cell. Biol. 2013, 33, 2918–2929. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Mattijssen, S.; Maraia, R.J. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol. Cell. Biol. 2013, 33, 4900–4908. [Google Scholar] [CrossRef] [PubMed]
- Urbonavicius, J. Transfer RNA modifications that alter +1 frameshifting in general fail to affect -1 frameshifting. RNA 2003, 9, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.Y.; Tomizawa, K. Development of type 2 diabetes caused by a deficiency of a tRNAlys modification. Islets 2012, 4, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Wei, F.Y.; Watanabe, S.; Hirayama, M.; Ohuchi, Y.; Fujimura, A.; Kaitsuka, T.; Ishii, I.; Sawa, T.; Nakayama, H.; et al. Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res. 2017, 45, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Waas, W.F.; Druzina, Z.; Hanan, M.; Schimmel, P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 2007, 282, 26026–26034. [Google Scholar] [CrossRef] [PubMed]
- De Crecy-Lagard, V.; Brochier-Armanet, C.; Urbonavicius, J.; Fernandez, B.; Phillips, G.; Lyons, B.; Noma, A.; Alvarez, S.; Droogmans, L.; Armengaud, J.; et al. Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 2010, 27, 2062–2077. [Google Scholar] [CrossRef] [PubMed]
- Sample, P.J.; Koreny, L.; Paris, Z.; Gaston, K.W.; Rubio, M.A.; Fleming, I.M.; Hinger, S.; Horakova, E.; Limbach, P.A.; Lukes, J.; et al. A common tRNA modification at an unusual location: The discovery of wyosine biosynthesis in mitochondria. Nucleic Acids Res. 2015, 43, 4262–4273. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Schmidt, P.G.; Petsch, J.; Agris, P.F. Nuclear magnetic resonance signal assignments of purified [13C]methyl-enriched yeast phenylalanine transfer ribonucleic acid. Biochemistry 1985, 24, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Matuszewski, M.; Sochacka, E. Stability studies on the newly discovered cyclic form of tRNA N6-threonylcarbamoyladenosine (ct6A). Bioorg. Med. Chem. Lett. 2014, 24, 2703–2706. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Kimura, S.; Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013, 9, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.-i.; Miyauchi, K.; Matuszewski, M.; D’Almeida, G.S.; Rubio, M.A.T.; Alfonzo, J.D.; Inoue, K.; Sakaguchi, Y.; Suzuki, T.; Sochacka, E.; et al. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 2017, 45, 2124–2136. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Sussman, J.L.; Suddath, F.L.; Quigley, G.J.; McPherson, A.; Wang, A.H.J.; Seeman, N.C.; Rich, A. The general structure of transfer RNA molecules. Proc. Natl. Acad. Sci. USA 1974, 71, 4970–4974. [Google Scholar] [CrossRef] [PubMed]
- Ponnuswamy, P.K.; Gromiha, M.M. On the conformational stability of oligonucleotide duplexes and tRNA molecules. J. Theor. Biol. 1994, 169, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Bjork, G.R.; Wikstrom, P.M.; Bystrom, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 1989, 244, 986–989. [Google Scholar] [CrossRef] [PubMed]
- Gamper, H.B.; Masuda, I.; Frenkel-Morgenstern, M.; Hou, Y.M. Maintenance of protein synthesis reading frame by EF-P and m1G37-tRNA. Nat. Commun. 2015, 6, 7226. [Google Scholar] [CrossRef] [PubMed]
- Brulé, H.; Holmes, W.M.; Keith, G.; Giegé, R.; Florentz, C. Effect of a mutation in the anticodon of human mitochondrial tRNAPro on its post-transcriptional modification pattern. Nucleic Acids Res. 1998, 26, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Swinehart, W.E.; Jackman, J.E. Diversity in mechanism and function of tRNA methyltransferases. RNA Biol. 2015, 12, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Brulé, H.; Elliott, M.; Redlak, M.; Zehner, Z.E.; Holmes, W.M. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 2004, 43, 9243–9255. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.A.; Kopajtich, R.; D’Souza, A.R.; Rorbach, J.; Kremer, L.S.; Husain, R.A.; Dallabona, C.; Donnini, C.; Alston, C.L.; Griffin, H.; et al. TRMT5 mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am. J. Hum. Genet. 2015, 97, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Yakovchuk, P.; Protozanova, E.; Frank-Kamenetskii, M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Butcher, S.E.; Pyle, A.M. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011, 44, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Bujalowski, W.; Graeser, E.; McLaughlin, L.W.; Porschke, D. Anticodon loop of tRNAPhe: Structure, dynamics, and Mg2+ binding. Biochemistry 1986, 25, 6365–6371. [Google Scholar] [CrossRef] [PubMed]
- Wells, B.D. The conformation of the tRNAPhe anticodon loop monitored by fluorescence. Nucleic Acids Res. 1984, 12, 2157–2170. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, J.M.; Kearns, D.R. Europium as a fluorescent probe of transfer RNA structure. Biochemistry 1975, 14, 1436–1444. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sierzputowska-Gracz, H.; Guenther, R.; Everett, K.; Agris, P.F. 5-methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry 1993, 32, 10249–10253. [Google Scholar] [CrossRef] [PubMed]
- Dao, V.; Guenther, R.H.; Agris, P.F. The role of 5-methylcytidine in the anticodon arm of yeast tRNAPhe: site-specific Mg2+ binding and coupled conformational transition in DNA analogs. Biochemistry 1992, 31, 11012–11019. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Kintanar, A.; Horowitz, J. Nucleoside modifications stabilize Mg2+ binding in Escherichia coli tRNAVal: An imino proton NMR investigation. Biochemistry 1994, 33, 8905–8911. [Google Scholar] [CrossRef] [PubMed]
- Maglott, E.J.; Deo, S.S.; Przykorska, A.; Glick, G.D. Conformational transitions of an unmodified tRNA: Implications for RNA folding. Biochemistry 1998, 37, 16349–16359. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.; Spencer, A.C.; Hsu, J.L.; Spremulli, L.; Martinis, S.A.; DeRider, M.; Agris, P.F. A counterintuitive Mg2+-dependent and modification-assisted functional folding of mitochondrial tRNAs. J. Mol. Biol. 2006, 362, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, M.P.; De, N.; Pulsipher, M.; Brown, M.; Reddy, P.R.; Petrie, C.R., 3rd; Chheda, G.B. Quantitative aspects of metal ion binding to certain transfer RNA anticodon loop modified nucleosides. Biochim. Biophys. Acta 1984, 802, 352–361. [Google Scholar] [CrossRef]
- Varnagy, K.; Jezowska-Bojczuk, M.; Swiatek, J.; Kozlowski, H.; Sovago, I.; Adamiak, R.W. Metal binding ability of hypermodified nucleosides of t-RNA. Potentiometric and spectroscopic studies on the metal complexes of N-[(9-β-d-ribofuranosylpurin-6-yl)-carbamoyl] threonine. J. Inorg. Biochem. 1990, 40, 357–363. [Google Scholar] [PubMed]
- Cabello-Villegas, J.; Tworowska, I.; Nikonowicz, E.P. Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem-loop of Escherichia coli tRNAPhe. Biochemistry 2004, 43, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, P.; Westhof, E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999, 292, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, M.; Uhlenbeck, O.C. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 2006, 88, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Baldi, M.I.; Mattoccia, E.; Bufardeci, E.; Fabbri, S.; Tocchini-Valentini, G.P. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 1992, 255, 1404–1408. [Google Scholar] [CrossRef] [PubMed]
- Ogle, J.M.; Brodersen, D.E.; Clemons, W.M., Jr.; Tarry, M.J.; Carter, A.P.; Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 2001, 292, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Bullock, T.L.; Uter, N.; Nissan, T.A.; Perona, J.J. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J. Mol. Biol. 2003, 328, 395–408. [Google Scholar] [CrossRef]
- Yaremchuk, A.; Tukalo, M.; Grotli, M.; Cusack, S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: Comparison with histidyl-tRNA synthetase. J. Mol. Biol. 2001, 309, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Bilbille, Y.; Kim, J.; Kaiser, R.; Leszczynska, G.; Malkiewicz, A.; Agris, P.F. Modifications modulate anticodon loop dynamics and codon recognition of E. coli tRNAArg1,2. J. Mol. Biol. 2012, 416, 579–597. [Google Scholar] [CrossRef] [PubMed]
- Vangaveti, S.; Ranganathan, S.V.; The RNA Institute, Albany, NY, USA. Personal communication, 2017.
- Ashraf, S.S.; Ansari, G.; Guenther, R.; Sochacka, E.; Malkiewicz, A.; Agris, P.F. The uridine in “U-turn”: Contributions to tRNA-ribosomal binding. RNA 1999, 5, 503–511. [Google Scholar] [CrossRef] [PubMed]
- von Ahsen, U.; Green, R.; Schroeder, R.; Noller, H.F. Identification of 2’-hydroxyl groups required for interaction of a tRNA anticodon stem-loop region with the ribosome. RNA 1997, 3, 49–56. [Google Scholar] [PubMed]
- Phelps, S.S.; Jerinic, O.; Joseph, S. Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol. Cell 2002, 10, 799–807. [Google Scholar] [CrossRef]
- Sundaram, M.; Durant, P.C.; Davis, D.R. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 2000, 39, 15652. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 1999, 286, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Björk, G.R.; Jacobsson, K.; Nilsson, K.; Johansson, M.J.; Bystrom, A.S.; Persson, O.P. A primordial tRNA modification required for the evolution of life? EMBO J. 2001, 20, 231–239. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Lyons, B.; Cruz, Y.; Reddy, R.; Nordin, B.; Agnelli, F.; Williamson, J.R.; Schimmel, P.; Swairjo, M.A.; de Crecy-Lagard, V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009, 37, 2894–2909. [Google Scholar] [CrossRef] [PubMed]
- Durant, P.C.; Davis, D.R. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999, 285, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Newby, M.I.; Greenbaum, N.L. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 2001, 7, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Astrom, S.U.; Bystrom, A.S. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell 1994, 79, 535–546. [Google Scholar] [CrossRef]
- Demeshkina, N.; Jenner, L.; Westhof, E.; Yusupov, M.; Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 2012, 484, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989, 53, 273–298. [Google Scholar] [PubMed]
- Roth, A.C. Decoding properties of tRNA leave a detectable signal in codon usage bias. Bioinformatics 2012, 28, i340–i348. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013, 194, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Chawla, M.; Oliva, R.; Bujnicki, J.M.; Cavallo, L. An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies. Nucleic Acids Res. 2015, 43, 6714–6729. [Google Scholar] [CrossRef] [PubMed]
- Vendeix, F.A.P.; Munoz, A.M.; Agris, P.F. Free energy calculation of modified base-pair formation in explicit solvent: A predictive model. RNA 2009, 15, 2278–2287. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Väre, V.Y.P.; Eruysal, E.R.; Narendran, A.; Sarachan, K.L.; Agris, P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules 2017, 7, 29. https://doi.org/10.3390/biom7010029
Väre VYP, Eruysal ER, Narendran A, Sarachan KL, Agris PF. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017; 7(1):29. https://doi.org/10.3390/biom7010029
Chicago/Turabian StyleVäre, Ville Y. P., Emily R. Eruysal, Amithi Narendran, Kathryn L. Sarachan, and Paul F. Agris. 2017. "Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function" Biomolecules 7, no. 1: 29. https://doi.org/10.3390/biom7010029