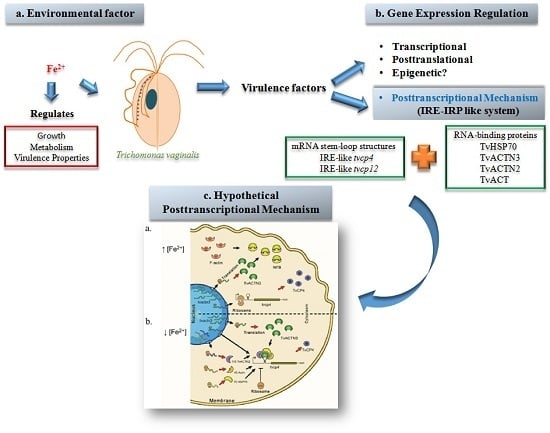
RNA-Binding Proteins in Trichomonas vaginalis: Atypical Multifunctional Proteins
Abstract
:1. Introduction
2. Iron Homeostasis: Intracelullar Regulation Mediated by the IRE/IRP System
2.1. IRPs and the Aconitase Family: General Characteristics and Functions
2.2. IREs, mRNA Stem-Loop Structures: Sequence and Location

2.3. IRE-IRP Interaction: A Key Step for the Posttranscriptional Regulation Mediated by Iron
2.4. Atypical IREs and IRPs
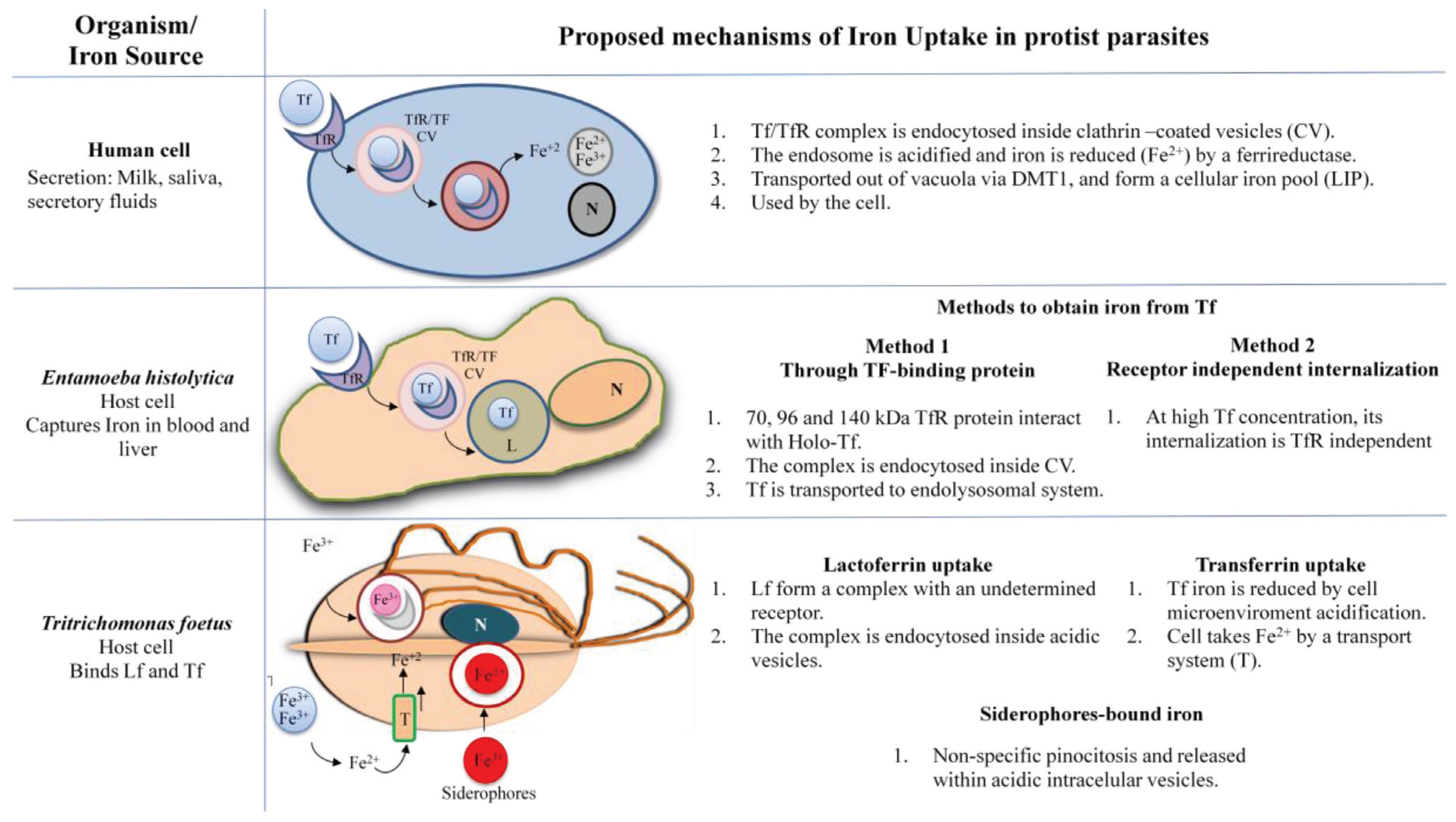
IRE-IRP Iron Regulation in Protists
3. Trichomonas vaginalis
3.1. Iron in the Biology of Trichomonas vaginalis
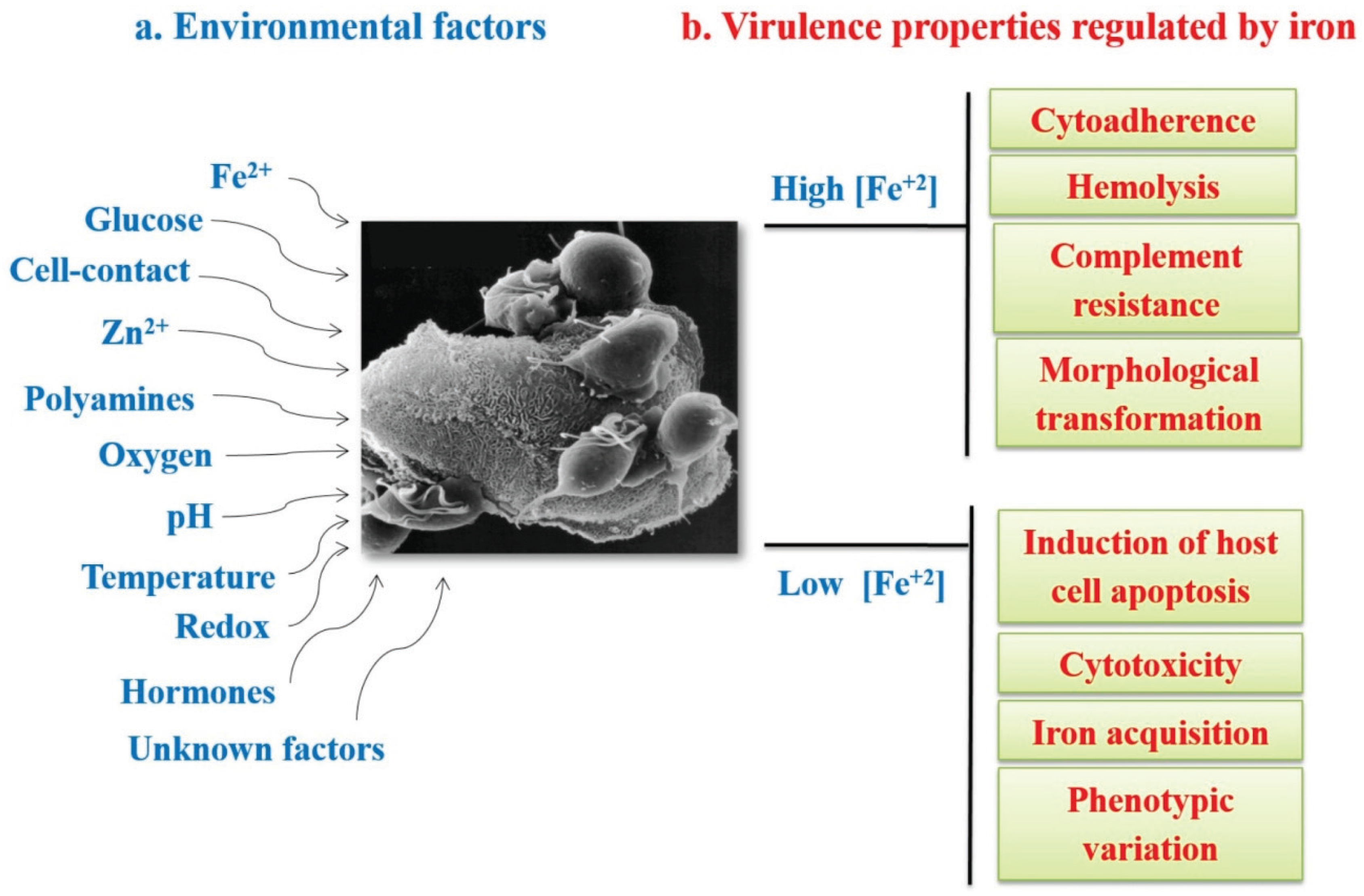
3.2. Iron Gene Expression Regulation in T. vaginalis
| Gene or Protein ID Number or NCBI Accession Number | Function | Type of Regulation | References |
|---|---|---|---|
| AP23 Uk | Adhesin/virulence factor involved in cytoadherence | ND | [92,110,111] |
| AP33 TVAG_047890 | Adhesin/ α-subunit of succinyl coenzyme synthase/virulence factor involved in cytoadherence | ND | [85,92,110,111,112] |
| AP51 TVAG_259190 | Adhesin/β-subunit of succinyl coenzyme synthase/virulence factor involved in cytoadherence | ND | [85,92,110,111,112] |
| AP65-1 TVAG_340290 | Adhesin Malic Enzyme/virulence factor involved in cytoadherence | Transcriptional level | [85,92,110,111,112,113,114,115,116,117,118] |
| AP120 TVAG_198110 | Adhesin/Pyruvate: Ferredoxin Oxidoreductase/virulence factor involved in cytoadherence | Transcriptional level Posttranslational level G | [85,86,111,112] |
| BspA-like TVAG_397210 TVAG_441420 | Adhesin/Cytoadherence Host immune evasion | Transcriptional level | [85,86,112] |
| TvCP4 TVAG_467970 | CP/Hemolysis | Posttranscriptional level IRE/IRP | [60,61,101,112] |
| TvCP62 Uk | CP/Cytoadherence | ND | [105] |
| TvENO-1 TVAG_464170 | FN receptor Enolase/Cytoadherence | Transcriptional level | [85,86,112] |
| TVF Uk | Cytolytic effector 250 kDa/Cytolytic activity | ND | [86] |
| TvGAPDH TVAG_146910 | Plasminogen receptor Glyceraldehyde-3-Phosphate Dehydrogenase/Cytoadherence | Transcriptional level | [80,84,86,112] |
| TvLEGU-1 TVAG_426660 | CP/Cytoadherence | Transcriptional level Posttranslational level PG | [85,96,108,109,112] |
| TvLIP AY870437 | Triacylglycerol Lipase/Hemolysis | ND | [85,86,112] |
| TvLPG Gl | Lipophosphoglycan/Cytoadherence and Cytotoxicity | Posttranslational level G | [85,86,119] |
| Ubiquitin TVAG_097660 TVAG_264700 | Ubiquitination | ND | [85,107,112] |
| EF1α TVAG_067400 TVAG_463940 | Elongation factor 1α/Translation | ND | [85,107,112] |
| Myb-3 TVAG_252420 | MYB-like transcription factor | Transcriptional level Posttranscriptional P level | [112,113,116,117,118] |
| TvACTN1 TVAG_156680 | α-Actinin 1/Actin binding protein | Transcriptional level | [85,120] |
| TvACTN2 TVAG_190450 | α-Actinin 2/Actin binding protein/ RNA-binding protein | Transcriptional level | [85,120] This article |
| TvACTN3 TVAG_239310 | α-Actinin-3/Actin binding protein/RNA-binding protein | Transcriptional level | [85,120] This article |
| TvHSP70 TVAG_044510 TVAG_139320 TVAG_092490 | Chaperone/RNA-binding protein | Transcriptional Level Posttranslational level U | [85,112] This article |
| Gene or Protein ID Number or NCBI accesion Number | Function | Type of Regulation | References |
|---|---|---|---|
| BspA-like TVAG_093850 TVAG_299910 | Adhesin/Cytoadherence Host immune evasion | Transcriptional level | [85,86,112] |
| P270 AAD40228.1 TVAG_379560 | Membrane Molecule/Phenotypic variation | Posttranslational P | [80,121] |
| TvCP12 TVAG_410260 | CP/Cytotoxicity | Posttranscriptional level IRE/IRP | ([85,86], our unpublished data) |
| TvCP30M | CP/Cytoadherence Protein Degradation | ND | [61,85,93,94,95,105] |
| TvCP39 TVAG_298080 | CP/Cytotoxicity Igs Degradation | Posttranslational level GC | [85,86,98,99,105] |
| TvCP65 TVAG_096740 | CP/Cytotoxicity | Transcriptional level Posttranslational level C | [85,86,97,105] |
| Myb-2 AY948337 TVAG_211210 | MYB-like transcription factor | Transcriptional level | [85,113,115] |
| TvHSP70 TVAG_163000 | Chaperone | ND | [85,107] |
Transcriptional Regulation by Iron in T. vaginalis
3.3. Posttranslational Regulation in T. vaginalis
4. Posttranscriptional Regulation by Iron in T. vaginalis: A Regulatory System Parallel to the IRE-IRP System in T. vaginalis
4.1. Atypical IREs in T. vaginalis
4.1.1. IRE-Like tvcp4
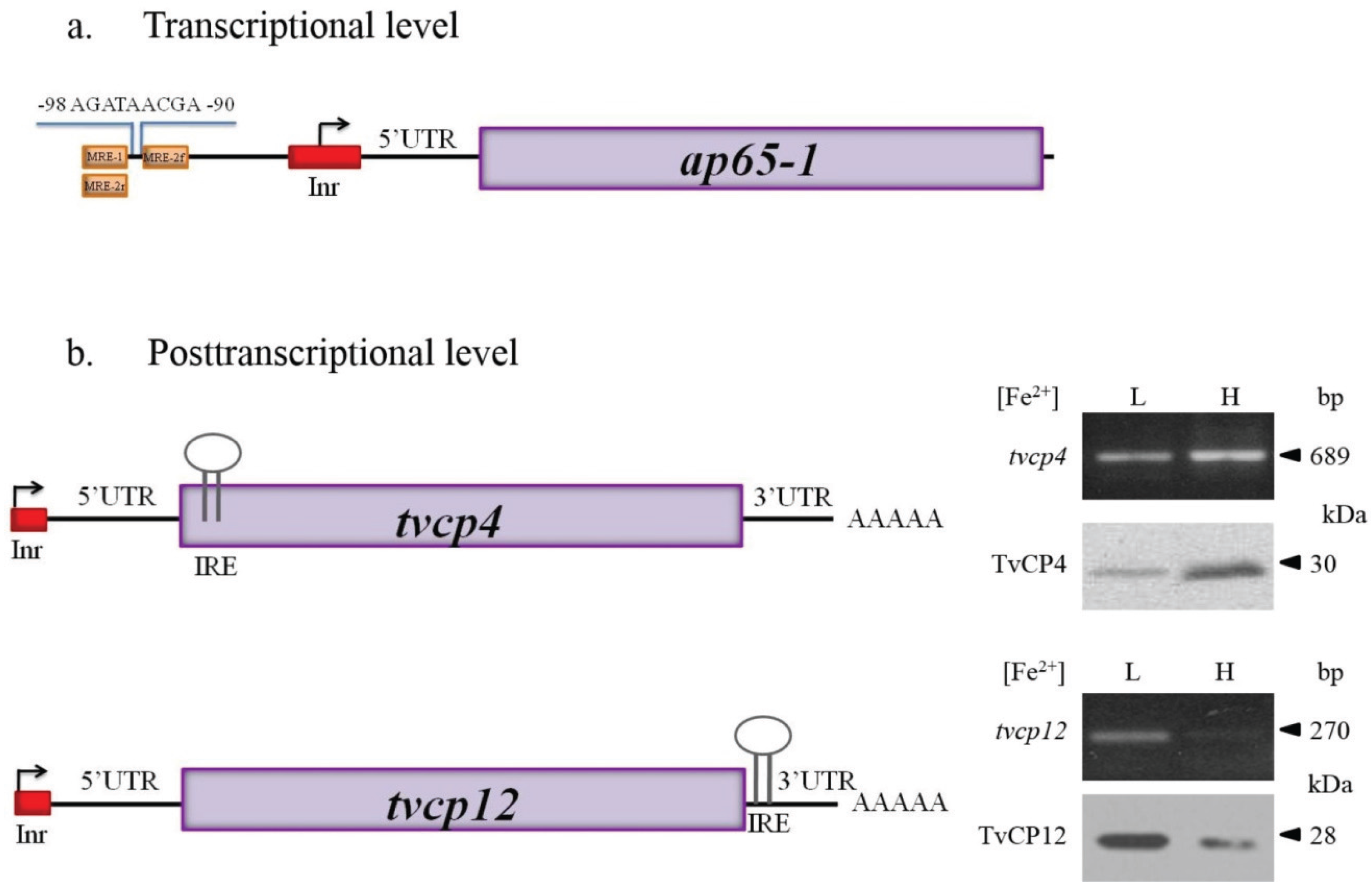
4.1.2. IRE-Like tvcp12
4.2. RNA-Binding Proteins in T. vaginalis
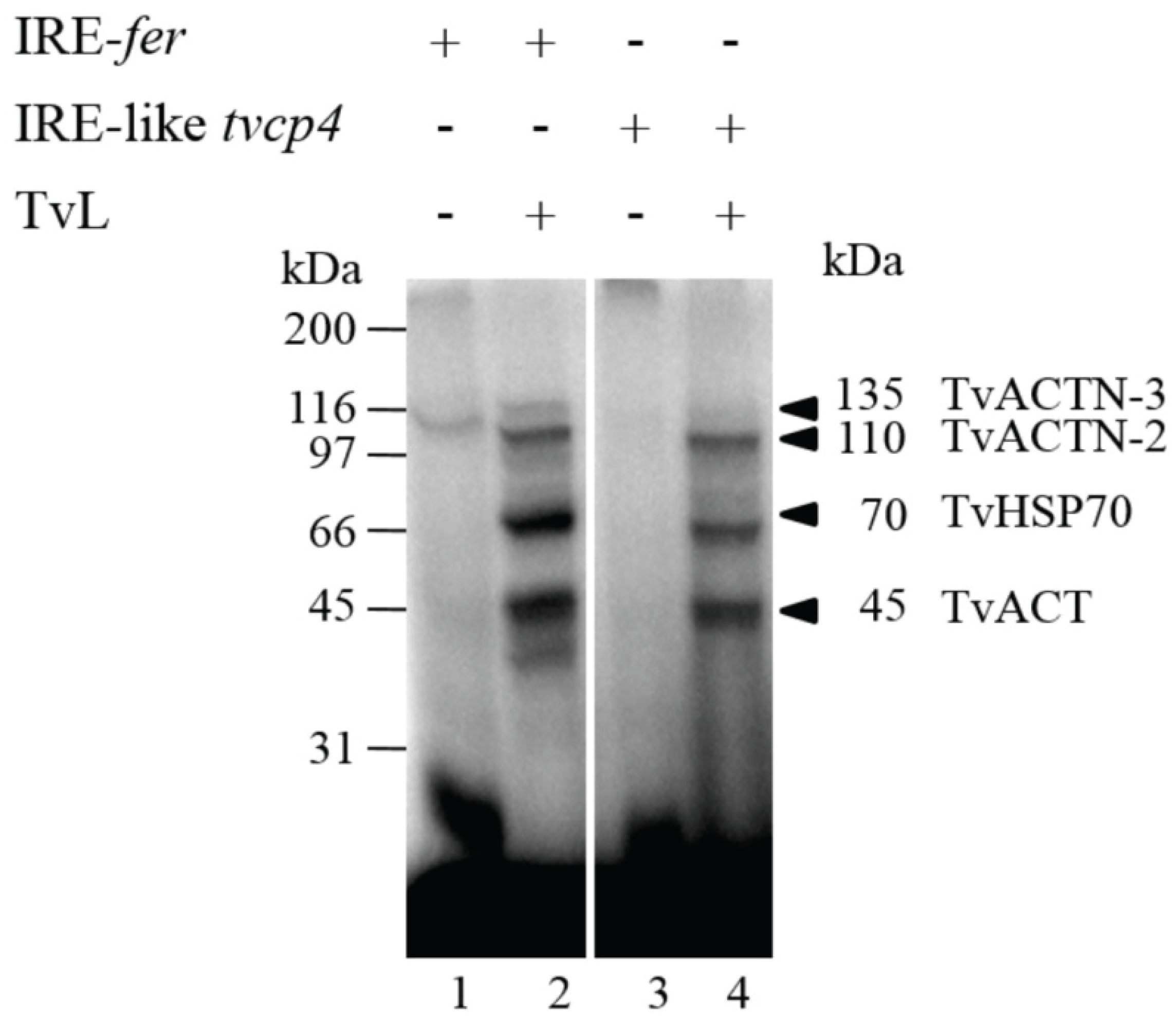
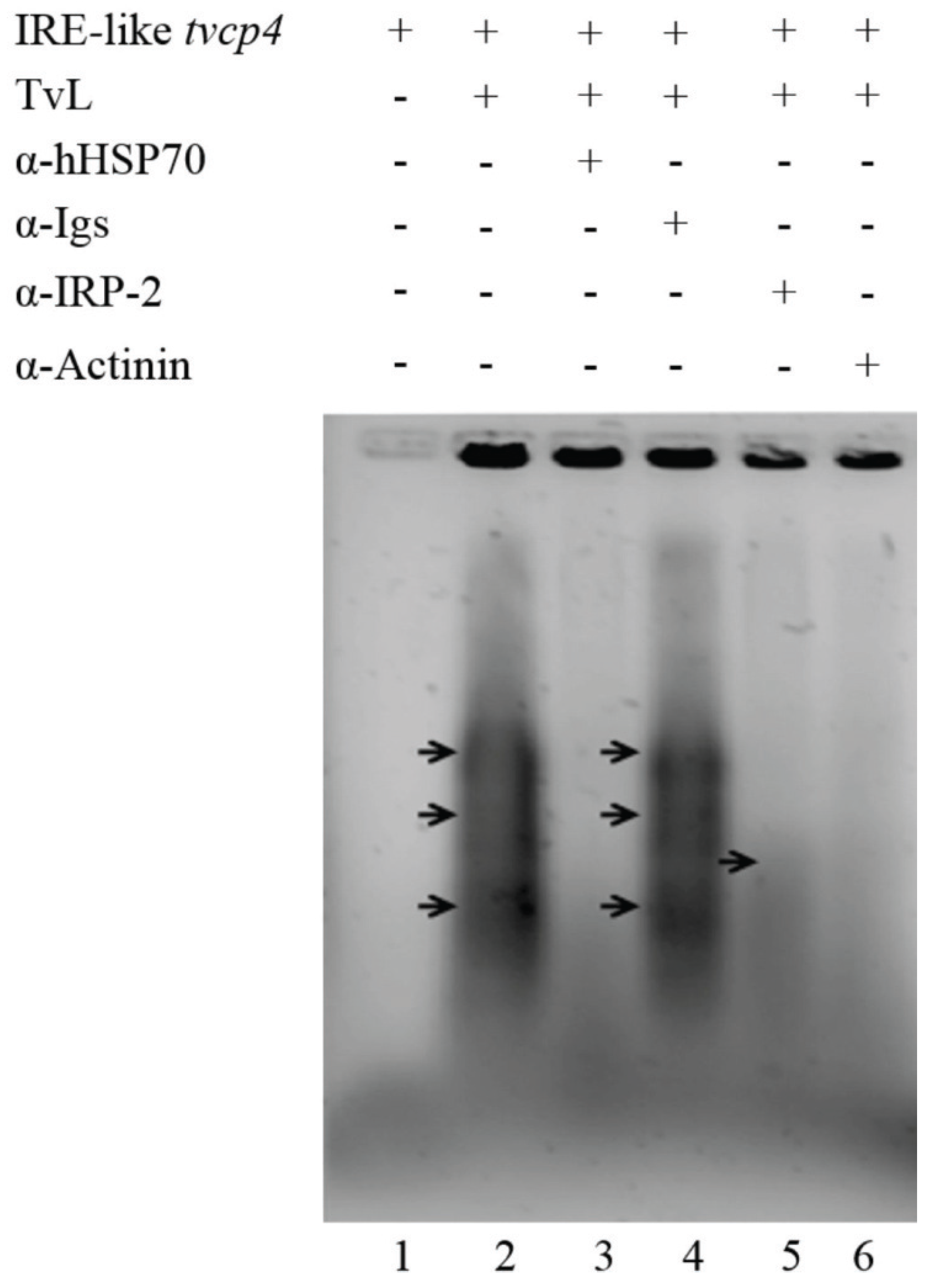
4.2.1. TvHSP70 as an RNA-Binding Protein in T. vaginalis
| ID | Name | Total EST a | Identity (%) |
|---|---|---|---|
| TVAG_044510 | HSP70-4 * | 66 | 100 |
| TVAG_161100 | HSP-70 putative | 16 | 96.52 |
| TVAG_151220 | HSP70-4 | 22 | 96.16 |
| TVAG_163000 | HSP putative | 17 | 96.00 |
| TVAG_137250 | HSP70-4 | 5 | 95.44 |
| TVAG_206270 | HSP putative | 14 | 95.80 |
| TVAG_300470 | HSP putative | 14 | 95.24 |
| TVAG_184320 | HSP putative | 12 | 95.39 |
| TVAG_171670 | HSP putative | 10 | 97.17 |
| TVAG_092490 | HSP-70 putative | 104 | 62.80 |
| TVAG_340390 | HSP70-4 | 25 | 57.06 |
| ID | Name | Total EST a | Identity (%) |
| TVAG_156680 | α-Actinin-1 | 112 | 51.66 |
| TVAG_190450 | α-Actinin-2 | 1086 | 61.96 |
| TVAG_239310 | α-Actinin-3* | 495 | 100 |
| TVAG_247460 | α-Actinin-4 | 2 | 41.92 |
| TVAG_260390 | α-Actinin-5 | 1 | 41.82 |
| ID | Name | Total EST a | Identity (%) |
| TVAG_054030 | actin putative * | 338 | 100 |
| TVAG_090470 | actin putative | 231 | 99.12 |
| TVAG_149090 | actin putative | 141 | 98.67 |
| TVAG_150270 | actin putative | 140 | 90.80 |
| TVAG_160060 | actin putative | 168 | 99.20 |
| TVAG_172680 | actin putative | 503 | 98.67 |
| TVAG_200190 | actin putative | 322 | 99.12 |
| TVAG_247170 | actin putative | 3 | 56.30 |
| TVAG_249200 | actin putative | 404 | 98.85 |
| TVAG_310030 | actin putative | 239 | 98.50 |
| TVAG_337240 | actin putative | 181 | 98.50 |
| TVAG_485210 | actin putative | 73 | 98.67 |
| TVAG_512800 | actin putative | 86 | 99.17 |
| TVAG_534990 | actin putative | 75 | 99.30 |
4.2.2. TvACTN3 and TvACTN2 as RNA-Binding Proteins in T. vaginalis
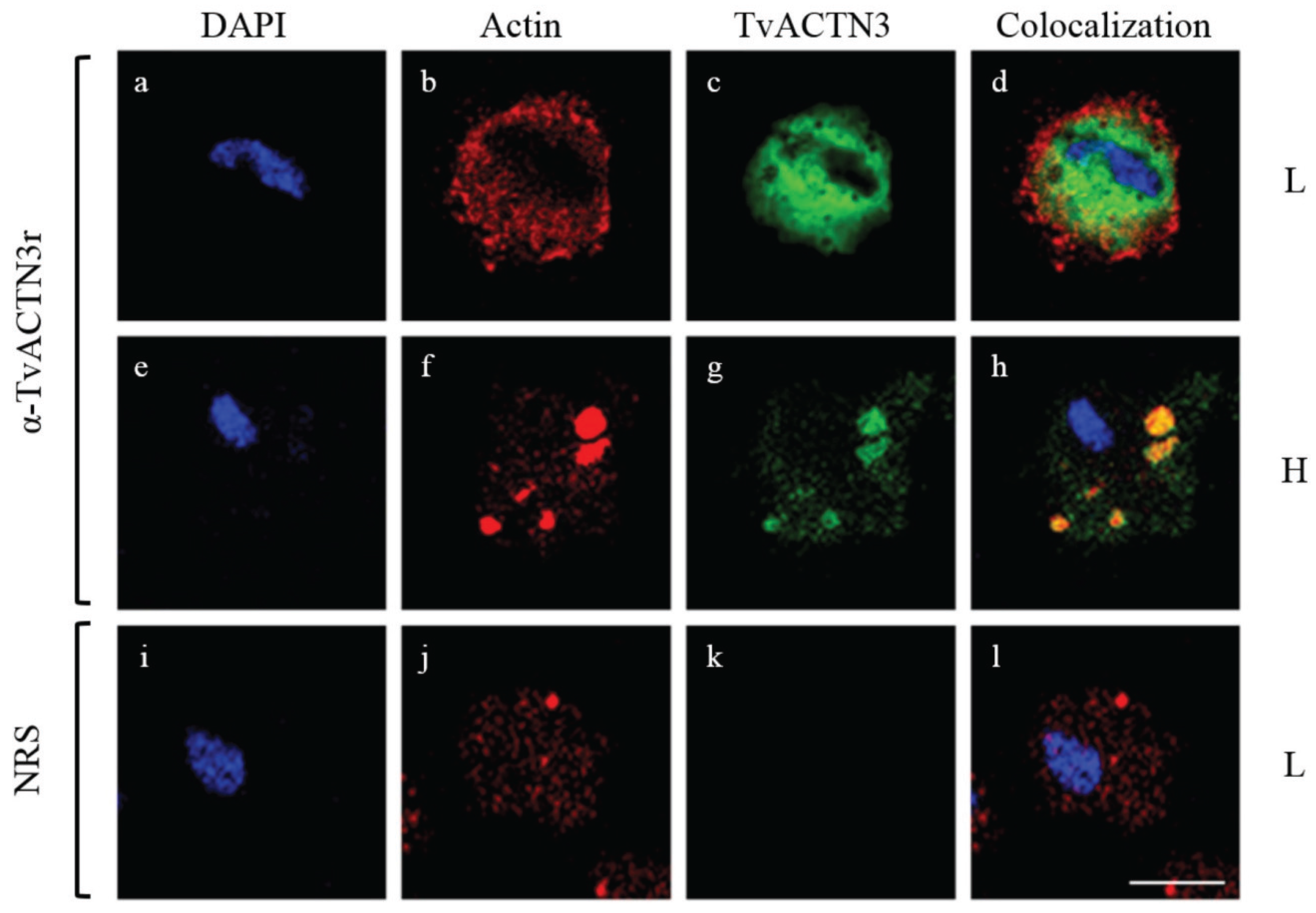
4.2.3. TvACT as an RNA-Binding Protein in T. vaginalis

5. A Posttranscriptional Iron Regulatory Mechanism in T. vaginalis Parallels the Typical IRP/IRP Mechanism
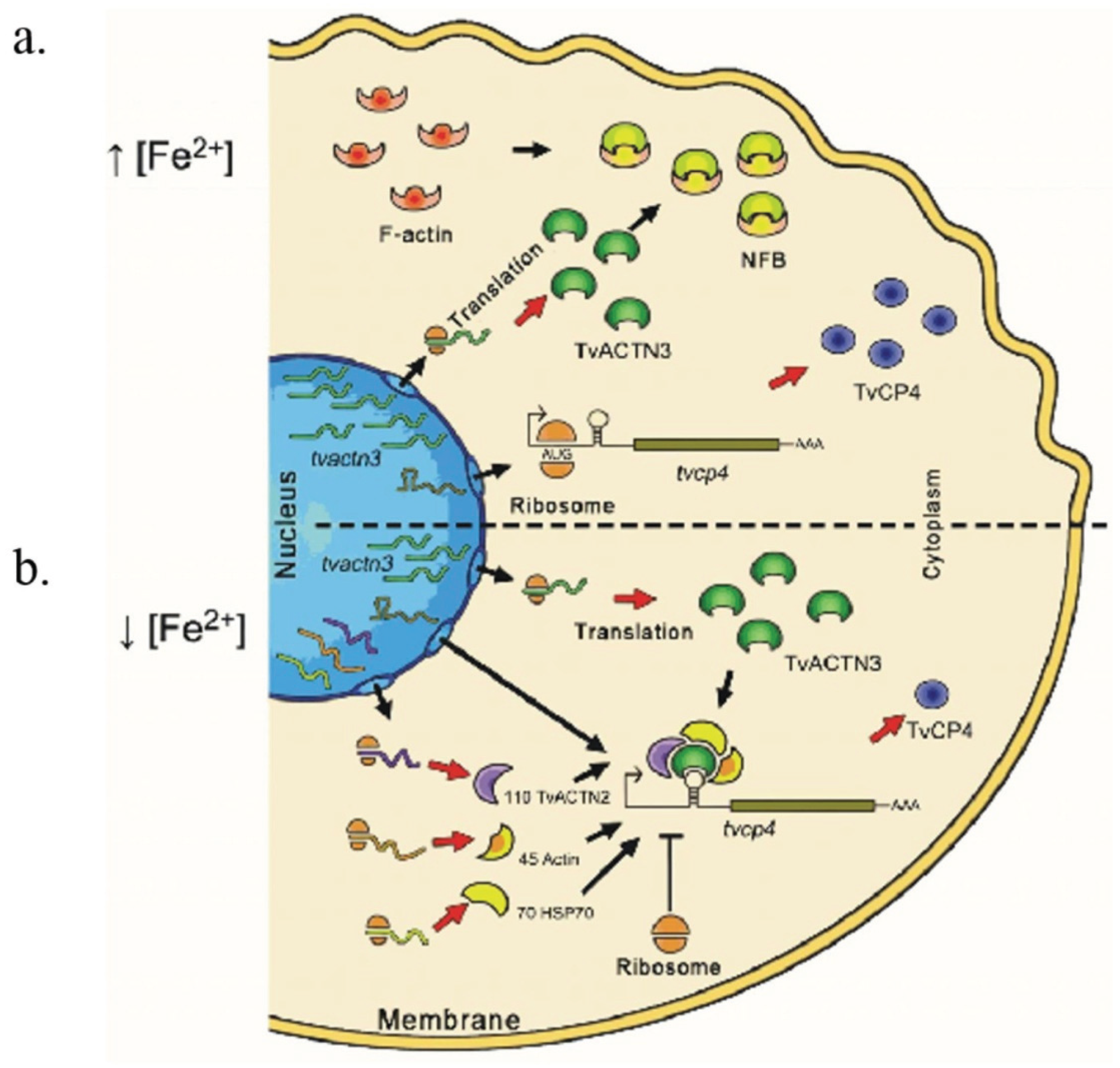
5.1. Possible Events Occurring under High Iron Conditions with tvcp4 Expression
5.2. Possible Events Occurring under Low Iron Conditions with tvcp4 Expression
5.3. Possible Events Occurring under Low and High Iron Conditions with Tvcp12 Expression
5.4. A Comparative Analysis of Trichomonad RNA-Binding Proteins with Typical IRPs
| Characteristic | IRP-1 | IRP-2 | pfIRPa a | TvHSP70 b | TvACTN3 c | TvACTN2 d | TvACT d |
|---|---|---|---|---|---|---|---|
| RNA-interaction under iron-depleted condition | Yes | Yes | Yes | Yes | Yes | ND | ND |
| Reducing agent dependence | Yes | No | Yes | No | No | No | No |
| Cytoplasmic localization | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Functions | RBP Aconitase | RBP NP | RBP Aconitase | RBP Chaperone | RBP Actin-binding protein | NC Actin-binding protein | NC Cytoskeleton filaments |
| Molecular Weight (kDa) | 98 | 105 | 103 | 70 | 135 | 110 | 45 |
6. Conclusions and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dlouhy, A.C.; Outten, C.E. The iron metallome in eukaryotic organisms. Met. Ions Life Sci. 2013, 12, 241–278. [Google Scholar] [PubMed]
- Heath, J.L.; Weiss, J.M.; Lavau, C.P.; Wechsler, D.S. Iron deprivation in cancer—Potential therapeutic implications. Nutrients 2013, 5, 2836–2859. [Google Scholar] [CrossRef] [PubMed]
- Testa, U. Proteins of Iron Metabolism; CRC Press: Boca Raton, FL, USA, 2002; p. 559. [Google Scholar]
- Pantopoulos, K. Iron metabolism and the IRE/IRP regulatory system: An update. Ann. N. Y. Acad. Sci. 2004, 1012, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.; McVey Ward, D.; Crisp, R.J.; Philpott, C.C. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim. Biophys. Acta 2006, 1763, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Richardson, D.R. Iron chelation and regulation of the cell cycle: 2 mechanisms of posttranscriptional regulation of the universal cyclin-dependent kinase inhibitor p21CIP1/WAF1 by iron depletion. Blood 2007, 110, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Siriwardana, G.; Seligman, P.A. Two cell cycle blocks caused by iron chelation of neuroblastoma cells: Separating cell cycle events associated with each block. Physiol. Rep. 2013. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Rouault, T.A. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 2010, 49, 4945–4956. [Google Scholar] [CrossRef] [PubMed]
- White, M.F.; Dillingham, M.S. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012, 22, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, G.; Huang, M. Ribonucleotide reductase metallocofactor: Assembly, maintenance and inhibition. Front. Biol. 2014, 9, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, H.; Zhang, C.; An, X.; Liu, L.; Stubbe, J.; Huang, M. Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proc. Natl. Acad. Sci. USA 2014, 111, E1695–E1704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C. Essential functions of iron-requiring proteins in DNA replication, repair and cell cycle control. Protein Cell 2014, 5, 750–760. [Google Scholar] [CrossRef] [PubMed]
- De la Garza Amaya, M.; Vaca Pacheco, S. The Struggle for Iron: Pathogen vs. Host. Available online: http://www.researchgate.net/publication/255980746 (accessed on 4 November 2015).
- Arroyo, R.; Ochoa, T.; Tai, J.H.; de la Garza, M. Iron and Parasites. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Estrada, G.; Luna-Castro, S.; Piña-Vazquez, C.; Samaniego-Barrón, L.; León-Sicairos, N.; Serrano-Luna, J.; de la Garza, M. Iron-satured lactoferrin and pathogenic protozoa: Could this protein be an iron source for their parasitic style of life? Future Microbiol. 2012, 7, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, S.; Goyal, K.; Sehgal, A. Trichomoniasis and lactoferrin: Future prospects. Infect. Dis. Obstet. Gynecol. 2012. [Google Scholar] [CrossRef]
- Arroyo, R.; Cárdenas-Guerra, R.E.; Figueroa-Angulo, E.E.; Puente-Rivera, J.; Zamudio-Prieto, O.; Ortega-López, J. Trichomonas vaginalis cysteine proteinases: Iron response in gene expression and proteolytic activity. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sutak, R.; Lesuisse, E.; Tachezy, J.; Richardson, D.R. Crusade for iron: Iron uptake in unicelular eukaryotes and its significance for virulence. Trends Mcirobiol. 2008, 16, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Reyes-López, M.; Piña-Vázquez, C.; Serrano-Luna, J. Transferrin: Endocytosis and cell signaling in parasitic protozoa. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Aisen, P.; Enns, C.; Wessling-Resnick, M. Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 2001, 33, 940–959. [Google Scholar] [CrossRef]
- Tandara, L.; Salamunic, I. Iron metabolism: Current facts and future directions. Biochem. Med. 2012, 22, 311–328. [Google Scholar] [CrossRef]
- Zhang, D.L.; Ghosh, M.C.; Rouault, T.A. The physiological functions of iron regulatory proteins in iron homeostasis—An update. Front. Pharmacol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Haile, D.J.; Rouault, T.A.; Harford, J.B.; Kennedy, M.C.; Blondin, G.A.; Beinert, H.; Klausner, R.D. Cellular regulation of the iron-responsive element binding protein: Disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc. Natl. Acad. Sci. USA 1992, 89, 11735–11739. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Kuhn, L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 1996, 93, 8175–8182. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006, 2, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. Iron metabolism in the CNS: Implications for neurodegenerative diseases. Nat. Rev. Neurosci. 2013, 14, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Wallander, M.L.; Leibold, E.A.; Eisenstein, R.S. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta 2006, 1763, 668–689. [Google Scholar] [CrossRef] [PubMed]
- Muckenthaler, M.U.; Galy, B.; Hentze, M.W. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 2008, 28, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Volz, K. The functional duality of iron regulatory protein 1. Curr. Opin. Struct. Biol. 2008, 18, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, L.C. How iron controls iron. Cell Metab. 2009, 10, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.A.; Zumbrennen, K.B.; Huang, X.; Powers, D.N.; Durazo, A.; Sun, D.; Bhaskaran, N.; Persson, A.; Uhlen, M.; Sangfelt, O.; et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 2009, 326, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Salahudeen, A.A.; Thompson, J.W.; Ruiz, J.C.; Ma, H.W.; Kinch, L.N.; Li, Q.; Grishin, N.V.; Bruick, R.K. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 2009, 326, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Gruer, M.J.; Artymiuk, P.J.; Guest, J.R. The aconitase family: Three structural variations on a common theme. Trends Biochem. Sci. 1997, 22, 3–6. [Google Scholar] [CrossRef]
- Artymiuk, P.J.; Green, J. The double life of aconitase. Structure 2006, 14, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, J.; Volbeda, A.; Carpentier, P.; Darnault, C.; Moulis, J.M.; Fontecilla-Camps, J.C. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure 2006, 14, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Yu, Y.; Leibold, E.A. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J. Biol. Chem. 1994, 269, 24252–24260. [Google Scholar] [PubMed]
- Guo, B.; Brown, F.M.; Phillips, J.D.; Yu, Y.; Leibold, E.A. Characterization and expression of iron regulatory protein 2 (IRP2). Presence of multiple IRP2 transcripts regulated by intracellular iron levels. J. Biol. Chem. 1995, 270, 16529–16535. [Google Scholar] [CrossRef] [PubMed]
- Iwai, K.; Klausner, R.D.; Rouault, T.A. Requirements for iron-regulated degradation of the RNA-binding protein, iron regulatory protein 2. EMBO J. 1995, 14, 5350–5357. [Google Scholar] [PubMed]
- Ke, Y.; Wu, J.; Leibold, E.A.; Walden, W.E.; Theil, E.C. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding. Fine-tuning of mRNA regulation? J. Biol. Chem. 1998, 273, 23637–23640. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.; Klausner, R. Regulation of iron metabolism in eukaryotes. Curr. Top. Cell. Regul. 1997, 35, 1–19. [Google Scholar] [PubMed]
- Smith, S.R.; Ghosh, M.C.; Ollivierre-Wilson, H.; Hang Tong, W.; Rouault, T.A. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol. Dis. 2006, 36, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Meyron-Holtz, E.G.; Ghosh, M.C.; Iwai, K.; LaVaute, T.; Brazzolotto, X.; Berger, U.V.; Land, W.; Ollivierre-Wilson, H.; Grinberg, A.; Love, P.; et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004, 23, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Galy, B.; Ferring-Appel, D.; Sauer, S.W.; Kaden, S.; Lyoumi, S.; Puy, H.; Kolker, S.; Grone, H.J.; Hentze, M.W. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab. 2010, 12, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.C.; Zhang, D.L.; Jeong, S.Y.; Kovtunovych, G.; Ollivierre-Wilson, H.; Noguchi, A.; Tu, T.; Senecal, T.; Robinson, G.; Crooks, D.R.; et al. Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2α. Cell Metab. 2013, 17, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, N.; Pantopoulos, K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2α mRNA translation. Blood 2013, 122, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Cooperman, S.S.; Meyron-Holtz, E.G.; Olivierre-Wilson, H.; Ghosh, M.C.; McConnell, J.P.; Rouault, T.A. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 2005, 106, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Galy, B.; Ferring, D.; Minana, B.; Bell, O.; Janser, H.G.; Muckenthaler, M.; Schumann, K.; Hentze, M.W. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood 2005, 106, 2580–2589. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.C.; Tong, W.H.; Zhang, D.; Ollivierre-Wilson, H.; Singh, A.; Krishna, M.C.; Mitchell, J.B.; Rouault, T.A. Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc. Natl. Acad. Sci. USA 2008, 105, 12028–12033. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, N.; Pantopoulos, K. The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Leibold, E.A.; Munro, H.N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5' untranslated region of ferritin heavy- and light-subunit mRNAs. Proc. Natl. Acad. Sci. USA 1988, 85, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Seiser, C.; Kuhn, L.C. Characterization of a second RNA-binding protein in rodents with specificity for iron-responsive elements. J. Biol. Chem. 1993, 268, 27327–27334. [Google Scholar] [PubMed]
- Dandekar, T.; Stripecke, R.; Gray, N.K.; Goossen, B.; Constable, A.; Johansson, H.E.; Hentze, M.W. Identification of a novel iron-responsive element in murine and human erythroid delta-aminolevulinic acid synthase mRNA. EMBO J. 1991, 10, 1903–1909. [Google Scholar] [PubMed]
- Addess, K.J.; Basilion, J.P.; Klausner, R.D.; Rouault, T.A.; Pardi, A. Structure and dynamics of the iron responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 1997, 274, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Menotti, E.; Bonnard, C.; Kuhn, L.C. Optimal sequence and structure of iron-responsive elements. Selection of RNA stem-loops with high affinity for iron regulatory factor. J. Biol. Chem. 1994, 269, 17481–17489. [Google Scholar] [PubMed]
- Piccinelli, P.; Samuelsson, T. Evolution of the iron-responsive element. RNA 2007, 13, 952–966. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Galy, B.; Schwanhaeusser, B.; Blake, J.; Bahr-Ivacevic, T.; Benes, V.; Selbach, M.; Muckenthaler, M.U.; Hentze, M.W. Iron regulatory protein-1 and -2: Transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood 2011, 118, e168–e179. [Google Scholar] [CrossRef] [PubMed]
- Menotti, E.; Henderson, B.R.; Kuhn, L.C. Translational regulation of mRNAs with distinct IRE sequences by iron regulatory proteins 1 and 2. J. Biol. Chem. 1998, 273, 1821–1824. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Parsels, L.A.; Voeller, D.M.; Allegra, C.J.; Maley, G.F.; Maley, F.; Chu, E. Characterization of a cis-acting regulatory element in the protein coding region of thymidylate synthase mRNA. Nucleic Acids Res. 2000, 28, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Tai, N.; Schmitz, J.C.; Chen, T.M.; Chu, E. Characterization of a cis-acting regulatory element in the protein-coding region of human dihydrofolate reductase mRNA. Biochem. J. 2004, 378, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Solano-González, E.; Burrola-Barraza, E.; León-Sicairos, C.; Avila-González, L.; Gutiérrez-Escolano, L.; Ortega-López, J.; Arroyo, R. The trichomonad cysteine proteinase tvcp4 transcript contains an iron-responsive element. FEBS Lett. 2007, 581, 2919–2928. [Google Scholar] [CrossRef] [PubMed]
- Torres-Romero, J.C.; Arroyo, R. Responsiveness of Trichomonas vaginalis to iron concentrations: Evidence for a post-transcriptional iron regulation by an IRE/IRP-like system. Infect. Genet. Evol. 2009, 9, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Laing, L.G.; Hall, K.B. A model of the iron responsive element RNA hairpin loop structure determined from NMR and thermodynamic data. Biochemistry 1996, 35, 13586–13596. [Google Scholar] [CrossRef] [PubMed]
- Walden, W.E.; Selezneva, A.I.; Dupuy, J.; Volbeda, A.; Fontecilla-Camps, J.C.; Theil, E.C.; Volz, K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 2006, 314, 1903–1908. [Google Scholar] [CrossRef] [PubMed]
- Walden, W.E.; Selezneva, A.; Volz, K. Accommodating variety in iron-responsive elements: Crystal structure of transferrin receptor 1 b IRE bound to iron regulatory protein 1. FEBS Lett. 2012, 586, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Selezneva, A.I.; Walden, W.E.; Volz, K.W. Nucleotide-specific recognition of iron-responsive elements by iron regulatory protein 1. J. Mol. Biol. 2013, 425, 3301–3310. [Google Scholar] [CrossRef] [PubMed]
- Goforth, J.B.; Anderson, S.A.; Nizzi, C.P.; Eisenstein, R.S. Multiple determinants within iron-responsive elements dictate iron regulatory protein binding and regulatory hierarchy. RNA 2010, 16, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Ma, J.; Walden, W.E.; Merrick, W.C.; Theil, E.C.; Goss, D.J. Rapid kinetics of iron responsive element (IRE) RNA/iron regulatory protein 1 and IRE-RNA/EIf4f complexes respond differently to metal ions. Nucleic Acids Res. 2014, 42, 6567–6577. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Graziano, J.H.; Freyer, G.A. Regulation of the 75-kDa subunit of mitochondrial complex I by iron. J. Biol. Chem. 2001, 276, 27685–27692. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A.; Tang, C.K.; Kaptain, S.; Burgess, W.H.; Haile, D.J.; Samaniego, F.; McBride, O.W.; Harford, J.B.; Klausner, R.D. Cloning of the cDNA encoding an RNA regulatory protein—The human iron-responsive element-binding protein. Proc. Natl. Acad. Sci. USA 1990, 87, 7958–7962. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.; Pantopoulos, K.; Kao, H.T.; Muckenthaler, M.; Youson, J.H.; Pieribone, V. Regulation of iron metabolism in the sanguivore lamprey Lampetra fluviatilis—Molecular cloning of two ferritin subunits and two iron-regulatory proteins (IRP) reveals evolutionary conservation of the iron-regulatory element (IRE)/IRP regulatory system. Eur. J. Biochem. 1998, 254, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Muckenthaler, M.; Gunkel, N.; Frishman, D.; Cyrklaff, A.; Tomancak, P.; Hentze, M.W. Iron-regulatory protein-1 (IRP-1) is highly conserved in two invertebrate species—Characterization of IRP-1 homologues in Drosophila melanogaster and Caenorhabditis elegans. Eur. J. Biochem. 1998, 254, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Guest, J.R. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 1999, 145 Pt 11, 3069–3079. [Google Scholar] [CrossRef] [PubMed]
- Meehan, H.A.; Lundberg, R.A.; Connell, G.J. A trypanosomatid protein specifically interacts with a mammalian iron-responsive element. Parasitol. Res. 2000, 86, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Saas, J.; Ziegelbauer, K.; von Haeseler, A.; Fast, B.; Boshart, M. A developmentally regulated aconitase related to iron-regulatory protein-1 is localized in the cytoplasm and in the mitochondrion of Trypanosoma brucei. J. Biol. Chem. 2000, 275, 2745–2755. [Google Scholar] [CrossRef] [PubMed]
- Loyevsky, M.; LaVaute, T.; Allerson, C.R.; Stearman, R.; Kassim, O.O.; Cooperman, S.; Gordeuk, V.R.; Rouault, T.A. An IRP-like protein from Plasmodium falciparum binds to a mammalian iron-responsive element. Blood 2001, 98, 2555–2562. [Google Scholar] [CrossRef] [PubMed]
- Hodges, M.; Yikilmaz, E.; Patterson, G.; Kasvosve, I.; Rouault, T.A.; Gordeuk, V.R.; Loyevsky, M. An iron regulatory-like protein expressed in Plasmodium falciparum displays aconitase activity. Mol. Biochem. Parasitol. 2005, 143, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Loyevsky, M.; Mompoint, F.; Yikilmaz, E.; Altschul, S.F.; Madden, T.; Wootton, J.C.; Kurantsin-Mills, J.; Kassim, O.O.; Gordeuk, V.R.; Rouault, T.A. Expression of a recombinant IRP-like Plasmodium falciparum protein that specifically binds putative plasmodial IREs. Mol. Biochem. Parasitol. 2003, 126, 231–238. [Google Scholar] [CrossRef]
- Coronado, L.M.; Nadovich, C.T.; Spadafora, C. Malarial hemozoin: From target to tool. Biochim. Biophys. Acta 2014, 1840, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Cruz, C.; Lopez-Casamichana, M.; Cruz-Castaneda, A.; de Jesus Olivares-Trejo, J. Transferrin regulates mRNA levels of a gene involved in iron utilization in Entamoeba histolytica. Mol. Biol. Rep. 2012, 39, 4545–4551. [Google Scholar] [CrossRef] [PubMed]
- Lehker, M.W.; Alderete, J.F. Biology of trichomonosis. Curr. Opin. Infect. Dis. 2000, 13, 37–45. [Google Scholar] [CrossRef] [PubMed]
- McClelland, R.S.; Sangare, L.; Hassan, W.M.; Lavreys, L.; Mandaliya, K.; Kiarie, J.; Ndinya-Achola, J.; Jaoko, W.; Baeten, J.M. Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J. Infect. Dis. 2007, 195, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Honigberg, B.M. Trichomonads found outside the urogenital tract of humans. In Trichomonads Parasitic in Humans; Honigberg, B.M., Ed.; Springer-Verlag: New York, NY, USA, 1990; Chapter 19; pp. 342–343. [Google Scholar]
- Mann, J.R.; McDermott, S.; Barnes, T.L.; Hardin, J.; Bao, H.; Zhou, L. Trichomoniasis in pregnancy and mental retardation in children. Ann. Epidemiol. 2009, 19, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Kucknoor, A.S.; Mundodi, V.; Alderete, J.F. Adherence to human vaginal epithelial cells signals for increased expression of Trichomonas vaginalis genes. Infect. Immun. 2005, 73, 6472–6478. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.M.; Hirt, R.P.; Silva, J.C.; Delcher, A.L.; Schatz, M.; Zhao, Q.; Wortman, J.R.; Bidwell, S.L.; Alsmark, U.C.; Besteiro, S.; et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 2007, 315, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Angulo, E.E.; Rendón-Gandarilla, F.J.; Puente-Rivera, J.; Calla-Choque, J.S.; Cárdenas-Guerra, R.E.; Ortega-López, J.; Quintas-Granados, L.I.; Alvarez-Sánchez, M.E.; Arroyo, R. The effects of environmental factors on the virulence of Trichomonas vaginalis. Microbes Infect. 2012, 14, 1411–1427. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.Y.; Chen, Y.Y.; Fang, Y.K.; Cheng, W.H.; Cheng, C.C.; Chen, Y.C.; Wu, T.E.; Ku, F.M.; Chen, S.C.; Lin, R.; et al. Adaptive responses to glucose restriction enhance cell survival, antioxidant capability, and autophagy of the protozoan parasite Trichomonas vaginalis. Biochim. Biophys. Acta 2014, 1840, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.B.; Woehle, C.; Kusdian, G.; Landan, G.; Tachezy, J.; Zimorski, V.; Martin, W.F. Deep sequencing of Trichomonas vaginalis during the early infection of vaginal epithelial cells and amoeboid transition. Int. J. Parasitol. 2013, 43, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Huang, K.Y.; Huang, P.J.; Hsu, J.H.; Fang, Y.K.; Chiu, C.H.; Tang, P. Nitric oxide maintains cell survival of Trichomonas vaginalis upon iron depletion. Parasites Vectors 2015. [Google Scholar] [CrossRef] [PubMed]
- Gorrell, T.E. Effect of culture medium iron content on the biochemical composition and metabolism of Trichomonas vaginalis. J. Bacteriol. 1985, 161, 1228–1230. [Google Scholar] [PubMed]
- Lehker, M.W.; Alderete, J.F. Iron regulates growth of Trichomonas vaginalis and the expression of immunogenic Trichomonas vaginalis. Mol. Microbiol. 1992, 6, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Lehker, M.W.; Arroyo, R.; Alderete, J.F. The regulation by iron of the synthesis of adhesins and cytoadherence levels in the protozoan Trichomonas vaginalis. J. Exp. Med. 1991, 174, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, R.; Alderete, J.F. Trichomonas vaginalis surface proteinase activity is necessary for parasite adherence to epithelial cells. Infect. Immun. 1989, 57, 2991–2997. [Google Scholar] [PubMed]
- Arroyo, R.; Alderete, J.F. Two Trichomonas vaginalis surface proteinases bind to host epithelial cells and are related to levels of cytoadherence and cytotoxicity. Arch. Med. Res. 1995, 26, 279–285. [Google Scholar] [PubMed]
- Mendoza-López, M.R.; Becerril-Garcia, C.; Fattel-Facenda, L.V.; Avila-González, L.; Ruiz-Tachiquín, M.E.; Ortega-López, J.; Arroyo, R. CP30, a cysteine proteinase involved in Trichomonas vaginalis cytoadherence. Infect. Immun. 2000, 68, 4907–4912. [Google Scholar] [CrossRef] [PubMed]
- Rendón-Gandarilla, F.J.; Ramón-Luing, L.A.; Ortega-López, J.; Rosa de Andrade, I.; Benchimol, M.; Arroyo, R. The TVLEGU-1, a legumain-like cysteine proteinase, plays a key role in Trichomonas vaginalis cytoadherence. Biomed. Res. Int. 2013. [Google Scholar] [CrossRef]
- Alvarez-Sánchez, M.E.; Avila-González, L.; Becerril-Garcia, C.; Fattel-Facenda, L.V.; Ortega-López, J.; Arroyo, R. A novel cysteine proteinase (CP65) of Trichomonas vaginalis involved in cytotoxicity. Microb. Pathog. 2000, 28, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Gutiérrez, R.; Avila-González, L.; Ortega-López, J.; Cruz-Talonia, F.; Gómez-Gutiérrez, G.; Arroyo, R. Trichomonas vaginalis: Characterization of a 39-kDa cysteine proteinase found in patient vaginal secretions. Exp. Parasitol. 2004, 107, 125–135. [Google Scholar] [PubMed]
- Ramon-Luing, L.A.; Rendón-Gandarilla, F.J.; Puente-Rivera, J.; Avila-González, L.; Arroyo, R. Identification and characterization of the immunogenic cytotoxic TvCP39 proteinase gene of Trichomonas vaginalis. Int. J. Biochem. Cell Biol. 2011, 43, 1500–1511. [Google Scholar] [CrossRef] [PubMed]
- Dailey, D.C.; Chang, T.H.; Alderete, J.F. Characterization of Trichomonas vaginalis haemolysis. Parasitology 1990, 101, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas-Guerra, R.E.; Arroyo, R.; Rosa de Andrade, I.; Benchimol, M.; Ortega-López, J. The iron-induced cysteine proteinase TvCP4 plays a key role in Trichomonas vaginalis haemolysis. Microbes Infect. 2013, 15, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Alderete, J.F.; Provenzano, D.; Lehker, M.W. Iron mediates Trichomonas vaginalis resistance to complement lysis. Microb. Pathog. 1995, 19, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Fiori, P.L.; Rappelli, P.; Addis, M.F. The flagellated parasite Trichomonas vaginalis: New insights into cytopathogenicity mechanisms. Microbes Infect. 1999, 1, 149–156. [Google Scholar] [CrossRef]
- Draper, D.; Donohoe, W.; Mortimer, L.; Heine, R.P. Cysteine proteases of Trichomonas vaginalis degrade secretory leukocyte protease inhibitor. J. Infect. Dis. 1998, 178, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, H.M.; Marcet, R.; Sarracent, J. Biological roles of cysteine proteinases in the pathogenesis of Trichomonas vaginalis. Parasite 2014. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, D.; Alderete, J.F. Analysis of human immunoglobulin-degrading cysteine proteinases of Trichomonas vaginalis. Infect. Immun. 1995, 63, 3388–3395. [Google Scholar] [PubMed]
- De Jesus, J.B.; Cuervo, P.; Junqueira, M.; Britto, C.; Silva-Filho, F.C.; Soares, M.J.; Cupolillo, E.; Fernandes, O.; Domont, G.B. A further proteomic study on the effect of iron in the human pathogen Trichomonas vaginalis. Proteomics 2007, 7, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Ramón-Luing, L.A.; Rendón-Gandarilla, F.J.; Cárdenas-Guerra, R.E.; Rodríguez-Cabrera, N.A.; Ortega-López, J.; Avila-González, L.; Angel-Ortiz, C.; Herrera-Sánchez, C.N.; Mendoza-García, M.; Arroyo, R. Immunoproteomics of the active degradome to identify biomarkers for Trichomonas vaginalis. Proteomics 2010, 10, 435–444. [Google Scholar] [CrossRef] [PubMed]
- León-Félix, J.; Ortega-López, J.; Orozco-Solis, R.; Arroyo, R. Two novel asparaginyl endopeptidase-like cysteine proteinases from the protist Trichomonas vaginalis: Their evolutionary relationship within the clan CD cysteine proteinases. Gene 2004, 335, 25–35. [Google Scholar] [PubMed]
- Alderete, J.F.; Arroyo, R.; Lehker, M.W. Analysis for adhesins and specific cytoadhesion of Trichomonas vaginalis. Methods Enzymol. 1995, 253, 407–414. [Google Scholar] [PubMed]
- Arroyo, R.; Engbring, J.; Alderete, J.F. Molecular basis of host epithelial cell recognition by Trichomonas vaginalis. Mol. Microbiol. 1992, 6, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Horvathova, L.; Safarikova, L.; Basler, M.; Hrdy, I.; Campo, N.B.; Shin, J.W.; Huang, K.Y.; Huang, P.J.; Lin, R.; Tang, P.; et al. Transcriptomic identification of iron-regulated and iron-independent gene copies within the heavily duplicated Trichomonas vaginalis genome. Genome Biol. Evol. 2012, 4, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.D.; Liu, H.W.; Tai, J.H. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 2002, 277, 5153–5162. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.J.; Hsu, H.M.; Liu, H.W.; Chu, C.H.; Tai, J.H. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel MYB1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 2006, 5, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.J.; Hsu, H.M.; Liu, H.W.; Chu, C.H.; Tai, J.H. Activation of multifarious transcription of an adhesion protein ap65-1 gene by a novel MYB2 protein in the protozoan parasite Trichomonas vaginalis. J. Biol. Chem. 2007, 282, 6716–6725. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.M.; Lee, Y.; Indra, D.; Wei, S.Y.; Liu, H.W.; Chang, L.C.; Chen, C.; Ong, S.J.; Tai, J.H. Iron-inducible nuclear translocation of a MYB3 transcription factor in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 2012, 11, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.M.; Lee, Y.; Hsu, P.H.; Liu, H.W.; Chu, C.H.; Chou, Y.W.; Chen, Y.R.; Chen, S.H.; Tai, J.H. Signal transduction triggered by iron to induce the nuclear importation of a MYB3 transcription factor in the parasitic protozoan Trichomonas vaginalis. J. Biol. Chem. 2014, 289, 29334–29349. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.M.; Ong, S.J.; Lee, M.C.; Tai, J.H. Transcriptional regulation of an iron-inducible gene by differential and alternate promoter entries of multiple MYB proteins in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 2009, 8, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Paschinger, K.; Hykollari, A.; Razzazi-Fazeli, E.; Greenwell, P.; Leitsch, D.; Walochnik, J.; Wilson, I.B. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology 2012, 22, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Calla-Choque, J.S.; Figueroa-Angulo, E.E.; Avila-Gonzalez, L.; Arroyo, R. Α-actinin TvACTN3 of Trichomonas vaginalis is an RNA-binding protein that could participate in its posttranscriptional iron regulatory mechanism. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Alderete, J.F. Iron modulates phenotypic variation and phosphorylation of P270 in double-stranded RNA virus-infected Trichomonas vaginalis. Infect. Immun. 1999, 67, 4298–4302. [Google Scholar] [PubMed]
- Puente-Rivera, J.; Ramón-Luing, L.A.; Figueroa-Angulo, E.E.; Ortega-López, J.; Arroyo, R. Trichocystatin-2 (TC-2): An endogenous inhibitor of cysteine proteinases in Trichomonas vaginalis is associated with TvCP39. Int. J. Biochem. Cell Biol. 2014, 54, 255–265. [Google Scholar] [PubMed]
- Smith, A.; Johnson, P. Gene expression in the unicellular eukaryote Trichomonas vaginalis. Res. Microbiol. 2011, 162, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Liston, D.R.; Johnson, P.J. Analysis of a ubiquitous promoter element in a primitive eukaryote: Early evolution of the initiator element. Mol. Cell. Biol. 1999, 19, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Chudnovsky, L.; Simoes-Barbosa, A.; Delgadillo-Correa, M.G.; Jonsson, Z.O.; Wohlschlegel, J.A.; Johnson, P.J. Novel core promoter elements and a cognate transcription factor in the divergent unicellular eukaryote Trichomonas vaginalis. Mol. Cell. Biol. 2011, 31, 1444–1458. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Lau, A.O.; Johnson, P.J. Structural basis of core promoter recognition in a primitive eukaryote. Cell 2003, 115, 413–424. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Salvesen, G.S. Regulating cysteine protease activity: Essential role of protease inhibitors as guardians and regulators. Curr. Pharm. Des. 2002, 8, 1623–1637. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, S.J.; Westrop, G.D.; Scharfstein, J.; Mottram, J.C.; Coombs, G.H. Functional conservation of a natural cysteine peptidase inhibitor in protozoan and bacterial pathogens. FEBS Lett. 2003, 542, 12–16. [Google Scholar] [CrossRef]
- Santamaria, M.E.; Hernandez-Crespo, P.; Ortego, F.; Grbic, V.; Grbic, M.; Diaz, I.; Martinez, M. Cysteine peptidases and their inhibitors in Tetranychus urticae: A comparative genomic approach. BMC Genom. 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y.; Song, S.M.; Moon, E.K.; Lee, Y.R.; Jha, B.K.; Danne, D.B.; Cha, H.J.; Yu, H.S.; Kong, H.H.; Chung, D.I.; et al. Cysteine protease inhibitor (acstefin) is required for complete cyst formation of Acanthamoeba. Eukaryot. Cell 2013, 12, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Galy, B.; Dandekar, T.; Bengert, P.; Vainshtein, Y.; Stolte, J.; Muckenthaler, M.U.; Hentze, M.W. Iron regulation and the cell cycle: Identification of an iron-responsive element in the 3'-untranslated region of human cell division cycle 14a mRNA by a refined microarray-based screening strategy. J. Biol. Chem. 2006, 281, 22865–22874. [Google Scholar] [CrossRef] [PubMed]
- Serio, A.W.; Pechter, K.B.; Sonenshein, A.L. Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 2006, 188, 6396–6405. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.O.; Dore, L.C.; Valentine, E.; Shelat, S.G.; Hardison, R.C.; Ghosh, M.; Wang, W.; Eisenstein, R.S.; Costa, F.F.; Weiss, M.J. An iron responsive element-like stem-loop regulates α-hemoglobin-stabilizing protein mRNA. J. Biol. Chem. 2008, 283, 26956–26964. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.F.; Feng, M.; Liu, P.; Millership, J.J.; Yount, B.; Baric, R.S.; Leibowitz, J.L. Effect of mutations in the mouse hepatitis virus 3'(+)42 protein binding element on RNA replication. J. Virol. 2005, 79, 14570–14585. [Google Scholar] [CrossRef] [PubMed]
- Nanda, S.K.; Leibowitz, J.L. Mitochondrial aconitase binds to the 3' untranslated region of the mouse hepatitis virus genome. J. Virol. 2001, 75, 3352–3362. [Google Scholar] [CrossRef] [PubMed]
- Muller, M. Energy metabolism of ancestral eukaryotes: A hypothesis based on the biochemistry of amitochondriate parasitic protists. Biosystems 1992, 28, 33–40. [Google Scholar] [CrossRef]
- Kim, Y.S.; Song, H.O.; Choi, I.H.; Park, S.J.; Ryu, J.S. Hydrogenosomal activity of Trichomonas vaginalis cultivated under different iron conditions. Korean J. Parasitol. 2006, 44, 373–378. [Google Scholar] [CrossRef] [PubMed]
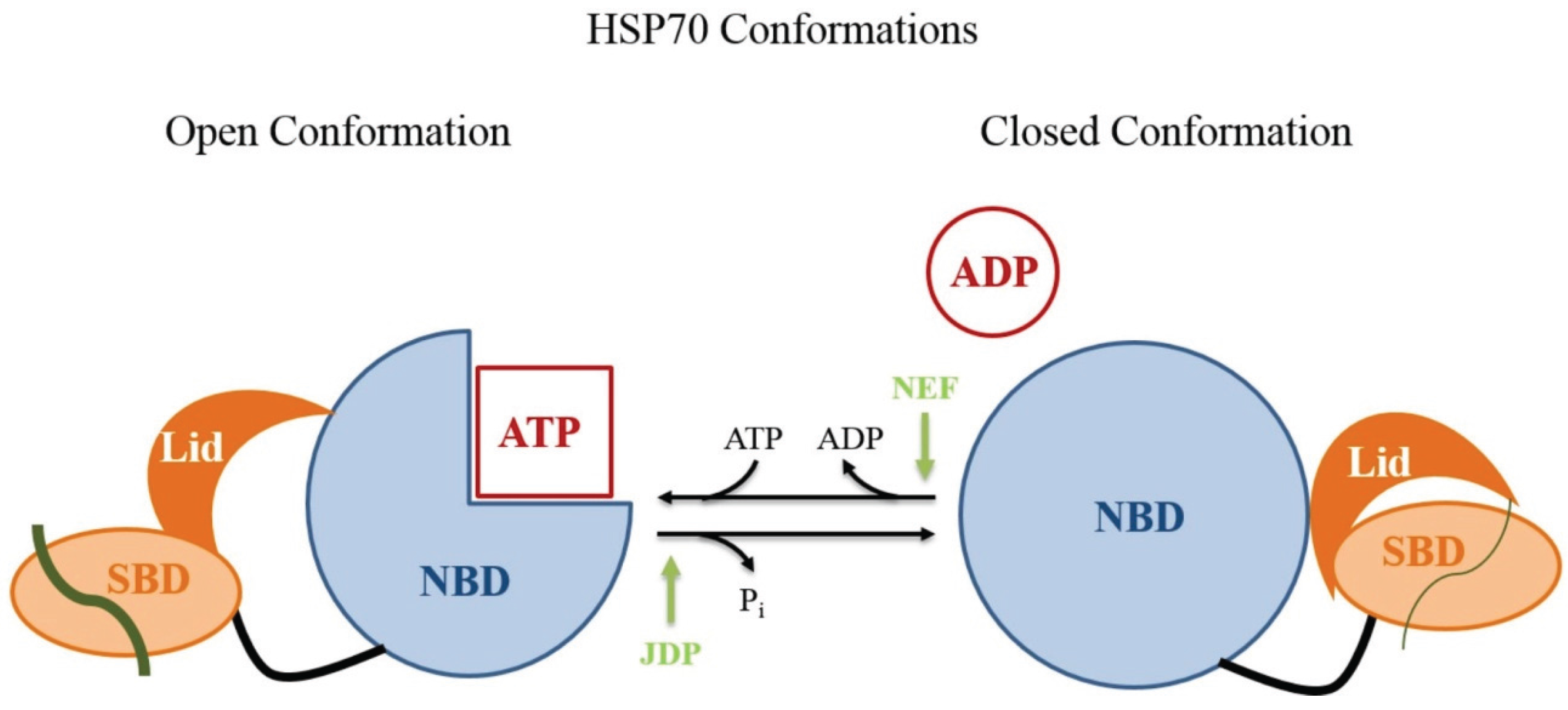
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. HSP70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Rudiger, S.; Germeroth, L.; Schneider-Mergener, J.; Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997, 16, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural analysis of substrate binding by the molecular chaperone DNAK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70 kDa heat-shock cognate protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, E.B.; Chang, L.; Gestwicki, J.E.; Zuiderweg, E.R. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA 2009, 106, 8471–8476. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Soss, S.E.; Rose, K.L.; Hill, S.; Jouan, S.; Chazin, W.J. Biochemical and proteomic analysis of ubiquitination of HSC70 and HSP70 by the E3 ligase chip. PLoS ONE 2015, 10, e0128240. [Google Scholar] [CrossRef] [PubMed]
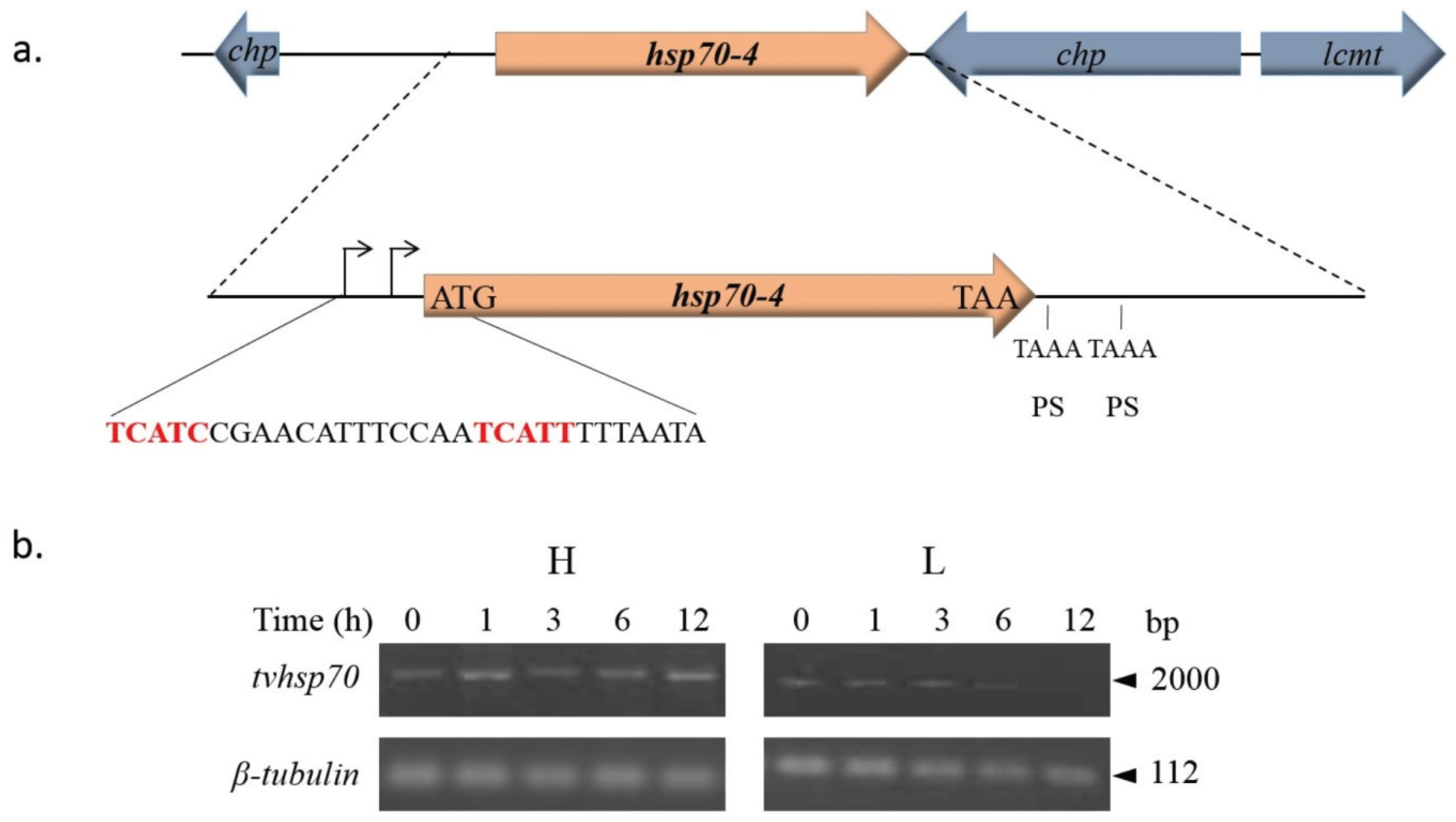
- Espinosa, N.; Hernandez, R.; Lopez-Griego, L.; Lopez-Villasenor, I. Separable putative polyadenylation and cleavage motifs in Trichomonas vaginalis mRNAs. Gene 2002, 289, 81–86. [Google Scholar] [CrossRef]
- Henics, T.; Nagy, E.; Oh, H.J.; Csermely, P.; von Gabain, A.; Subjeck, J.R. Mammalian HSP70 and HSP110 proteins bind to rna motifs involved in mRNA stability. J. Biol. Chem. 1999, 274, 17318–17324. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.; von Gabain, A.; Henics, T. Analysis of sequence-specific binding of RNA to HSP70 and its various homologs indicates the involvement of N- and C-terminal interactions. RNA 2001, 7, 1628–1637. [Google Scholar] [PubMed]
- Sjoblom, B.; Salmazo, A.; Djinovic-Carugo, K. Α-actinin structure and regulation. Cell. Mol. Life Sci. 2008, 65, 2688–2701. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, A.; Ohanian, V.; Critchley, D. The structure and function of α-actinin. J. Muscle Res. Cell Motil. 1989, 10, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Norwood, F.L.; Sutherland-Smith, A.J.; Keep, N.H.; Kendrick-Jones, J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure 2000, 8, 481–491. [Google Scholar] [CrossRef]
- Witke, W.; Hofmann, A.; Koppel, B.; Schleicher, M.; Noegel, A.A. The Ca(2+)-binding domains in non-muscle type α-actinin: Biochemical and genetic analysis. J. Cell Biol. 1993, 121, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Djinovic-Carugo, K.; Gautel, M.; Ylanne, J.; Young, P. The spectrin repeat: A structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002, 513, 119–123. [Google Scholar] [CrossRef]
- Addis, M.F.; Rappelli, P.; Delogu, G.; Carta, F.; Cappuccinelli, P.; Fiori, P.L. Cloning and molecular characterization of a cDNA clone coding for Trichomonas vaginalis α-actinin and intracellular localization of the protein. Infect. Immun. 1998, 66, 4924–4931. [Google Scholar] [PubMed]
- Bricheux, G.; Coffe, G.; Pradel, N.; Brugerolle, G. Evidence for an uncommon α-actinin protein in Trichomonas vaginalis. Mol. Biochem. Parasitol. 1998, 95, 241–249. [Google Scholar] [CrossRef]
- Virel, A.; Backman, L. Molecular evolution and structure of α-actinin. Mol. Biol. Evol. 2004, 21, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Treisman, R. Shedding light on nuclear actin dynamics and function. Trends Biochem. Sci. 2013, 38, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Gurdon, J.B. Transcriptional regulation and nuclear reprogramming: Roles of nuclear actin and actin-binding proteins. Cell. Mol. Life Sci. 2013, 70, 3289–3302. [Google Scholar] [CrossRef] [PubMed]
- Müller, M. Biochemistry of Trichomonas vaginalis. In Trichomonads Parasitic in Humans; Honigberg, B.M., Ed.; Springer-Verlag: New York, NY, USA, 1990; Chapter 5; pp. 71–72. [Google Scholar]
- Bricheux, G.; Coffe, G.; Bayle, D.; Brugerolle, G. Characterization, cloning and immunolocalization of a coronin homologue in Trichomonas vaginalis. Eur. J. Cell Biol. 2000, 79, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E.; Britigan, B.E. Iron acquisition by parasitic protozoa. Parasitol. Today 1998, 14, 348–353. [Google Scholar] [CrossRef]
- Schaible, U.E.; Kaufmann, S.H. Iron and microbial infection. Nat. Rev. Microbiol. 2004, 2, 946–953. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figueroa-Angulo, E.E.; Calla-Choque, J.S.; Mancilla-Olea, M.I.; Arroyo, R. RNA-Binding Proteins in Trichomonas vaginalis: Atypical Multifunctional Proteins. Biomolecules 2015, 5, 3354-3395. https://doi.org/10.3390/biom5043354
Figueroa-Angulo EE, Calla-Choque JS, Mancilla-Olea MI, Arroyo R. RNA-Binding Proteins in Trichomonas vaginalis: Atypical Multifunctional Proteins. Biomolecules. 2015; 5(4):3354-3395. https://doi.org/10.3390/biom5043354
Chicago/Turabian StyleFigueroa-Angulo, Elisa E., Jaeson S. Calla-Choque, Maria Inocente Mancilla-Olea, and Rossana Arroyo. 2015. "RNA-Binding Proteins in Trichomonas vaginalis: Atypical Multifunctional Proteins" Biomolecules 5, no. 4: 3354-3395. https://doi.org/10.3390/biom5043354