Comparative Geometrical Analysis of Leucine-Rich Repeat Structures in the Nod-Like and Toll-Like Receptors in Vertebrate Innate Immunity
Abstract
:1. Introduction
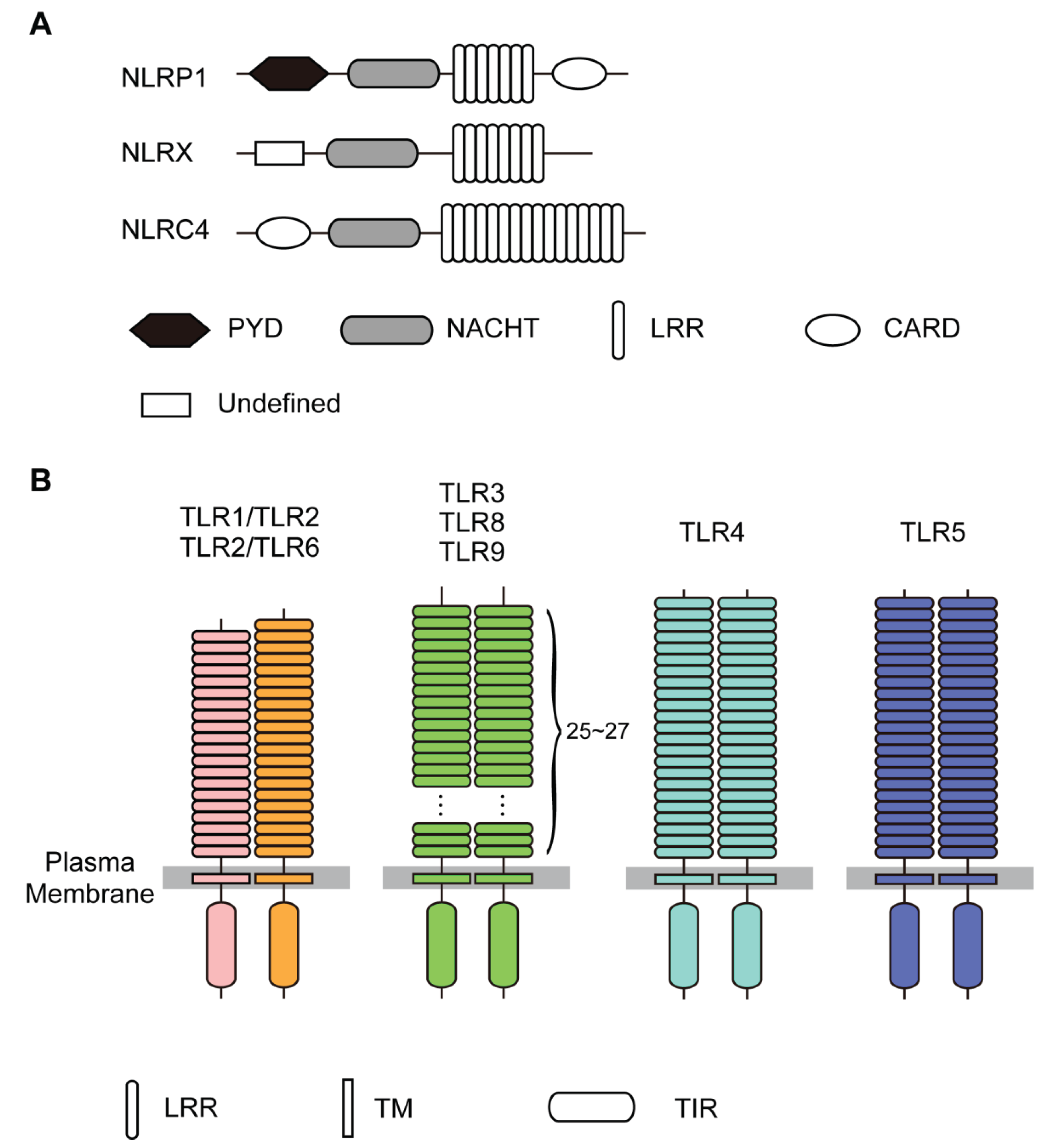
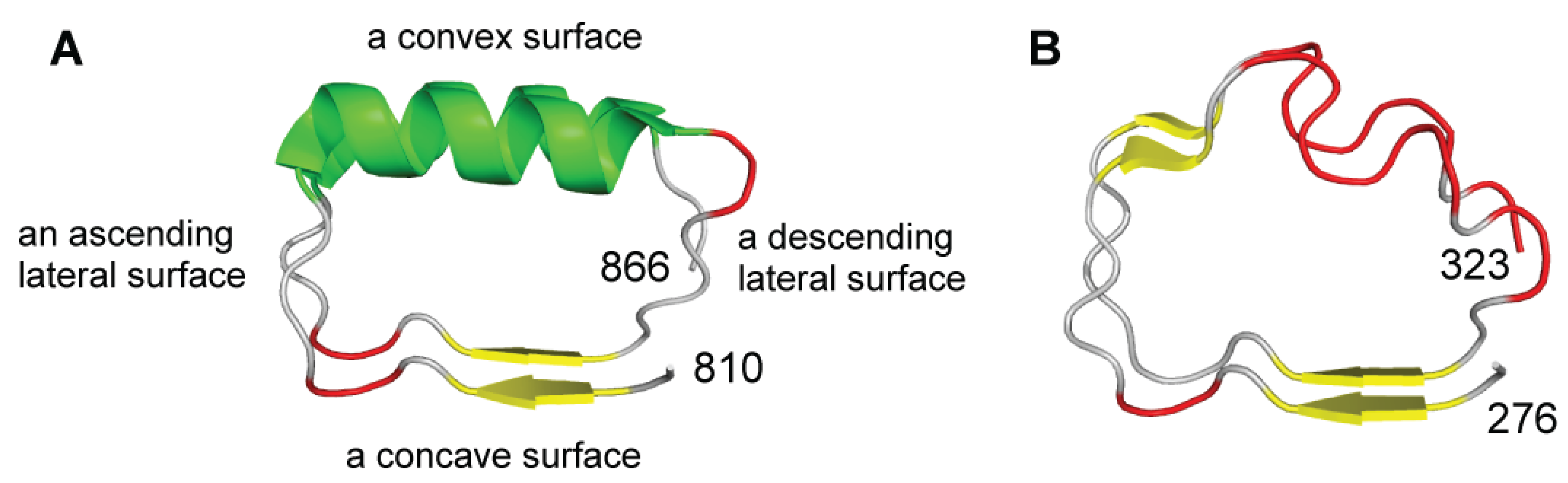
2. Analysis Methods
2.1. Structural Data
2.2. Structural Secondary Structure Analysis of LRRs
2.3. Geometrical Analysis
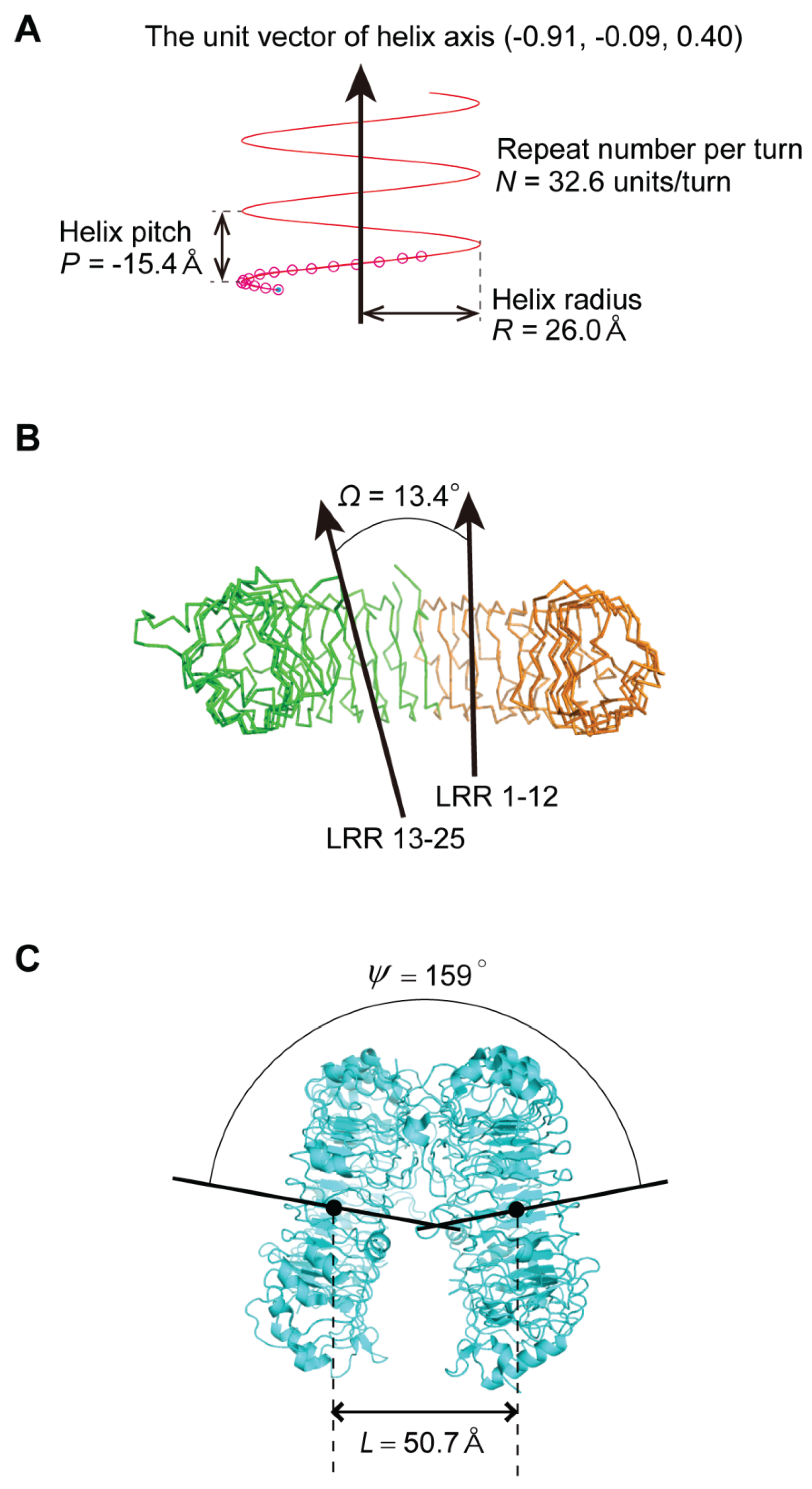
3. Structures of NLRs
3.1. The LRRs in NLRs
3.2. Human NLRP1
3.3. Human NLRX1
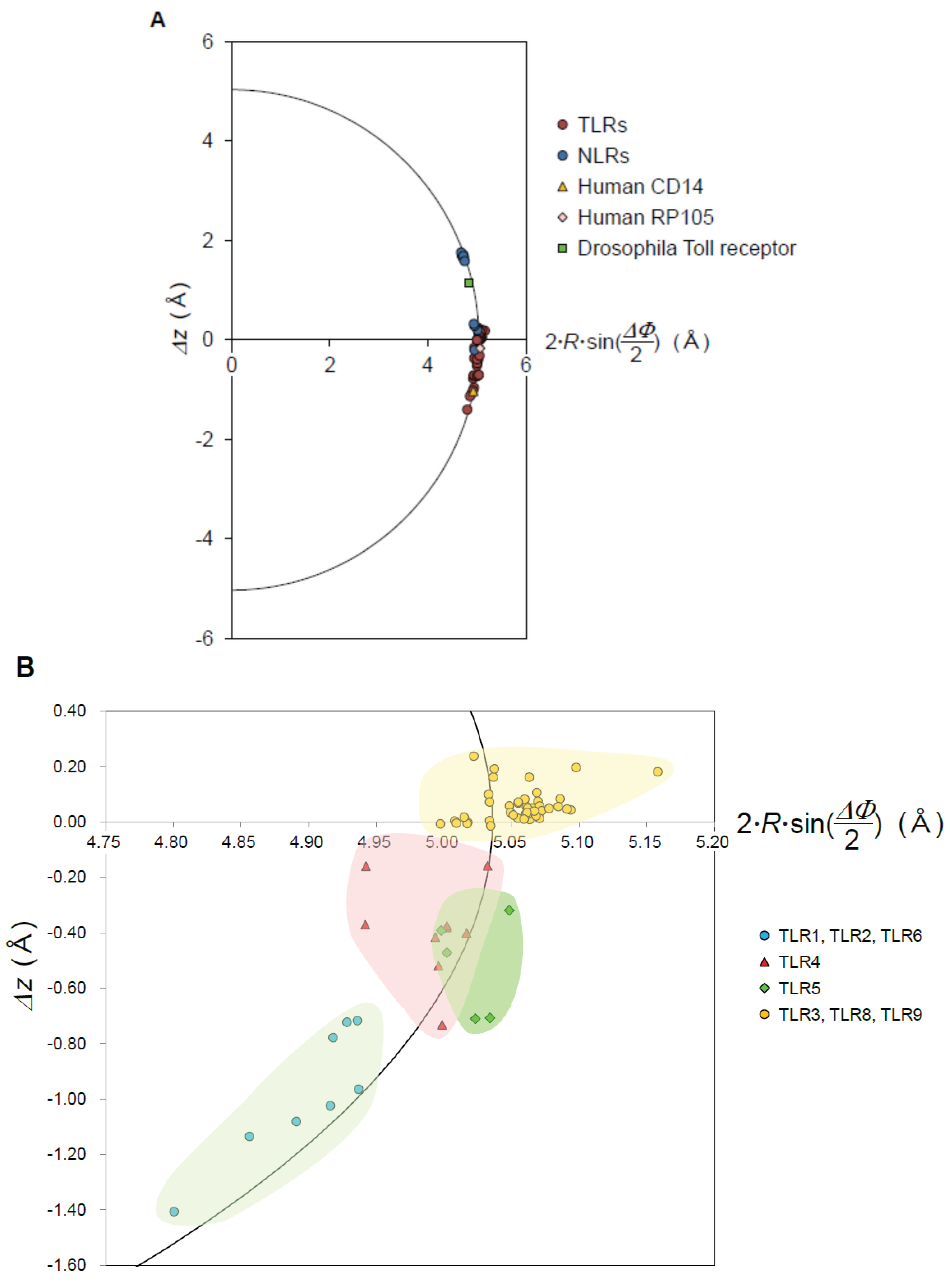
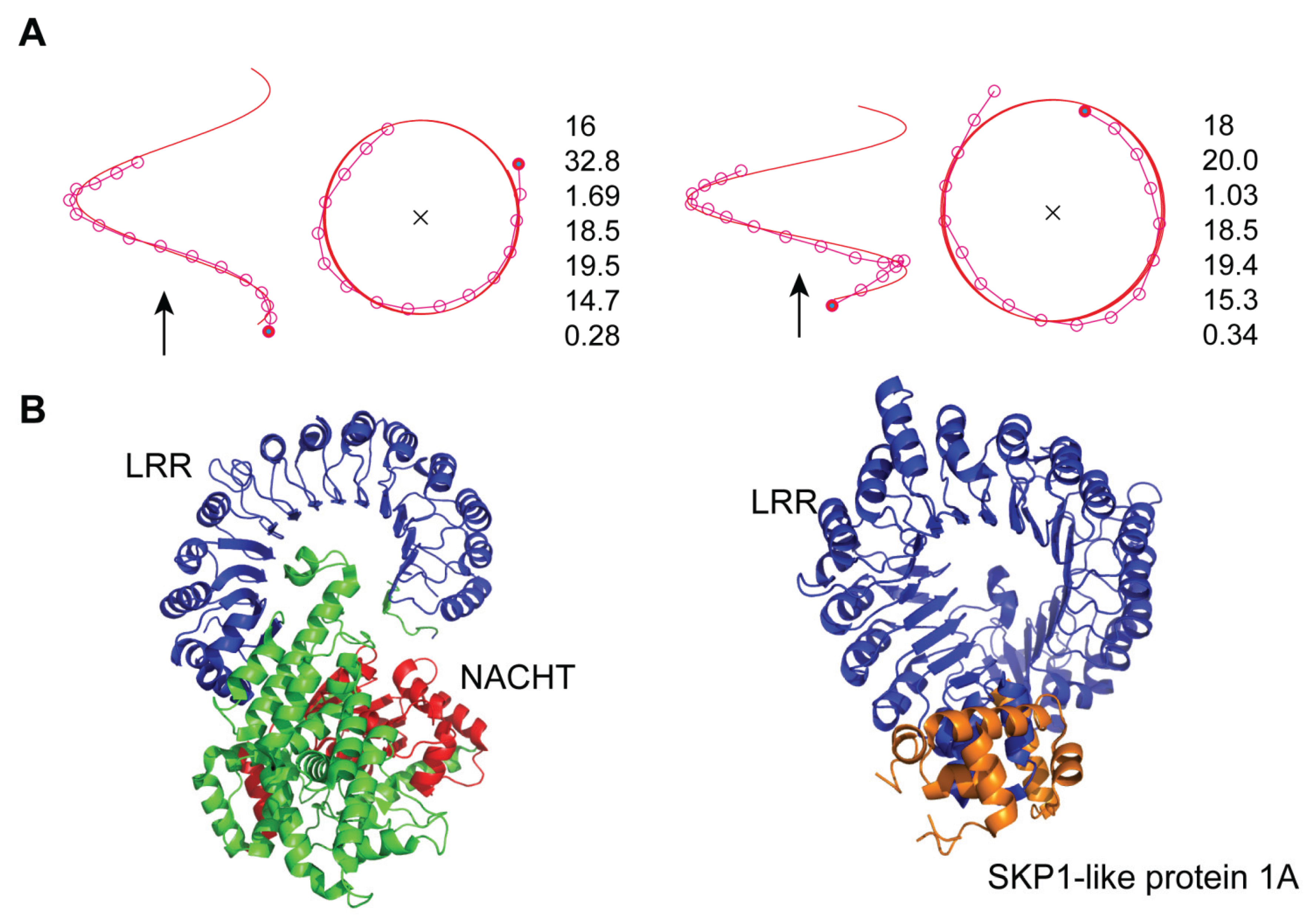
3.4. Mouse NLRC4
4. Structures of TLRs
4.1. The LRRs in TLRs
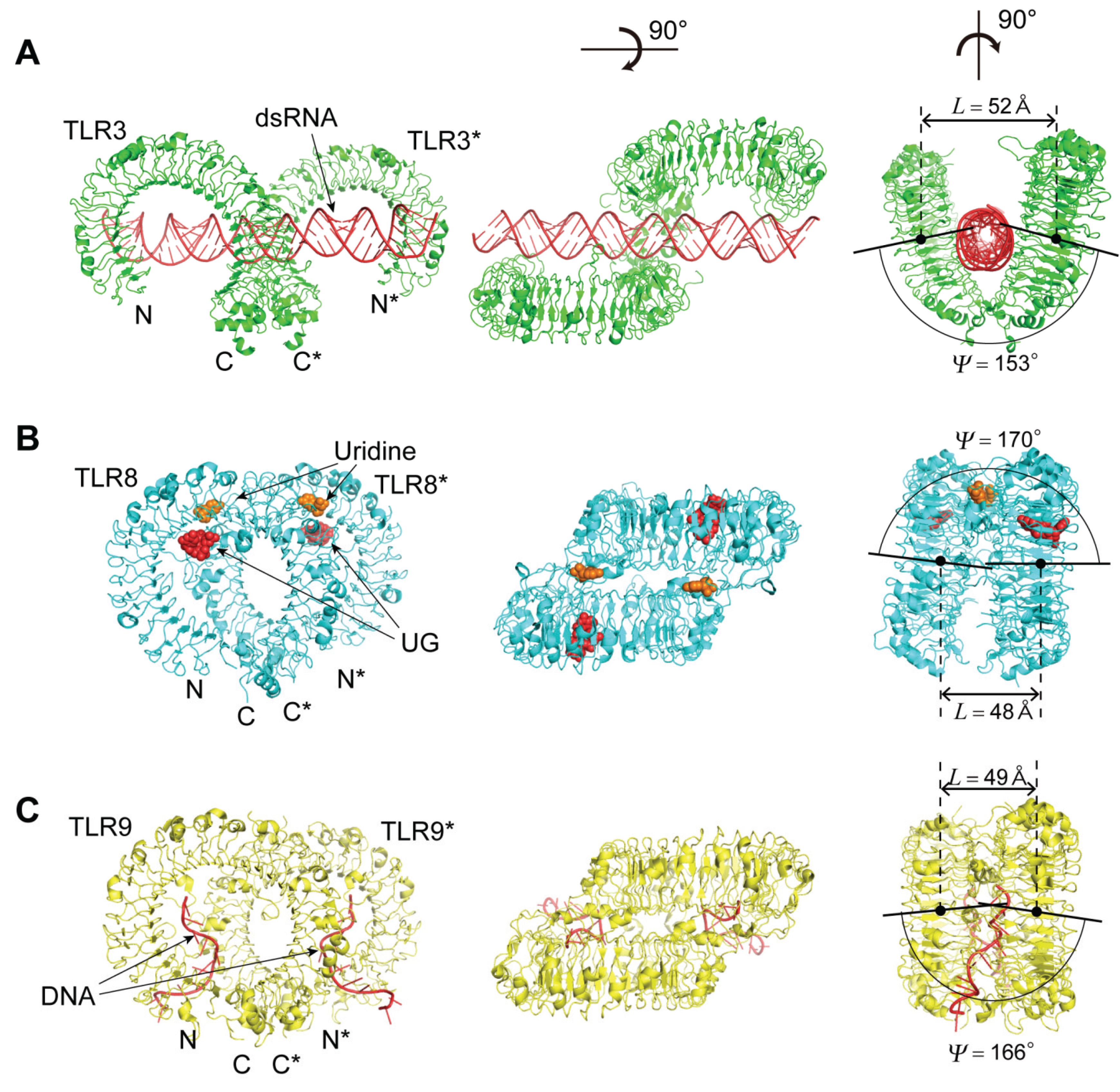
4.2. TLR3, TLR8, and TLR9
4.2.1. The Human and Mouse TLR3; Human TLR8; and Horse, Cow and Mouse TLR9
4.2.2. The TLR3—dsRNA Complex
4.2.3. The TLR3—Fab Complex
4.2.4. The TLR8—Complexes with Agonistic Ligands
4.2.5. The TLR8—Complexes with Degradation Products of ssRNA
4.2.6. The TLR9—CpG-DNA Complex
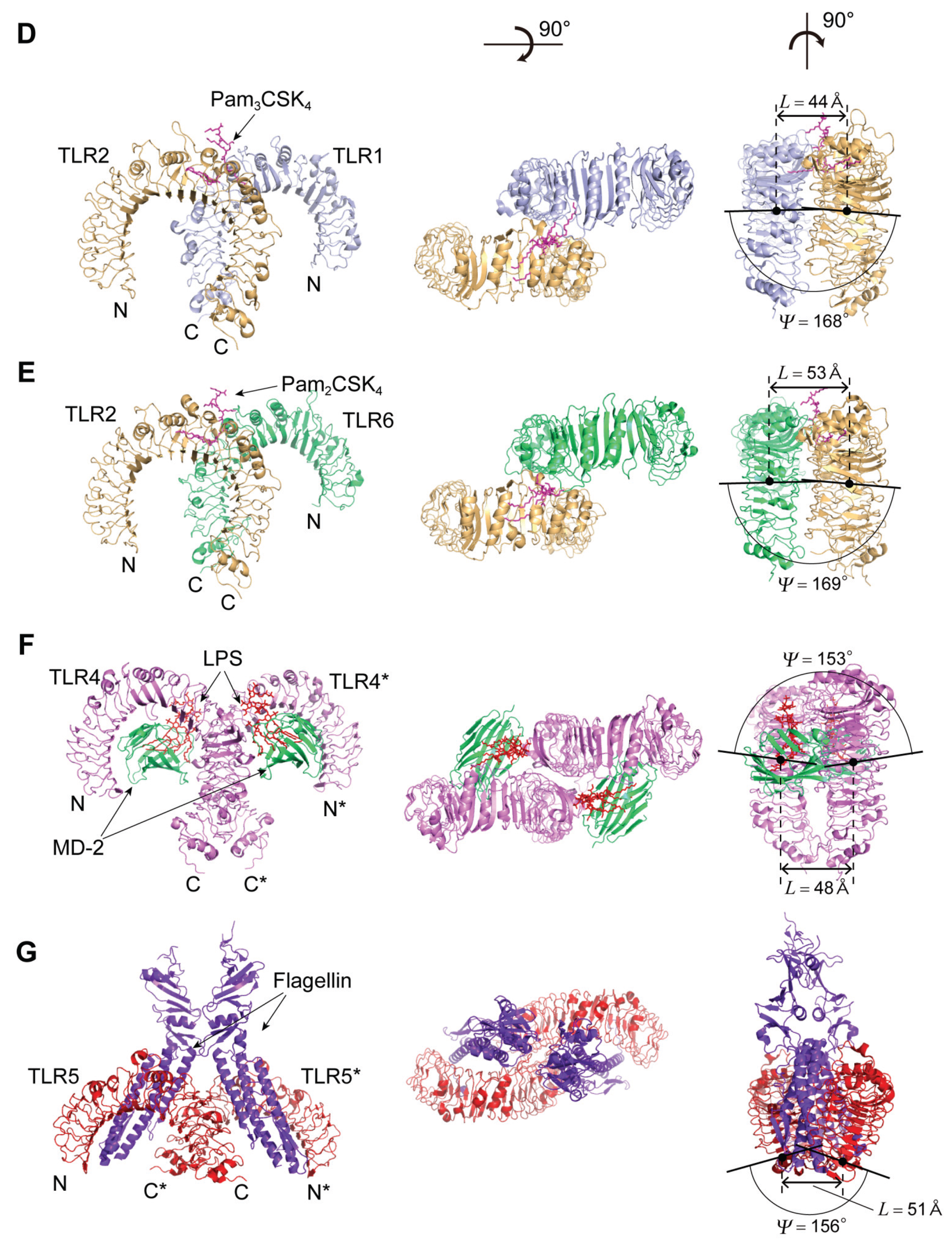
4.3. TLR1, TLR2, and TLR6
The Human TLR1/2 Heterodimer and the Mouse TLR2/6 Heterodimer
4.4. TLR4
4.4.1. Human and Mouse TLR4
4.4.2. TLR4 Complexes with MD-2/LPS
4.4.3. TLR4 Complexes with Antagonistic Ligands
4.5. TLR5
4.5.1. Human and Zebrafish TLR5
4.5.2. TLR5-Flagellin Complex
5. Structures of RP105, CD14, and Drosophila Toll Receptor
5.1. Human, Mouse, and Cow RP105
5.2. Human and Mouse CD14
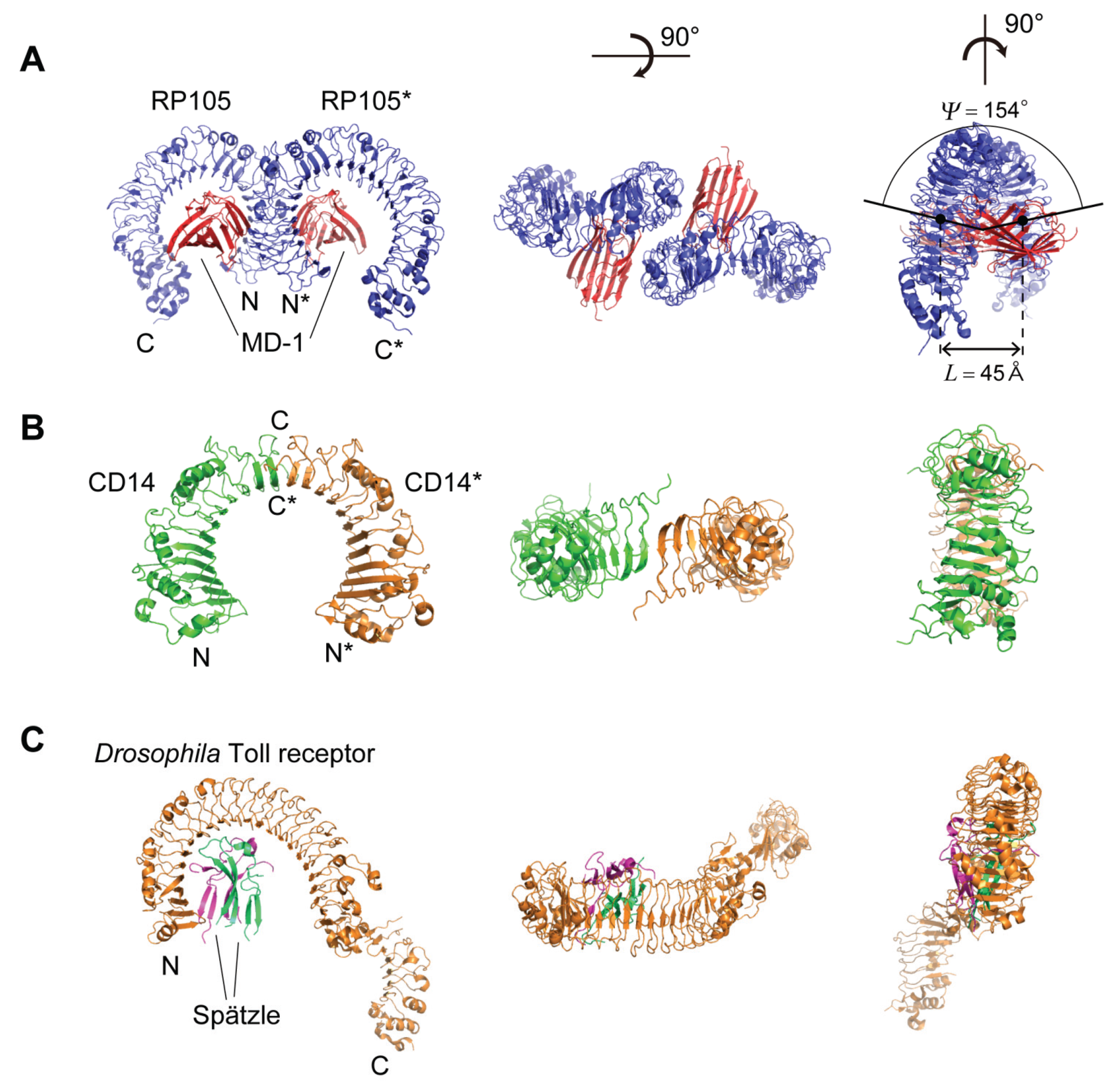
5.3. Drosophila Toll Receptor
6. Diversity of Protein Protein/Ligand Interactions
7. Conclusions
- (1)
- The LRRs in NLRs and TLRs belong to the “RI-like” and “Typical” class, respectively. The LRR units of the two classes show secondary structure motifs characteristic of the class.
- (2)
- The nine geometrical parameters—P, Δz, ΔΦ, N, R, p, Ω, L, and ψ—characterize the LRR structures in NLRs and TLRs. The LRRs in NLRC4, NLRP1, and NLRX1 adopt a right-handed helix, while TLR1-5, TLR6, TLR8, and TLR9 adopt either a left-handed helix or are nearly flat. Moreover, RP105 and CD14 also adopt a left-handed helix.
- (3)
- (4)
- The LRRs in NLRC4 and plant TIR1/COI1 adopt a distorted right-handed helix with one full turn. All or most of the individual LRR units adopt a β-α structural motif.
- (5)
- Consequently, the spatial arrangement of the NACHT domain in mNLRC4 is similar to that of SKP1A in the AtTIR1/AtCOI1 complex.
- (6)
- In the TLRs the ascending lateral surface are involved in protein, protein/ligand interactions. The diversity of the ascending loop conformations and of the dimerization conformations enables interactions with various targets that range from small molecules to proteins.
Supplementary Files
Supplementary File 1Conflicts of Interest
References
- Motta, V.; Soares, F.; Sun, T.; Philpott, D.J. NOD-like receptors: Versatile cytosolic sentinels. Physiol. Rev. 2015, 95, 149–178. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Kanneganti, T.D. Unsolved mysteries in NLR biology. Front. Immunol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Oviedo-Boyso, J.; Bravo-Patiño, A.; Baizabal-Aguirre, V. Collaborative action of Toll-like and NOD-like receptors as modulators of the inflammatory response to pathogenic bacteria. Mediat. Inflamm. 2014. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q. Nucleotide-binding oligomerization domain containing 2: Structure, function, and diseases. Semin. Arthritis Rheum. 2013, 43, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Kawai, T.; Akira, S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, J.O. Structural biology of the Toll-like receptor family. Annu. Rev. Biochem. 2011, 80, 917–941. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Moehle, K. Structural aspects of molecular recognition in the immune system. Part II: Pattern recognition receptors. Pure Appl. Chem. 2014, 86, 1483–1538. [Google Scholar] [CrossRef]
- Berglund, N.; Kargas, V.; Ortiz-Suarez, M.L.; Bond, P.J. The role of protein-protein interactions in Toll-like receptor function. Prog. Biophys. Mol. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Gay, N.J.; Gangloff, M. Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 2007, 76, 141–165. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.N.; Tauszig-Delamasure, S.; Hoffmann, J.A.; Lelievre, E.; Gascan, H.; Ray, K.P.; Morse, M.A.; Imler, J.L.; Gay, N.J. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat. Immunol. 2003, 4, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Zhang, J.; Kong, X.; Zhou, C.; Li, L.; Nie, G.; Li, X. Toll-like receptor recognition of bacteria in fish: Ligand specificity and signal pathways. Fish Shellfish Immunol. 2014, 41, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; Parker, A.E.; O’Neill, L.A. Targeting Toll-like receptors: Emerging therapeutics? Nat. Rev. Drug Discov. 2010, 9, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Matsushima, N.; Enkhbayar, P.; Kamiya, M.; Osaki, M.; Kretsinger, R. Leucine-Rich Repeats (LRRs): Structure, Function, Evolution and Interaction with Ligands. Drug Des. Rev. 2005, 2, 305–322. [Google Scholar] [CrossRef]
- Matsushima, N.; Tachi, N.; Kuroki, Y.; Enkhbayar, P.; Osaki, M.; Kamiya, M.; Kretsinger, R.H. Structural analysis of leucine-rich-repeat variants in proteins associated with human diseases. Cell. Mol. Life Sci. 2005, 62, 2771–2791. [Google Scholar] [CrossRef] [PubMed]
- Bella, J.; Hindle, K.L.; McEwan, P.A.; Lovell, S.C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008, 65, 2307–2333. [Google Scholar] [CrossRef] [PubMed]
- Enkhbayar, P.; Kamiya, M.; Osaki, M.; Matsumoto, T.; Matsushima, N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins 2004, 54, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Kajava, A.V.; Anisimova, M.; Peeters, N. Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: From plants to bacteria? PLoS ONE 2008, 3, e1694. [Google Scholar] [CrossRef] [PubMed]
- Kajava, A.V. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998, 277, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, N.; Miyashita, H.; Mikami, T.; Kuroki, Y. A nested leucine rich repeat (LRR) domain: The precursor of LRRs is a ten or eleven residue motif. BMC Microbiol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Enkhbayar, P.; Miyashita, H.; Kretsinger, R.; Matsushima, N. Helical parameters and correlations of tandem leucine rich repeats in proteins. J. Proteom. Bioinform. 2014, 7, 139–150. [Google Scholar] [CrossRef]
- Enkhbayar, P.; Damdinsuren, S.; Osaki, M.; Matsushima, N. HELFIT: Helix fitting by a total least squares method. Comput. Biol. Chem. 2008, 32, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Enkhbayar, P.; Matsushima, N. HELFIT: Algorithm and applications. AIP Conf. Proc. 2012, 1479, 1942–1947. [Google Scholar]
- Song, D.H.; Lee, J.O. Sensing of microbial molecular patterns by Toll-like receptors. Immunol. Rev. 2012, 250, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Lee, J.O. Structures of the toll-like receptor family and its ligand complexes. Immunity 2008, 29, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Lee, J.O. Structures of TLR-ligand complexes. Curr. Opin. Immunol. 2008, 20, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Segal, D.M.; Davies, D.R. The structural biology of Toll-like receptors. Structure 2011, 19, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; O’Neill, L.A. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem. J. 2009, 422, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Liu, L.; Wang, Y.; Segal, D.M.; Davies, D.R. The toll-like receptor 3:dsRNA signaling complex. Biochim. Biophys. Acta 2009, 1789, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Lechtenberg, B.C.; Mace, P.D.; Riedl, S.J. Structural mechanisms in NLR inflammasome signaling. Curr. Opin. Struct. Biol. 2014, 29, 17–25. [Google Scholar] [CrossRef] [PubMed]
- ccPDB. Available online: http://crdd.osdd.net/raghava/ccpdb/beta2_up.php (accessed on 2 March 2015).
- Reubold, T.F.; Hahne, G.; Wohlgemuth, S.; Eschenburg, S. Crystal structure of the leucine-rich repeat domain of the NOD-like receptor NLRP1: Implications for binding of muramyl dipeptide. FEBS Lett. 2014, 588, 3327–3332. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Yoon, S.I.; Wilson, I.A. Structure and functional characterization of the RNA-binding element of the NLRX1 innate immune modulator. Immunity 2012, 36, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yan, C.; Liu, P.; Huang, Z.; Ma, R.; Zhang, C.; Wang, R.; Zhang, Y.; Martinon, F.; Miao, D.; et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 2013, 341, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Faustin, B.; Lartigue, L.; Bruey, J.M.; Luciano, F.; Sergienko, E.; Bailly-Maitre, B.; Volkmann, N.; Hanein, D.; Rouiller, I.; Reed, J.C.; et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 2007, 25, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Tan, X.; Zheng, N.; Hatate, T.; Kimura, Y.; Kepinski, S.; Nozaki, H. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 5632–5637. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.G.; Lee, H.Y.; Lee, J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Nan, X.; Jin, M.S.; Youn, S.J.; Ryu, Y.H.; Mah, S.; Han, S.H.; Lee, H.; Paik, S.G.; Lee, J.O.; et al. Recognition of Lipopeptide Patterns by Toll-like Receptor 2-Toll-like Receptor 6 Heterodimer. Immunity 2009, 31, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Kelker, M.S.; Wilson, I.A. Crystal structure of human Toll-like receptor 3 (TLR3) ectodomain. Science 2005, 309, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.K.; Botos, I.; Hall, P.R.; Askins, J.; Shiloach, J.; Segal, D.M.; Davies, D.R. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc. Natl. Acad. Sci. USA 2005, 102, 10976–10980. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.Q.; Obmolova, G.; Malia, T.J.; Wu, S.J.; Duffy, K.E.; Marion, J.D.; Bell, J.K.; Ge, P.; Zhou, Z.H.; Teplyakov, A.; et al. Lateral clustering of TLR3:dsRNA signaling units revealed by TLR3ecd:3Fabs quaternary structure. J. Mol. Biol. 2012, 421, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Botos, I.; Wang, Y.; Leonard, J.N.; Shiloach, J.; Segal, D.M.; Davies, D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 2008, 320, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Park, B.S.; Kim, J.I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Fukase, K.; Miyake, K.; Shimizu, T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl. Acad. Sci. USA 2012, 109, 7421–7426. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Yamakawa, N.; Akashi-Takamura, S.; Miyake, K.; Shimizu, T. Structural Analyses of Human Toll-like Receptor 4 Polymorphisms D299G and T399I. J. Biol. Chem. 2012, 287, 40611–40617. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.F.; Kanai, R.; Lee, P.; Wang, H.W.; Modis, Y. Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin. J. Struct. Biol. 2012, 177, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.I.; Kurnasov, O.; Natarajan, V.; Hong, M.S.; Gudkov, A.V.; Osterman, A.L.; Wilson, I.A. Structural Basis of TLR5-Flagellin Recognition and Signaling. Science 2012, 335, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Tanji, H.; Ohto, U.; Shibata, T.; Miyake, K.; Shimizu, T. Structural Reorganization of the Toll-Like Receptor 8 Dimer Induced by Agonistic Ligands. Science 2013, 339, 1426–1429. [Google Scholar] [CrossRef] [PubMed]
- Yoo, E.; Salunke, D.; Sil, D.; Guo, X.; Salyer, A.; Hermanson, A.; Kumar, M.; Malladi, S.; Balakrishna, R.; Thompson, W.; et al. Determinants of activity at human Toll-like receptors 7 and 8: Quantitative structure-activity relationship (QSAR) of diverse heterocyclic scaffolds. J. Med. Chem. 2014, 57, 7955–7970. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.; Wilson, I.A. Crystal structure of the C-terminal domain of mouse TLR9. Proteins 2014, 82, 2874–2878. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Fukase, K.; Miyake, K.; Satow, Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science 2007, 316, 1632–1634. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Shibata, T.; Tanji, H.; Ishida, H.; Krayukhina, E.; Uchiyama, S.; Miyake, K.; Shimizu, T. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature 2015, 520, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Tanji, H.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Isobe, T.; Miyake, K.; Shimizu, T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015, 22, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Kokatla, H.P.; Sil, D.; Tanji, H.; Ohto, U.; Malladi, S.S.; Fox, L.M.; Shimizu, T.; David, S.A. Structure-based design of novel human Toll-like receptor 8 agonists. ChemMedChem 2014, 9, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, N.; Tanaka, T.; Enkhbayar, P.; Mikami, T.; Taga, M.; Yamada, K.; Kuroki, Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genom. 2007. [Google Scholar] [CrossRef] [PubMed]
- Roach, J.C.; Glusman, G.; Rowen, L.; Kaur, A.; Purcell, M.K.; Smith, K.D.; Hood, L.E.; Aderem, A. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 9577–9582. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gu, J.; Lamont, S.J.; Gu, X. Evolutionary analysis for functional divergence of the toll-like receptor gene family and altered functional constraints. J. Mol. Evol. 2007, 65, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Mikami, T.; Miyashita, H.; Takatsuka, S.; Kuroki, Y.; Matsushima, N. Molecular evolution of vertebrate Toll-like receptors: Evolutionary rate difference between their leucine-rich repeats and their TIR domains. Gene 2012, 503, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, N.; Ohyanagi, T.; Tanaka, T.; Kretsinger, R.H. Super-motifs and evolution of tandem leucine-rich repeats within the small proteoglycans—Biglycan, decorin, lumican, fibromodulin, PRELP, keratocan, osteoadherin, epiphycan, and osteoglycin. Proteins 2000, 38, 210–225. [Google Scholar] [CrossRef]
- Matsushima, N.; Kamiya, M.; Suzuki, N.; Tanaka, T. Super-motifs of leucine-rich repeats (LRRs) proteins. Genome Inform. 2000, 11, 343–345. [Google Scholar]
- Scott, P.G.; Dodd, C.M.; Bergmann, E.M.; Sheehan, J.K.; Bishop, P.N. Crystal structure of the biglycan dimer and evidence that dimerization is essential for folding and stability of class I small leucine-rich repeat proteoglycans. J. Biol. Chem. 2006, 281, 13324–13332. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.G.; McEwan, P.A.; Dodd, C.M.; Bergmann, E.M.; Bishop, P.N.; Bella, J. Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan. Proc. Natl. Acad. Sci. USA 2004, 101, 15633–15638. [Google Scholar] [CrossRef] [PubMed]
- Seiradake, E.; del Toro, D.; Nagel, D.; Cop, F.; Hartl, R.; Ruff, T.; Seyit-Bremer, G.; Harlos, K.; Border, E.C.; Acker-Palmer, A.; et al. FLRT structure: Balancing repulsion and cell adhesion in cortical and vascular development. Neuron 2014, 84, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Kirschning, C.J.; Hacker, H.; Redecke, V.; Hausmann, S.; Akira, S.; Wagner, H.; Lipford, G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 2001, 98, 9237–9242. [Google Scholar] [CrossRef] [PubMed]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [PubMed]
- Miyake, K.; Yamashita, Y.; Ogata, M.; Sudo, T.; Kimoto, M. RP105, a novel B cell surface molecule implicated in B cell activation, is a member of the leucine-rich repeat protein family. J. Immunol. 1995, 154, 3333–3340. [Google Scholar] [PubMed]
- Divanovic, S.; Trompette, A.; Atabani, S.F.; Madan, R.; Golenbock, D.T.; Visintin, A.; Finberg, R.W.; Tarakhovsky, A.; Vogel, S.N.; Belkaid, Y.; et al. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat. Immunol. 2005, 6, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Shimazu, R.; Kondo, J.; Niki, T.; Akashi, S.; Ogata, H.; Yamashita, Y.; Miura, Y.; Kimoto, M. Mouse MD-1, a molecule that is physically associated with RP105 and positively regulates its expression. J. Immunol. 1998, 161, 1348–1353. [Google Scholar] [PubMed]
- Inohara, N.; Nunez, G. ML—A conserved domain involved in innate immunity and lipid metabolism. Trends Biochem. Sci. 2002, 27, 219–221. [Google Scholar] [CrossRef]
- Ohto, U.; Miyake, K.; Shimizu, T. Crystal Structures of Mouse and Human RP105/MD-1 Complexes Reveal Unique Dimer Organization of the Toll-Like Receptor Family. J. Mol. Biol. 2011, 413, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.I.; Hong, M.S.; Wilson, I.A. An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1. Nat. Struct. Mol. Biol. 2011, 18, U1028–U1088. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, M.; Muroi, M.; Tanamoto, K.; Suzuki, T.; Azuma, H.; Ikeda, H. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: Roles of the receptor complex. Pharmacol. Ther. 2003, 100, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K. Innate recognition of lipopolysaccharide by CD14 and toll-like receptor 4-MD-2: Unique roles for MD-2. Int. Immunopharmacol. 2003, 3, 119–128. [Google Scholar] [CrossRef]
- Kelley, S.L.; Lukk, T.; Nair, S.K.; Tapping, R.I. The Crystal Structure of Human Soluble CD14 Reveals a Bent Solenoid with a Hydrophobic Amino-Terminal Pocket. J. Immunol. 2013, 190, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Lee, C.J.; Jin, M.S.; Lee, C.H.; Paik, S.G.; Lee, H.; Lee, J.O. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J. Biol. Chem. 2005, 280, 11347–11351. [Google Scholar] [CrossRef] [PubMed]
- Parthier, C.; Stelter, M.; Ursel, C.; Fandrich, U.; Lilie, H.; Breithaupt, C.; Stubbs, M.T. Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity. Proc. Natl. Acad. Sci. USA 2014, 111, 6281–6286. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsushima, N.; Miyashita, H.; Enkhbayar, P.; Kretsinger, R.H. Comparative Geometrical Analysis of Leucine-Rich Repeat Structures in the Nod-Like and Toll-Like Receptors in Vertebrate Innate Immunity. Biomolecules 2015, 5, 1955-1978. https://doi.org/10.3390/biom5031955
Matsushima N, Miyashita H, Enkhbayar P, Kretsinger RH. Comparative Geometrical Analysis of Leucine-Rich Repeat Structures in the Nod-Like and Toll-Like Receptors in Vertebrate Innate Immunity. Biomolecules. 2015; 5(3):1955-1978. https://doi.org/10.3390/biom5031955
Chicago/Turabian StyleMatsushima, Norio, Hiroki Miyashita, Purevjav Enkhbayar, and Robert H. Kretsinger. 2015. "Comparative Geometrical Analysis of Leucine-Rich Repeat Structures in the Nod-Like and Toll-Like Receptors in Vertebrate Innate Immunity" Biomolecules 5, no. 3: 1955-1978. https://doi.org/10.3390/biom5031955




