Immunomodulatory Potential of Fungal Extracellular Vesicles: Insights for Therapeutic Applications
Abstract
:1. Introduction
2. Fungal EVs: Discovery, Composition and Release
2.1. Discovery and Seminal Studies
2.2. Biogenesis and Release of Fungal EVs
2.3. Composition of Fungal EVs
2.3.1. Proteins
2.3.2. Lipids
2.3.3. Nucleic Acids
2.3.4. Other Molecules
3. Comparisons between Bacterial, Mycobacterial and Fungal EVs
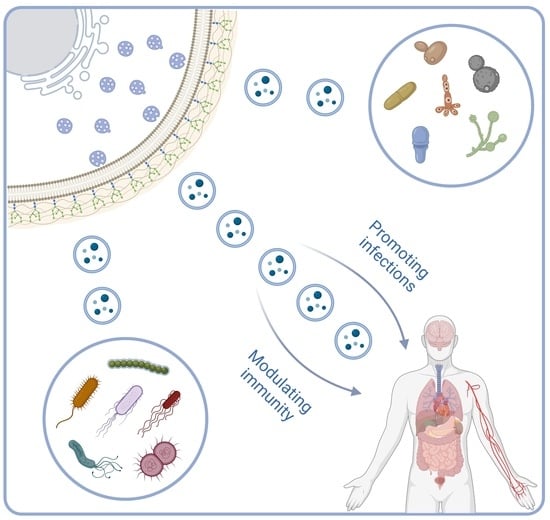
4. Fungal EVs and Interactions with Host Immunity
4.1. Promoting Infections
4.2. Modulating Immunity
4.2.1. Cryptococcus
4.2.2. Candida
4.2.3. Histoplasma capsulatum
4.2.4. Malassezia
4.2.5. Paracoccidioides
4.2.6. Aspergillus
4.2.7. Sporothrix
4.2.8. Trichophyton
5. Fungal EVs for Therapeutical Applications
6. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Mateescu, B.; Kowal, E.J.K.; Van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef]
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2019, 43, 273–303. [Google Scholar] [CrossRef]
- Liebana-Jordan, M.; Brotons, B.; Falcon-Perez, J.M.; Gonzalez, E. Extracellular Vesicles in the Fungi Kingdom. Int. J. Mol. Sci. 2021, 22, 7221. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Donoso-Quezada, J.; Gallo-Villanueva, R.C.; Perez-Gonzalez, V.H.; González-Valdez, J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis 2019, 40, 3036–3049. [Google Scholar] [CrossRef]
- Hartjes, T.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; van Royen, M. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6, 7. [Google Scholar] [CrossRef]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef]
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Oliveira, D.L.; Vargas, G.; Girard-Dias, W.; Franzen, A.J.; Frasés, S.; Miranda, K.; Nimrichter, L. Analysis of Yeast Extracellular Vesicles. In Unconventional Protein Secretion; Pompa, A., De Marchis, F., Eds.; Springer: New York, NY, USA, 2016; pp. 175–190. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef]
- Wolf, P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Wiklander, O.P.B.; Brennan, M.Á.; Lötvall, J.; Breakefield, X.O.; EL Andaloussi, S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019, 11, eaav8521. [Google Scholar] [CrossRef]
- Agrahari, V.; Agrahari, V.; Burnouf, P.-A.; Chew, C.H.; Burnouf, T. Extracellular Microvesicles as New Industrial Therapeutic Frontiers. Trends Biotechnol. 2019, 37, 707–729. [Google Scholar] [CrossRef]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; De Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular Polysaccharide Export in Cryptococcus neoformans Is a Eukaryotic Solution to the Problem of Fungal Trans-Cell Wall Transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef]
- Rizzo, J.; Rodrigues, M.L.; Janbon, G. Extracellular Vesicles in Fungi: Past, Present, and Future Perspectives. Front. Cell. Infect. Microbiol. 2020, 10, 346. [Google Scholar] [CrossRef]
- Leone, F.; Bellani, L.; Mucciflora, S.; Giorgetti, L.; Bongioanni, P.; Simili, M.; Maserti, B.; Del Carratore, R. Analysis of extracellular vesicles produced in the biofilm by the dimorphic yeast Pichia fermentans. J. Cell. Physiol. 2017, 233, 2759–2767. [Google Scholar] [CrossRef]
- Zarnowski, R.; Sanchez, H.; Covelli, A.S.; Dominguez, E.; Jaromin, A.; Bernhardt, J.; Mitchell, K.F.; Heiss, C.; Azadi, P.; Mitchell, A.; et al. Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018, 16, e2006872. [Google Scholar] [CrossRef]
- Voelz, K.; Johnston, S.A.; Smith, L.M.; Hall, R.A.; Idnurm, A.; May, R.C. ‘Division of labour’ in response to host oxidative burst drives a fatal Cryptococcus gattii outbreak. Nat. Commun. 2014, 5, 5194. [Google Scholar] [CrossRef]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.-S.; et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2019, 2, 305. [Google Scholar] [CrossRef]
- Kabani, M.; Melki, R. Sup35p in Its Soluble and Prion States Is Packaged inside Extracellular Vesicles. mBio 2015, 6, e01017-15. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Weiberg, A.; Buck, A.H.; Jin, H. Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog. 2019, 15, e1008090. [Google Scholar] [CrossRef]
- Freitas, M.S.; Bonato, V.L.D.; Pessoni, A.M.; Rodrigues, M.L.; Casadevall, A.; Almeida, F. Fungal Extracellular Vesicles as Potential Targets for Immune Interventions. mSphere 2019, 4, e00747-19. [Google Scholar] [CrossRef]
- Bielska, E.; May, R.C. Extracellular vesicles of human pathogenic fungi. Curr. Opin. Microbiol. 2019, 52, 90–99. [Google Scholar] [CrossRef]
- Marina, C.L.; Bürgel, P.H.; Agostinho, D.P.; Zamith-Miranda, D.; Las-Casas, L.O.; Tavares, A.H.; Nosanchuk, J.D.; Bocca, A.L. Nutritional Conditions Modulate C. neoformans Extracellular Vesicles’ Capacity to Elicit Host Immune Response. Microorganisms 2020, 8, 1815. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Freire-De-Lima, C.G.; Nosanchuk, J.D.; Casadevall, A.; Rodrigues, M.L.; Nimrichter, L. Extracellular Vesicles from Cryptococcus neoformans Modulate Macrophage Functions. Infect. Immun. 2010, 78, 1601–1609. [Google Scholar] [CrossRef]
- Da Silva, T.A.; Roque-Barreira, M.C.; Casadevall, A.; Almeida, F. Extracellular vesicles from Paracoccidioides brasiliensis induced M1 polarization in vitro. Sci. Rep. 2016, 6, 35867. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Rezende, C.P.; Quaresemin, N.R.; Moreno, P.; Hatanaka, O.; Rossi, A.; Martinez-Rossi, N.M.; Almeida, F. Extracellular Vesicles from the Dermatophyte Trichophyton interdigitale Modulate Macrophage and Keratinocyte Functions. Front. Immunol. 2018, 9, 2343. [Google Scholar] [CrossRef]
- Ikeda, M.A.K.; De Almeida, J.R.F.; Jannuzzi, G.P.; Cronemberger-Andrade, A.; Torrecilhas, A.C.T.; Moretti, N.S.; da Cunha, J.P.C.; De Almeida, S.R.; Ferreira, K.S. Extracellular Vesicles from Sporothrix brasiliensis Are an Important Virulence Factor That Induce an Increase in Fungal Burden in Experimental Sporotrichosis. Front. Microbiol. 2018, 9, 2286. [Google Scholar] [CrossRef] [PubMed]
- Brauer, V.S.; Pessoni, A.M.; Bitencourt, T.A.; de Paula, R.G.; de Oliveira Rocha, L.; Goldman, G.H.; Almeida, F. Extracellular Vesicles from Aspergillus flavus Induce M1 Polarization In Vitro. mSphere 2020, 5, e00190-20. [Google Scholar] [CrossRef] [PubMed]
- Kaparakis-Liaskos, M.; Ferrero, R.L. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 2015, 15, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Macia, L.; Nanan, R.; Hosseini-Beheshti, E.; Grau, G.E. Host- and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int. J. Mol. Sci. 2019, 21, 107. [Google Scholar] [CrossRef] [PubMed]
- Sartorio, M.G.; Pardue, E.J.; Feldman, M.F.; Haurat, M.F. Bacterial Outer Membrane Vesicles: From Discovery to Applications. Annu. Rev. Microbiol. 2021, 75, 609–630. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Defourny, K.A.Y.; Smid, E.J.; Abee, T. Gram-Positive Bacterial Extracellular Vesicles and Their Impact on Health and Disease. Front. Microbiol. 2018, 9, 1502. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Coelho, C.; Casadevall, A. Answers to naysayers regarding microbial extracellular vesicles. Biochem. Soc. Trans. 2019, 47, 1005–1012. [Google Scholar] [CrossRef]
- Honorato, L.; Bonilla, J.J.A.; Piffer, A.C.; Nimrichter, L. Fungal Extracellular Vesicles as a Potential Strategy for Vaccine Development. In Fungal Extracellular Vesicles; Rodrigues, M., Janbon, G., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 121–138. [Google Scholar]
- Köhler, J.R.; Hube, B.; Puccia, R.; Casadevall, A.; Perfect, J.R. Fungi that Infect Humans. Microbiol. Spectr. 2017, 5, 813–843. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Kontoyiannis, D.P.; Robert, V. On the Emergence of Candida auris: Climate Change, Azoles, Swamps, and Birds. mBio 2019, 10, e01397-19. [Google Scholar] [CrossRef] [PubMed]
- Gremião, I.D.F.; Miranda, L.H.M.; Reis, E.G.; Rodrigues, A.M.; Pereira, S.A. Zoonotic Epidemic of Sporotrichosis: Cat to Human Transmission. PLoS Pathog. 2017, 13, e1006077. [Google Scholar] [CrossRef] [PubMed]
- Siscar-Lewin, S.; Hube, B.; Brunke, S. Emergence and evolution of virulence in human pathogenic fungi. Trends Microbiol. 2022, 30, 693–704. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Farmakiotis, D.; Kontoyiannis, D.P. Epidemiology of antifungal resistance in human pathogenic yeasts: Current viewpoint and practical recommendations for management. Int. J. Antimicrob. Agents 2017, 50, 318–324. [Google Scholar] [CrossRef]
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular Evolution of Antifungal Drug Resistance. Annu. Rev. Microbiol. 2017, 71, 753–775. [Google Scholar] [CrossRef]
- Nimrichter, L.; de Souza, M.M.; Del Poeta, M.; Nosanchuk, J.D.; Joffe, L.; Tavares, P.d.M.; Rodrigues, M.L. Extracellular Vesicle-Associated Transitory Cell Wall Components and Their Impact on the Interaction of Fungi with Host Cells. Front. Microbiol. 2016, 7, 1034. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Almeida, I.C.; Nimrichter, L. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J. Proteom. 2014, 97, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Nosanchuk, J.D.; Williamson, P.; Rodrigues, M.L. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009, 17, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Casadevall, A. Challenges posed by extracellular vesicles from eukaryotic microbes. Curr. Opin. Microbiol. 2014, 22, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Godinho, R.M.C.; Zamith-Miranda, D.; Nimrichter, L. Traveling into Outer Space: Unanswered Questions about Fungal Extracellular Vesicles. PLoS Pathog. 2015, 11, e1005240. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, H.C.; Kato, A.F.; Sena, B.A.G.; Duarte, I.; Jozefowicz, L.J.; Castelli, R.F.; Kuczera, D.; Reis, F.C.G.; Alves, L.R.; Rodrigues, M.L. Biogenesis of Fungal Extracellular Vesicles: What Do We Know? In Fungal Extracellular Vesicles; Rodrigues, M., Janbon, G., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–11. [Google Scholar]
- Rodrigues, M.L.; Nimrichter, L. From fundamental biology to the search for innovation: The story of fungal extracellular vesicles. Eur. J. Cell Biol. 2022, 101, 151205. [Google Scholar] [CrossRef]
- Chigaleĭchik, A.G.; Belova, L.A.; Grishchenko, V.M.; Rylkin, S.S. Several properties of the extracellular vesicles of Candida tropicalis yeasts grown on n-alkanes. Mikrobiologiia 1977, 46, 467–471. [Google Scholar] [PubMed]
- Gibson, R.K.; Peberdy, J.F. Fine Structure of Protoplasts of Aspergillus nidulans. J. Gen. Microbiol. 1972, 72, 529–538. [Google Scholar] [CrossRef]
- Takeo, K.; Uesaka, I.; Uehira, K.; Nishiura, M. Fine Structure of Cryptococcus neoformans Grown In Vitro as Observed by Freeze-Etching. J. Bacteriol. 1973, 113, 1442–1448. [Google Scholar] [CrossRef]
- Anderson, J.; Mihalik, R.; Soll, D.R. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J. Bacteriol. 1990, 172, 224–235. [Google Scholar] [CrossRef]
- Rodrigues, M.; Janbon, G. Fungal Extracellular Vesicles: Biological Roles; Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar]
- Rodrigues, M.L.; Travassos, L.R.; Miranda, K.R.; Franzen, A.J.; Rozental, S.; de Souza, W.; Alviano, C.S.; Barreto-Bergter, E. Human Antibodies against a Purified Glucosylceramide from Cryptococcus neoformans Inhibit Cell Budding and Fungal Growth. Infect. Immun. 2000, 68, 7049–7060. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular Vesicles Produced by Cryptococcus neoformans Contain Protein Components Associated with Virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Matsuo, A.L.; Ganiko, L.; Medeiros, L.C.S.; Miranda, K.; Silva, L.S.; Freymüller-Haapalainen, E.; Sinigaglia-Coimbra, R.; Almeida, I.C.; Puccia, R. The Pathogenic Fungus Paracoccidioides brasiliensis Exports Extracellular Vesicles Containing Highly Immunogenic α-Galactosyl Epitopes. Eukaryot. Cell 2011, 10, 343–351. [Google Scholar] [CrossRef]
- Zamith-Miranda, D.; Heyman, H.M.; Couvillion, S.P.; Cordero, R.J.B.; Rodrigues, M.L.; Nimrichter, L.; Casadevall, A.; Amatuzzi, R.F.; Alves, L.R.; Nakayasu, E.S.; et al. Comparative Molecular and Immunoregulatory Analysis of Extracellular Vesicles from Candida albicans and Candida auris. mSystems 2021, 6, e0082221. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.; Chow, F.W.-N.; Tsang, C.-C.; Liu, X.; Yao, W.; Chan, T.T.-Y.; Siu, G.K.-H.; Ho, A.Y.-M.; Luk, K.S.; Lau, S.K.-P.; et al. Induction of amphotericin B resistance in susceptible Candida auris by extracellular vesicles. Emerg. Microbes Infect. 2022, 11, 1900–1909. [Google Scholar] [CrossRef] [PubMed]
- Amatuzzi, R.F.; Zamith-Miranda, D.; da Rocha, I.F.M.; Lucena, A.C.R.; de Toledo Martins, S.; Streit, R.; Staats, C.C.; Trentin, G.; Almeida, F.; Rodrigues, M.L.; et al. Caspofungin Affects Extracellular Vesicle Production and Cargo in Candida auris. J. Fungi 2022, 8, 990. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.G.T.; Pathan, M.; Zhao, K.; Ang, C.-S.; Mathivanan, S.; Anderson, M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2020, 10, 1610. [Google Scholar] [CrossRef]
- Costa, J.H.; Bazioli, J.M.; Barbosa, L.D.; dos Santos Júnior, P.L.T.; Reis, F.C.G.; Klimeck, T.; Crnkovic, C.M.; Berlinck, R.G.S.; Sussulini, A.; Rodrigues, M.L.; et al. Phytotoxic Tryptoquialanines Produced In Vivo by Penicillium digitatum Are Exported in Extracellular Vesicles. mBio 2021, 12, e03393-20. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, J.; Albuquerque, P.C.; Wolf, J.M.; Nascimento, R.; Pereira, M.D.; Nosanchuk, J.D.; Rodrigues, M.L. Analysis of multiple components involved in the interaction between Cryptococcus neoformans and Acanthamoeba castellanii. Fungal Biol. 2017, 121, 602–614. [Google Scholar] [CrossRef]
- Rizzo, J.; Chaze, T.; Miranda, K.; Roberson, R.W.; Gorgette, O.; Nimrichter, L.; Matondo, M.; Latgé, J.-P.; Beauvais, A.; Rodrigues, M.L. Characterization of Extracellular Vesicles Produced by Aspergillus fumigatus Protoplasts. mSphere 2020, 5, e00476-20. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, P.C.; Nakayasu, E.S.; Rodrigues, M.L.; Frases, S.; Casadevall, A.; Zancope-Oliveira, R.M.; Almeida, I.C.; Nosanchuk, J.D. Vesicular transport in Histoplasma capsulatum: An effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 2008, 10, 1695–1710. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Hatanaka, O.; Pessoni, A.M.; Freitas, M.S.; Trentin, G.; Santos, P.; Rossi, A.; Martinez-Rossi, N.M.; Alves, L.L.; Casadevall, A.; et al. Fungal Extracellular Vesicles Are Involved in Intraspecies Intracellular Communication. mBio 2022, 13, e0327221. [Google Scholar] [CrossRef] [PubMed]
- Giardina, B.J.; Stein, K.; Chiang, H.-L. The endocytosis gene END3 is essential for the glucose-induced rapid decline of small vesicles in the extracellular fraction in Saccharomyces cerevisiae. J. Extracell. Vesicles 2014, 3, 23497. [Google Scholar] [CrossRef] [PubMed]
- Stein, K.; Chiang, H.-L. Exocytosis and Endocytosis of Small Vesicles across the Plasma Membrane in Saccharomyces cerevisiae. Membranes 2014, 4, 608–629. [Google Scholar] [CrossRef]
- Stein, K.; Winters, C.; Chiang, H.-L. Vps15p regulates the distribution of cup-shaped organelles containing the major eisosome protein Pil1p to the extracellular fraction required for endocytosis of extracellular vesicles carrying metabolic enzymes: Roles of Vps15p and Pil1p in vesicle endocytosis. Biol. Cell 2017, 109, 190–209. [Google Scholar] [CrossRef] [PubMed]
- Kenno, S.; Speth, C.; Rambach, G.; Binder, U.; Chatterjee, S.; Caramalho, R.; Haas, H.; Lass-Flörl, C.; Shaughnessy, J.; Ram, S.; et al. Candida albicans Factor H Binding Molecule Hgt1p–A Low Glucose-Induced Transmembrane Protein Is Trafficked to the Cell Wall and Impairs Phagocytosis and Killing by Human Neutrophils. Front. Microbiol. 2019, 9, 3319. [Google Scholar] [CrossRef] [PubMed]
- Dawson, C.S.; Garcia-Ceron, D.; Rajapaksha, H.; Faou, P.; Bleackley, M.R.; Anderson, M.A. Protein markers for Candida albicans EVs include claudin-like Sur7 family proteins. J. Extracell. Vesicles 2020, 9, 1750810. [Google Scholar] [CrossRef]
- Reis, F.C.G.; Borges, B.S.; Jozefowicz, L.J.; Sena, B.A.G.; Garcia, A.W.A.; Medeiros, L.C.; Martins, S.T.; Honorato, L.; Schrank, A.; Vainstein, M.H.; et al. A Novel Protocol for the Isolation of Fungal Extracellular Vesicles Reveals the Participation of a Putative Scramblase in Polysaccharide Export and Capsule Construction in Cryptococcus gattii. mSphere 2019, 4, e00080-19. [Google Scholar] [CrossRef] [PubMed]
- Cleare, L.G.; Zamith, D.; Heyman, H.M.; Couvillion, S.P.; Nimrichter, L.; Rodrigues, M.L.; Nakayasu, E.S.; Nosanchuk, J.D. Media matters! Alterations in the loading and release of Histoplasma capsulatum extracellular vesicles in response to different nutritional milieus. Cell. Microbiol. 2020, 22, e13217. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Biogenesis of extracellular vesicles in yeast: Many questions with few answers. Commun. Integr. Biol. 2010, 3, 533–535. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nosanchuk, J.D.; Schrank, A.; Vainstein, M.H.; Casadevall, A.; Nimrichter, L.; Lai, R.C.; Chen, T.S.; Lim, S.K.; Hodge, T.W.; et al. Vesicular transport systems in fungi. Future Microbiol. 2011, 6, 1371–1381. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Rizzo, J.; Joffe, L.S.; Godinho, R.M.C.; Rodrigues, M.L. Where Do They Come from and Where Do They Go: Candidates for Regulating Extracellular Vesicle Formation in Fungi. Int. J. Mol. Sci. 2013, 14, 9581–9603. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Latifkar, A.; Hur, Y.H.; Sanchez, J.C.; Cerione, R.A.; Antonyak, M.A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. 2019, 132, jcs222406. [Google Scholar] [CrossRef] [PubMed]
- Novick, P.; Field, C.; Schekman, R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 1980, 21, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Schekman, R. SEC mutants and the secretory apparatus. Nat. Med. 2002, 8, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of Yeast Extracellular Vesicles: Evidence for the Participation of Different Pathways of Cellular Traffic in Vesicle Biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef]
- Panepinto, J.; Komperda, K.; Frases, S.; Park, Y.-D.; Djordjevic, J.T.; Casadevall, A.; Williamson, P.R. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol. Microbiol. 2009, 71, 1165–1176. [Google Scholar] [CrossRef]
- Da Silva, R.P.; Martins, S.D.T.; Rizzo, J.; Dos Reis, F.C.G.; Joffe, L.S.; Vainstein, M.; Kmetzsch, L.; Oliveira, D.L.; Puccia, R.; Goldenberg, S.; et al. Golgi Reassembly and Stacking Protein (GRASP) Participates in Vesicle-Mediated RNA Export in Cryptococcus neoformans. Genes 2018, 9, 400. [Google Scholar] [CrossRef]
- Piper, R.C.; Cooper, A.A.; Yang, H.; Stevens, T.H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 1995, 131, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Emr, S.D. The Escrt Complexes: Structure and Mechanism of a Membrane-Trafficking Network. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, T.; Onishi, M.; Ikeuchi, M.; Kita, A.; Sugiura, R.; Giga-Hama, Y.; Fukui, Y.; Takegawa, K. Essential roles of class E Vps proteins for sorting into multivesicular bodies in Schizosaccharomyces pombe. Microbiology 2007, 153, 2753–2764. [Google Scholar] [CrossRef] [PubMed]
- Chiaruttini, N.; Redondo-Morata, L.; Colom, A.; Humbert, F.; Lenz, M.; Scheuring, S.; Roux, A. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 2015, 163, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Caza, M.; Cadieux, B.; Chan, V.; Liu, V.; Kronstad, J. Cryptococcus neoformans Requires the ESCRT Protein Vps23 for Iron Acquisition from Heme, for Capsule Formation, and for Virulence. Infect. Immun. 2013, 81, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Caza, M.; Cadieux, B.; Bakkeren, E.; Do, E.; Jung, W.H.; Kronstad, J.W. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in C ryptococcus neoformans: ESCRT machinery and virulence in Cryptococcus neoformans. Mol. Microbiol. 2015, 96, 973–992. [Google Scholar] [CrossRef] [PubMed]
- Da Godinho, R.M.C.; Crestani, J.; Kmetzsch, L.; de Araujo, G.S.; Frases, S.; Staats, C.C.; Schrank, A.; Vainstein, M.H.; Rodrigues, M.L. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci. Rep. 2014, 4, 6198. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-D.; Chen, S.H.; Camacho, E.; Casadevall, A.; Williamson, P.R. Role of the ESCRT Pathway in Laccase Trafficking and Virulence of Cryptococcus neoformans. Infect. Immun. 2020, 88, e00954-19. [Google Scholar] [CrossRef]
- Bruckmann, A.; Künkel, W.; Augsten, K.; Wetzker, R.; Eck, R. The deletion of CaVPS34 in the human pathogenic yeast Candida albicans causes defects in vesicle-mediated protein sorting and nuclear segregation. Yeast 2001, 18, 343–353. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Franzen, A.J.; Nimrichter, L.; Miranda, K. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr. Opin. Microbiol. 2013, 16, 414–420. [Google Scholar] [CrossRef]
- Rizzo, J.; Oliveira, D.L.; Joffe, L.S.; Hu, G.; Gazos-Lopes, F.; Fonseca, F.L.; Almeida, I.C.; Frases, S.; Kronstad, J.W.; Rodrigues, M.L. Role of the Apt1 Protein in Polysaccharide Secretion by Cryptococcus neoformans. Eukaryot. Cell 2014, 13, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Espadas, J.; Luque-Garcia, J.; Reynolds, T.; Casadevall, A. Lipid Biosynthetic Genes Affect Candida albicans Extracellular Vesicle Morphology, Cargo, and Immunostimulatory Properties. Eukaryot. Cell 2015, 14, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, J.; Colombo, A.C.; Zamith-Miranda, D.; Silva, V.K.; Allegood, J.C.; Casadevall, A.; Del Poeta, M.; Nosanchuk, J.D.; Kronstad, J.W.; Rodrigues, M.L. The putative flippase Apt1 is required for intracellular membrane architecture and biosynthesis of polysaccharide and lipids in Cryptococcus neoformans. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2018, 1865, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Coronado, J.E.; Mneimneh, S.; Epstein, S.L.; Qiu, W.-G.; Lipke, P.N. Conserved Processes and Lineage-Specific Proteins in Fungal Cell Wall Evolution. Eukaryot. Cell 2007, 6, 2269–2277. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 28513415. [Google Scholar] [CrossRef] [PubMed]
- De Souza Pereira, R.; Geibel, J. Direct observation of oxidative stress on the cell wall of Saccharomyces cerevisiae strains with atomic force microscopy. Mol. Cell. Biochem. 1999, 201, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Espadas-Moreno, J.; Luque-Garcia, J.L.; Casadevall, A. Interaction of Cryptococcus neoformans Extracellular Vesicles with the Cell Wall. Eukaryot. Cell 2014, 13, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.; Sood, P.; Lenardon, M.D.; Milne, G.; Olson, J.; Jensen, G.; Wolf, J.; Casadevall, A.; Adler-Moore, J.; Gow, N.A.R. The Viscoelastic Properties of the Fungal Cell Wall Allow Traffic of AmBisome as Intact Liposome Vesicles. mBio 2018, 9, e02383-17. [Google Scholar] [CrossRef]
- De Toledo Martins, S.; Szwarc, P.; Goldenberg, S.; Alves, L.R. Extracellular Vesicles in Fungi: Composition and Functions. In Fungal Physiology and Immunopathogenesis; Rodrigues, M.L., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 45–59. [Google Scholar]
- Zamith-Miranda, D.; da Silva, R.P.; Couvillion, S.P.; Bredeweg, E.L.; Burnet, M.C.; Coelho, C.; Camacho, E.; Nimrichter, L.; Puccia, R.; Almeida, I.C.; et al. Omics Approaches for Understanding Biogenesis, Composition and Functions of Fungal Extracellular Vesicles. Front. Genet. 2021, 12, 648524. [Google Scholar] [CrossRef]
- Peres da Silva, R.; Puccia, R.; Rodrigues, M.L.; Oliveira, D.L.; Joffe, L.S.; César, G.V.; Nimrichter, L.; Goldenberg, S.; Alves, L.R. Extracellular vesicle-mediated export of fungal RNA. Sci. Rep. 2015, 5, 7763. [Google Scholar] [CrossRef]
- Rayner, S.; Bruhn, S.; Vallhov, H.; Andersson, A.; Billmyre, R.B.; Scheynius, A. Identification of small RNAs in extracellular vesicles from the commensal yeast Malassezia sympodialis. Sci. Rep. 2017, 7, 39742. [Google Scholar] [CrossRef] [PubMed]
- Alves, L.R.; da Silva, R.P.; Sanchez, D.A.; Zamith-Miranda, D.; Rodrigues, M.L.; Goldenberg, S.; Puccia, R.; Nosanchuk, J.D. Extracellular Vesicle-Mediated RNA Release in Histoplasma capsulatum. mSphere 2019, 4, e00176-19. [Google Scholar] [CrossRef] [PubMed]
- Peres da Silva, R.; Longo, L.G.V.; da Cunha, J.P.C.; Sobreira, T.J.P.; Rodrigues, M.L.; Faoro, H.; Goldenberg, S.; Alves, L.R.; Puccia, R. Comparison of the RNA Content of Extracellular Vesicles Derived from Paracoccidioides brasiliensis and Paracoccidioides lutzii. Cells 2019, 8, 765. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.P.; Longo, L.V.G.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and Vesicle-Free Extracellular Proteome of Paracoccidioides brasiliensis: Comparative Analysis with Other Pathogenic Fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Mencher, A.; Morales, P.; Valero, E.; Tronchoni, J.; Patil, K.R.; Gonzalez, R. Proteomic characterization of extracellular vesicles produced by several wine yeast species. Microb. Biotechnol. 2020, 13, 1581–1596. [Google Scholar] [CrossRef]
- Morales, P.; Mencher, A.; Tronchoni, J.; Gonzalez, R. Fungal Extracellular Vesicles; Rodrigues, M., Janbon, G., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 161–170. [Google Scholar]
- Johansson, H.J.; Vallhov, H.; Holm, T.; Gehrmann, U.; Andersson, A.; Johansson, C.; Blom, H.; Carroni, M.; Lehtiö, J.; Scheynius, A. Extracellular nanovesicles released from the commensal yeast Malassezia sympodialis are enriched in allergens and interact with cells in human skin. Sci. Rep. 2018, 8, 9182. [Google Scholar] [CrossRef] [PubMed]
- Gil-Bona, A.; Llama-Palacios, A.; Parra, C.M.; Vivanco, F.; Nombela, C.; Monteoliva, L.; Gil, C. Proteomics Unravels Extracellular Vesicles as Carriers of Classical Cytoplasmic Proteins in Candida albicans. J. Proteome Res. 2015, 14, 142–153. [Google Scholar] [CrossRef]
- Vargas, G.; Rocha, J.D.B.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.O.; Medeiros, L.C.A.S.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans: Extracellular vesicles from Candida albicans. Cell. Microbiol. 2014, 17, 389–407. [Google Scholar] [CrossRef]
- Kabani, M.; Pilard, M.; Melki, R. Glucose availability dictates the export of the soluble and prion forms of Sup35p via periplasmic or extracellular vesicles. Mol. Microbiol. 2020, 114, 322–332. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Nakayasu, E.S.; Longo, L.V.G.; Ganiko, L.; Lopes, F.G.; Matsuo, A.L.; Almeida, I.C.; Puccia, R. Lipidomic Analysis of Extracellular Vesicles from the Pathogenic Phase of Paracoccidioides brasiliensis. PLoS ONE 2012, 7, e39463. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Nosanchuk, J.D.; Casadevall, A. Vesicular Trans-Cell Wall Transport in Fungi: A Mechanism for the Delivery of Virulence-Associated Macromolecules? Lipid Insights 2008, 2, LPI.S1000. [Google Scholar] [CrossRef] [PubMed]
- Matos Baltazar, L.; Nakayasu, E.S.; Sobreira, T.J.P.; Choi, H.; Casadevall, A.; Nimrichter, L.; Nosanchuk, J.D. Antibody Binding Alters the Characteristics and Contents of Extracellular Vesicles Released by Histoplasma capsulatum. mSphere 2016, 1, e00085-15. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O’donoghue, E.J.; May, R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 2018, 9, 1556. [Google Scholar] [CrossRef] [PubMed]
- da Silva, R.P.; Heiss, C.; Black, I.; Azadi, P.; Gerlach, J.Q.; Travassos, L.R.; Joshi, L.; Kilcoyne, M.; Puccia, R. Extracellular vesicles from Paracoccidioides pathogenic species transport polysaccharide and expose ligands for DC-SIGN receptors. Sci. Rep. 2015, 5, 14213. [Google Scholar] [CrossRef] [PubMed]
- Rella, A.; Farnoud, A.M.; Del Poeta, M. Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 2016, 61, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Tsatsaronis, J.A.; Franch-Arroyo, S.; Resch, U.; Charpentier, E. Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol. 2018, 26, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Pessoni, A.M.; Oliveira, B.T.M.; Alves, L.R.; Almeida, F. The RNA Content of Fungal Extracellular Vesicles: At the “Cutting-Edge” of Pathophysiology Regulation. Cells 2022, 11, 2184. [Google Scholar] [CrossRef] [PubMed]
- Munhoz da Rocha, I.F.; Amatuzzi, R.F.; Lucena, A.C.R.; Faoro, H.; Alves, L.R. Cross-Kingdom Extracellular Vesicles EV-RNA Communication as a Mechanism for Host–Pathogen Interaction. Front. Cell. Infect. Microbiol. 2020, 10, 593160. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Weiberg, A.; Bellinger, M.; Jin, H. Conversations between kingdoms: Small RNAs. Curr. Opin. Biotechnol. 2015, 32, 207–215. [Google Scholar] [CrossRef]
- Lee, H.-J. Microbe-Host Communication by Small RNAs in Extracellular Vesicles: Vehicles for Transkingdom RNA Transportation. Int. J. Mol. Sci. 2019, 20, 1487. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, Z.; Liu, X.; Tyler, B.M. Biogenesis and Biological Functions of Extracellular Vesicles in Cellular and Organismal Communication With Microbes. Front. Microbiol. 2022, 13, 817844. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Tisserant, C.; Tulinski, M.; Weiberg, A.; Feldbrügge, M. Inside-out: From endosomes to extracellular vesicles in fungal RNA transport. Fungal Biol. Rev. 2020, 34, 89–99. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. tritici mi croRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.; Liang, X. One Small RNA of Fusarium graminearum Targets and Silences CEBiP Gene in Common Wheat. Microorganisms 2019, 7, 425. [Google Scholar] [CrossRef] [PubMed]
- Munhoz da Rocha, I.F.; Martins, S.T.; Amatuzzi, R.F.; Zamith-Miranda, D.; Nosanchuk, J.D.; Rodrigues, M.L.; Alves, L.R. Cellular and Extracellular Vesicle RNA Analysis in the Global Threat Fungus Candida auris. Microbiol. Spectr. 2021, 9, e0153821. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Mues, M.; Weber, S.E.; Frases, S.; Chaskes, S.; Gerfen, G.; Casadevall, A. Cryptococcus neoformans laccase catalyses melanin synthesis from both d- and l-DOPA. Microbiology 2007, 153, 3954–3962. [Google Scholar] [CrossRef]
- Kabani, M. Extracellular Vesicles-Encapsulated Yeast Prions and What They Can Tell Us about the Physical Nature of Propagons. Int. J. Mol. Sci. 2020, 22, 90. [Google Scholar] [CrossRef]
- Bishop, D.; Work, E. An extracellular glycolipid produced by Escherichia coli grown under lysine-limiting conditions. Biochem. J. 1965, 96, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Knox, K.W.; Vesk, M.; Work, E. Relation Between Excreted Lipopolysaccharide Complexes and Surface Structures of a Lysine-Limited Culture of Escherichia coli. J. Bacteriol. 1966, 92, 1206–1217. [Google Scholar] [CrossRef] [PubMed]
- Work, E.; Knox, K.W.; Vesk, M. The Chemistry and Electron Microscopy of an Extracellular Lipopolysaccharide from Escherichia coli. Ann. N. Y. Acad. Sci. 1966, 133, 438–449. [Google Scholar] [CrossRef]
- Knox, K.; Cullen, J.; Work, E. An extracellular lipopolysaccharide-phospholipid-protein complex produced by Escherichia coli grown under lysine-limiting conditions. Biochem. J. 1967, 103, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Dorward, D.W.; Garon, C.F. DNA Is Packaged within Membrane-Derived Vesicles of Gram-Negative but Not Gram-Positive Bacteria. Appl. Environ. Microbiol. 1990, 56, 1960–1962. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Genet. 2015, 13, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host–pathogen interaction. J. Bone Jt. Surg. 2005, 19, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, T.J. Structures of Gram-Negative Cell Walls and Their Derived Membrane Vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.N.; Kuehn, M.J. Virulence and Immunomodulatory Roles of Bacterial Outer Membrane Vesicles. Microbiol. Mol. Biol. Rev. 2010, 74, 81–94. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological Functions and Biogenesis of Secreted Bacterial Outer Membrane Vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef]
- Costerton, J.W.; Ingram, J.M.; Cheng, K.J. Structure and function of the cell envelope of gram-negative bacteria. Microbiol. Mol. Biol. Rev. 1974, 38, 87–110. [Google Scholar] [CrossRef]
- Kadurugamuwa, J.L.; Beveridge, T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. J. Bacteriol. 1995, 177, 3998–4008. [Google Scholar] [CrossRef] [PubMed]
- Kadurugamuwa, J.L.; Beveridge, T.J. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: Conceptually new antibiotics. J. Bacteriol. 1996, 178, 2767–2774. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.M.; Whiteley, M. Special delivery: Vesicle trafficking in prokaryotes. Mol. Microbiol. 2006, 61, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Schertzer, J.W.; Whiteley, M. A Bilayer-Couple Model of Bacterial Outer Membrane Vesicle Biogenesis. Mbio 2012, 3, e00297-11. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, H.M.; Jagannadham, M.V. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 2014, 160, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Kolling, G.L.; Matthews, K.R. Export of Virulence Genes and Shiga Toxin by Membrane Vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999, 65, 1843–1848. [Google Scholar] [CrossRef] [PubMed]
- Mashburn, L.M.; Whiteley, M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 2005, 437, 422–425. [Google Scholar] [CrossRef]
- Schooling, S.R.; Beveridge, T.J. Membrane Vesicles: An Overlooked Component of the Matrices of Biofilms. J. Bacteriol. 2006, 188, 5945–5957. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Rivera, J.; Cordero, R.J.B.; Nakouzi, A.S.; Frases, S.; Nicola, A.; Casadevall, A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. USA 2010, 107, 19002–19007. [Google Scholar] [CrossRef]
- Brown, L.; Kessler, A.; Cabezas-Sanchez, P.; Luque-Garcia, J.L.; Casadevall, A. Extracellular vesicles produced by the Gram-positive bacterium B acillus subtilis are disrupted by the lipopeptide surfactin: Vesicle production and disruption in Bacillus subtilis. Mol. Microbiol. 2014, 93, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Kong, Q.; Roland, K.L.; Curtiss, R. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int. J. Med. Microbiol. 2014, 304, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Schrempf, H.; Koebsch, I.; Walter, S.; Engelhardt, H.; Meschke, H. Extracellular Streptomyces vesicles: Amphorae for survival and defence. Microb. Biotechnol. 2011, 4, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Shockman, G.D.; Barren, J.F. Structure, Function, and Assembly of Cell Walls of Gram-Positive Bacteria. Annu. Rev. Microbiol. 1983, 37, 501–527. [Google Scholar] [CrossRef] [PubMed]
- Marsollier, L.; Brodin, P.; Jackson, M.; Korduláková, J.; Tafelmeyer, P.; Carbonnelle, E.; Aubry, J.; Milon, G.; Legras, P.; André, J.-P.S.; et al. Impact of Mycobacterium ulcerans Biofilm on Transmissibility to Ecological Niches and Buruli Ulcer Pathogenesis. PLoS Pathog. 2007, 3, e62. [Google Scholar] [CrossRef] [PubMed]
- Prados-Rosales, R.; Brown, L.; Casadevall, A.; Montalvo-Quirós, S.; Luque-Garcia, J.L. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX 2014, 1, 124–129. [Google Scholar] [CrossRef]
- Rath, P.; Huang, C.; Wang, T.; Wang, T.; Li, H.; Prados-Rosales, R.; Elemento, O.; Casadevall, A.; Nathan, C.F. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2013, 110, E4790–E4797. [Google Scholar] [CrossRef]
- Prados-Rosales, R.; Carreño, L.J.; Batista-Gonzalez, A.; Baena, A.; Venkataswamy, M.M.; Xu, J.; Yu, X.; Wallstrom, G.; Magee, D.M.; LaBaer, J.; et al. Mycobacterial Membrane Vesicles Administered Systemically in Mice Induce a Protective Immune Response to Surface Compartments of Mycobacterium tuberculosis. mBio 2014, 5, e01921-14. [Google Scholar] [CrossRef]
- Prados-Rosales, R.; Baena, A.; Martinez, L.R.; Luque-Garcia, J.; Kalscheuer, R.; Veeraraghavan, U.; Camara, C.; Nosanchuk, J.D.; Besra, G.S.; Chen, B.; et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Investig. 2011, 121, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 2003, 83, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Furuyama, N.; Sircili, M.P. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. BioMed Res. Int. 2021, 2021, 1490732. [Google Scholar] [CrossRef] [PubMed]
- Prados-Rosales, R.; Weinrick, B.C.; Piqué, D.G.; Jacobs, W.R.; Casadevall, A.; Rodriguez, G.M. Role for Mycobacterium tuberculosis Membrane Vesicles in Iron Acquisition. J. Bacteriol. 2014, 196, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Joffe, L.S.; Nimrichter, L.; Rodrigues, M.L.; Del Poeta, M. Potential Roles of Fungal Extracellular Vesicles during Infection. mSphere 2016, 1, e00099-16. [Google Scholar] [CrossRef]
- Freitas, M.S.; Pessoni, A.M.; Coelho, C.; Bonato, V.L.D.; Rodrigues, M.L.; Casadevall, A.; Almeida, F. Interactions of Extracellular Vesicles from Pathogenic Fungi with Innate Leukocytes. In Fungal Extracellular Vesicles; Rodrigues, M., Janbon, G., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 89–120. [Google Scholar]
- Piffer, A.C.; Kuczera, D.; Rodrigues, M.L.; Nimrichter, L. The paradoxical and still obscure properties of fungal extracellular vesicles. Mol. Immunol. 2021, 135, 137–146. [Google Scholar] [CrossRef]
- Patin, E.C.; Thompson, A.; Orr, S.J. Pattern recognition receptors in fungal immunity. Semin. Cell Dev. Biol. 2019, 89, 24–33. [Google Scholar] [CrossRef]
- Baltazar, L.M.; Zamith-Miranda, D.; Burnet, M.C.; Choi, H.; Nimrichter, L.; Nakayasu, E.S.; Nosanchuk, J.D. Concentration-dependent protein loading of extracellular vesicles released by Histoplasma capsulatum after antibody treatment and its modulatory action upon macrophages. Sci. Rep. 2018, 8, 8065. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.A.M.; Baltazar, L.d.M.; Carregal, V.M.; Gouveia-Eufrasio, L.; de Oliveira, A.G.; Dias, W.G.; Rocha, M.C.; de Miranda, K.R.; Malavazi, I.; Santos, D.d.A.; et al. Characterization of Aspergillus fumigatus Extracellular Vesicles and Their Effects on Macrophages and Neutrophils Functions. Front. Microbiol. 2019, 10, 2008. [Google Scholar] [CrossRef]
- Gehrmann, U.; Qazi, K.R.; Johansson, C.; Hultenby, K.; Karlsson, M.; Lundeberg, L.; Gabrielsson, S.; Scheynius, A. Nanovesicles from Malassezia sympodialis and Host Exosomes Induce Cytokine Responses–Novel Mechanisms for Host-Microbe Interactions in Atopic Eczema. PLoS ONE 2011, 6, e21480. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Honorato, L.; Guimarães, A.J.; Rodrigues, M.L.; Reis, F.C.G.; Vale, A.M.; Ray, A.; Nosanchuk, J.D.; Nimrichter, L. Protective effect of fungal extracellular vesicles against murine candidiasis. Cell. Microbiol. 2020, 22, e13238. [Google Scholar] [CrossRef]
- Zaragoza, O. Basic principles of the virulence of Cryptococcus. Virulence 2019, 10, 490–501. [Google Scholar] [CrossRef]
- Hai, T.P.; Tuan, T.L.; Van Anh, D.; Mai, T.N.; Phu Huong, L.N.; Thwaites, G.E.; Johnson, E.; Van Vinh Chau, N.; Baker, S.; Ashton, P.M.; et al. The virulence of the Cryptococcus neoformans VNIa-5 lineage is highly plastic and associated with isolate background. Microbiology 2020. [Google Scholar] [CrossRef]
- Höfs, S.; Mogavero, S.; Hube, B. Interaction of Candida albicans with host cells: Virulence factors, host defense, escape strategies, and the microbiota. J. Microbiol. 2016, 54, 149–169. [Google Scholar] [CrossRef]
- Boral, H.; Metin, B.; Döğen, A.; Seyedmousavi, S.; Ilkit, M. Overview of selected virulence attributes in Aspergillus fumigatus, Candida albicans, Cryptococcus neoformans, Trichophyton rubrum, and Exophiala dermatitidis. Fungal Genet. Biol. 2018, 111, 92–107. [Google Scholar] [CrossRef]
- Santos, L.A.; Grisolia, J.C.; Burger, E.; de Araujo Paula, F.B.; Dias, A.L.T.; Malaquias, L.C.C. Virulence factors of Paracoccidioides brasiliensis as therapeutic targets: A review. Antonie Leeuwenhoek 2020, 113, 593–604. [Google Scholar] [CrossRef]
- Mihu, M.R.; Nosanchuk, J.D. HistoplasmaVirulence and Host Responses. Int. J. Microbiol. 2011, 2012, 268123. [Google Scholar] [CrossRef]
- Moyes, D.L.; Richardson, J.P.; Naglik, J.R. Candida albicans-epithelial interactions and pathogenicity mechanisms: Scratching the surface. Virulence 2015, 6, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Tóth, R.; Gácser, A. Mechanisms of Pathogenic Candida Species to Evade the Host Complement Attack. Front. Cell. Infect. Microbiol. 2020, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a Multifunctional Adhesin and Invasin. Eukaryot. Cell 2011, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Coelho, C.; Cordero, R.J.B.; Dragotakes, Q.; Jung, E.; Vij, R.; Wear, M.P. The capsule of Cryptococcus neoformans. Virulence 2019, 10, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Monari, C.; Bistoni, F.; Vecchiarelli, A. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res. 2006, 6, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Villena, S.N.; Pinheiro, R.O.; Pinheiro, C.S.; Nunes, M.P.; Takiya, C.M.; DosReis, G.A.; Previato, J.O.; Mendonça-Previato, L.; Freire-De-Lima, C.G. Capsular polysaccharides galactoxylomannan and glucuronoxylomannan from Cryptococcus neoformans induce macrophage apoptosis mediated by Fas ligand. Cell. Microbiol. 2008, 10, 1274–1285. [Google Scholar] [CrossRef]
- Decote-Ricardo, D.; LaRocque-De-Freitas, I.F.; Rocha, J.D.B.; Nascimento, D.O.; Nunes, M.P.; Morrot, A.; Freire-De-Lima, L.; Previato, J.O.; Mendonça-Previato, L.; Freire-De-Lima, C.G. Immunomodulatory Role of Capsular Polysaccharides Constituents of Cryptococcus neoformans. Front. Med. 2019, 6, 129. [Google Scholar] [CrossRef]
- Esher, S.K.; Zaragoza, O.; Alspaugh, J.A. Cryptococcal pathogenic mechanisms: A dangerous trip from the environment to the brain. Memórias Inst. Oswaldo Cruz 2018, 113, e180057. [Google Scholar] [CrossRef]
- Long, K.H.; Gomez, F.J.; Morris, R.E.; Newman, S.L. Identification of Heat Shock Protein 60 as the Ligand on Histoplasma capsulatum That Mediates Binding to CD18 Receptors on Human Macrophages. J. Immunol. 2003, 170, 487–494. [Google Scholar] [CrossRef]
- Ehlers, M.R.W. CR3: A general purpose adhesion-recognition receptor essential for innate immunity. Microbes Infect. 2000, 2, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Rappleye, C.A. Differentiation of the fungus Histoplasma capsulatum into a pathogen of phagocytes. Curr. Opin. Microbiol. 2017, 40, 1–7. [Google Scholar] [CrossRef]
- Puccia, R.; Travassos, L.R. 43-kilodalton glycoprotein from Paracoccidioides brasiliensis: Immunochemical reactions with sera from patients with paracoccidioidomycosis, histoplasmosis, or Jorge Lobo’s disease. J. Clin. Microbiol. 1991, 29, 1610–1615. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, A.P.; Gesztesi, J.L.; Franco, M.F.; de Souza, W.; de Moraes, J.Z.; Travassos, L.R.; Lopes, J.D. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 1994, 62, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Giannini, M.J.; Taylor, M.L.; Bouchara, J.B.; Burger, E.; Calich, V.L.; Escalante, E.D.; Hanna, S.A.; Lenzi, H.L.; Machado, M.P.; Miyaji, M.; et al. Pathogenesis II: Fungal responses to host responses: Interaction of host cells with fungi. Med. Mycol. 2000, 38 (Suppl. S1), 113–123. [Google Scholar] [CrossRef] [PubMed]
- Flavia Popi, A.; Daniel Lopes, J.; Mariano, M. GP43 from Paracoccidioides brasiliensis inhibits macrophage functions. An evasion mechanism of the fungus. Cell. Immunol. 2002, 218, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-H.; Wu, C.-H.; Chang, Y.C.; Kwon-Chung, K.J.; Brown, R.J.; Jong, A. Cryptococcus neoformans-Derived Microvesicles Enhance the Pathogenesis of Fungal Brain Infection. PLoS ONE 2012, 7, e48570. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.C.; Rella, A.; Normile, T.; Joffe, L.S.; Tavares, P.M.; de S. Araújo, G.R.; Frases, S.; Orner, E.P.; Farnoud, A.M.; Fries, B.C.; et al. Cryptococcus neoformans Glucuronoxylomannan and Sterylglucoside Are Required for Host Protection in an Animal Vaccination Model. mBio 2019, 10, e02909-18. [Google Scholar] [CrossRef] [PubMed]
- Retini, C.; Kozel, T.R.; Pietrella, D.; Monari, C.; Bistoni, F.; Vecchiarelli, A. Interdependency of Interleukin-10 and Interleukin-12 in Regulation of T-Cell Differentiation and Effector Function of Monocytes in Response to Stimulation with Cryptococcus neoformans. Infect. Immun. 2001, 69, 6064–6073. [Google Scholar] [CrossRef]
- Witherden, E.A.; Shoaie, S.; Hall, R.A.; Moyes, D.L. The Human Mucosal Mycobiome and Fungal Community Interactions. J. Fungi 2017, 3, 56. [Google Scholar] [CrossRef]
- Polke, M.; Hube, B.; Jacobsen, I.D. Candida Survival Strategies. Adv. Appl. Microbiol. 2015, 91, 139–235. [Google Scholar] [CrossRef]
- Kumamoto, C.A.; Gresnigt, M.S.; Hube, B. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr. Opin. Microbiol. 2020, 56, 7–15. [Google Scholar] [CrossRef]
- Guimarães, A.J.; Frases, S.; Gomez, F.J.; Zancopé-Oliveira, R.M.; Nosanchuk, J.D. Monoclonal Antibodies to Heat Shock Protein 60 Alter the Pathogenesis of Histoplasma capsulatum. Infect. Immun. 2009, 77, 1357–1367. [Google Scholar] [CrossRef]
- Harada, K.; Saito, M.; Sugita, T.; Tsuboi, R. Malassezia species and their associated skin diseases. J. Dermatol. 2015, 42, 250–257. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Han, Y.; Sun, Y.-Z.; Jiang, H.-H.; Liu, M.; Qi, R.-Q.; Gao, X.-H. Extracellular vesicles derived from Malassezia furfur stimulate IL-6 production in keratinocytes as demonstrated in in vitro and in vivo models. J. Dermatol. Sci. 2019, 93, 168–175. [Google Scholar] [CrossRef]
- Vallhov, H.; Johansson, C.; Veerman, R.E.; Scheynius, A. Extracellular Vesicles Released from the Skin Commensal Yeast Malassezia sympodialis Activate Human Primary Keratinocytes. Front. Cell. Infect. Microbiol. 2020, 10, 6. [Google Scholar] [CrossRef]
- Walling, B.L.; Kim, M. LFA-1 in T Cell Migration and Differentiation. Front. Immunol. 2018, 9, 952. [Google Scholar] [CrossRef]
- Lyck, R.; Enzmann, G. The physiological roles of ICAM-1 and ICAM-2 in neutrophil migration into tissues. Curr. Opin. Hematol. 2015, 22, 53–59. [Google Scholar] [CrossRef]
- Martinez, R. Epidemiology of Paracoccidioidomycosis. Rev. Inst. Med. Trop. São Paulo 2015, 57 (Suppl. S19), 11–20. [Google Scholar] [CrossRef]
- Mendes, R.P.; Cavalcante, R.d.S.; Marques, S.A.; Marques, M.E.A.; Venturini, J.; Sylvestre, T.F.; Paniago, A.M.M.; Pereira, A.C.; da Silva, J.d.F.; Fabro, A.T.; et al. Paracoccidioidomycosis: Current Perspectives from Brazil. Open Microbiol. J. 2017, 11, 224–282. [Google Scholar] [CrossRef]
- Leopold Wager, C.M.; Wormley, F.L. Classical versus alternative macrophage activation: The Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol. 2014, 7, 1023–1035. [Google Scholar] [CrossRef]
- Pasqualotto, A.C. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med. Mycol. 2009, 47, S261–S270. [Google Scholar] [CrossRef]
- Almeida, F.; Rodrigues, M.L.; Coelho, C. The Still Underestimated Problem of Fungal Diseases Worldwide. Front. Microbiol. 2019, 10, 214. [Google Scholar] [CrossRef]
- Zhan, P.; Liu, W. The Changing Face of Dermatophytic Infections Worldwide. Mycopathologia 2017, 182, 77–86. [Google Scholar] [CrossRef]
- Nami, S.; Mohammadi, R.; Vakili, M.; Khezripour, K.; Mirzaei, H.; Morovati, H. Fungal vaccines, mechanism of actions and immunology: A comprehensive review. Biomed. Pharmacother. 2019, 109, 333–344. [Google Scholar] [CrossRef]
- Nicola, A.M.; Albuquerque, P.; Paes, H.C.; Fernandes, L.; Costa, F.F.; Kioshima, E.S.; Abadio, A.K.R.; Bocca, A.L.; Felipe, M.S. Antifungal drugs: New insights in research & development. Pharmacol. Ther. 2019, 195, 21–38. [Google Scholar] [CrossRef]
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol. Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef]
- Nguyen, M.H.; Wissel, M.C.; Salomoni, M.A.; Hao, B.; Press, E.G.; Shields, R.M.; Cheng, S.; Mitsani, D.; Vadnerkar, A.; Silveira, F.P.; et al. Performance of Candida Real-time Polymerase Chain Reaction, -D-Glucan Assay, and Blood Cultures in the Diagnosis of Invasive Candidiasis. Clin. Infect. Dis. 2012, 54, 1240–1248. [Google Scholar] [CrossRef]
- Baddley, J.W.; Andes, D.R.; Marr, K.A.; Kontoyiannis, D.P.; Alexander, B.D.; Kauffman, C.A.; Oster, R.A.; Anaissie, E.J.; Walsh, T.J.; Schuster, M.G.; et al. Factors Associated with Mortality in Transplant Patients with Invasive Aspergillosis. Clin. Infect. Dis. 2010, 50, 1559–1567. [Google Scholar] [CrossRef]
- Park, B.J.; Pappas, P.G.; Wannemuehler, K.A.; Alexander, B.D.; Anaissie, E.J.; Andes, D.R.; Baddley, J.W.; Brown, J.M.; Brumble, L.M.; Freifeld, A.G.; et al. Invasive Non-AspergillusMold Infections in Transplant Recipients, United States, 2001–2006. Emerg. Infect. Dis. 2011, 17, 1855–1864. [Google Scholar] [CrossRef]
- Thaler, M.; Pastakia, B.; Shawker, T.H.; O’Leary, T.; Pizzo, P.A. Hepatic Candidiasis in Cancer Patients: The Evolving Picture of the Syndrome. Ann. Intern. Med. 1988, 108, 88–100. [Google Scholar] [CrossRef]
- Berenguer, J.; Buck, M.; Witebsky, F.; Stock, F.; Pizzo, P.A.; Walsh, T.J. Lysis—Centrifugation blood cultures in the detection of tissue-proven invasive candidiasis disseminated versus single-organ infection. Diagn. Microbiol. Infect. Dis. 1993, 17, 103–109. [Google Scholar] [CrossRef]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.B.; Caliendo, A.M.; Mylonakis, E. Molecular and Nonmolecular Diagnostic Methods for Invasive Fungal Infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef]
- Lamoth, F. Galactomannan and 1,3-β-d-Glucan Testing for the Diagnosis of Invasive Aspergillosis. J. Fungi 2016, 2, 22. [Google Scholar] [CrossRef]
- Wall, G.; Lopez-Ribot, J.L. Current Antimycotics, New Prospects, and Future Approaches to Antifungal Therapy. Antibiotics 2020, 9, 445. [Google Scholar] [CrossRef]
- Nanjappa, S.G.; Klein, B.S. Vaccine immunity against fungal infections. Curr. Opin. Immunol. 2014, 28, 27–33. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Arvanitis, M.; Glavis-Bloom, J.; Mylonakis, E. Invertebrate models of fungal infection. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2013, 1832, 1378–1383. [Google Scholar] [CrossRef]
- Jemel, S.; Guillot, J.; Kallel, K.; Botterel, F.; Dannaoui, E. Galleria mellonella for the Evaluation of Antifungal Efficacy against Medically Important Fungi, a Narrative Review. Microorganisms 2020, 8, 390. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Zaragoza, O. Immune Response of Galleria mellonella against Human Fungal Pathogens. J. Fungi 2018, 5, 3. [Google Scholar] [CrossRef]
- Rizzo, J.; Wong, S.S.W.; Gazi, A.D.; Moyrand, F.; Chaze, T.; Commere, P.; Novault, S.; Matondo, M.; Péhau-Arnaudet, G.; Reis, F.C.G.; et al. Cryptococcus extracellular vesicles properties and their use as vaccine platforms. J. Extracell. Vesicles 2021, 10, e12129. [Google Scholar] [CrossRef]
- Swartz, M. The physiology of the lymphatic system. Adv. Drug Deliv. Rev. 2001, 50, 3–20. [Google Scholar] [CrossRef]
- Specht, C.A.; Lee, C.K.; Huang, H.; Hester, M.M.; Liu, J.; Luckie, B.A.; Santana, M.A.T.; Mirza, Z.; Khoshkenar, P.; Abraham, A.; et al. Vaccination with Recombinant Cryptococcus Proteins in Glucan Particles Protects Mice against Cryptococcosis in a Manner Dependent upon Mouse Strain and Cryptococcal Species. mBio 2017, 8, e01872-17. [Google Scholar] [CrossRef]
- Hester, M.M.; Lee, C.K.; Abraham, A.; Khoshkenar, P.; Ostroff, G.R.; Levitz, S.M.; Specht, C.A. Protection of mice against experimental cryptococcosis using glucan particle-based vaccines containing novel recombinant antigens. Vaccine 2020, 38, 620–626. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Lane, R.E.; Korbie, D.; Hill, M.M.; Trau, M. Extracellular vesicles as circulating cancer biomarkers: Opportunities and challenges. Clin. Transl. Med. 2018, 7, 14. [Google Scholar] [CrossRef]
- Schorey, J.S.; Harding, C.V. Extracellular vesicles and infectious diseases: New complexity to an old story. J. Clin. Investig. 2016, 126, 1181–1189. [Google Scholar] [CrossRef]
- Fuhrmann, G.; Neuer, A.L.; Herrmann, I.K. Extracellular vesicles–A promising avenue for the detection and treatment of infectious diseases? Eur. J. Pharm. Biopharm. 2017, 118, 56–61. [Google Scholar] [CrossRef]
- Kuipers, M.E.; Hokke, C.H.; Smits, H.H.; Hoen, E.N.M.N. Pathogen-Derived Extracellular Vesicle-Associated Molecules ThatAffect the Host Immune System: An Overview. Front. Microbiol. 2018, 9, 2182. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nenciarini, S.; Cavalieri, D. Immunomodulatory Potential of Fungal Extracellular Vesicles: Insights for Therapeutic Applications. Biomolecules 2023, 13, 1487. https://doi.org/10.3390/biom13101487
Nenciarini S, Cavalieri D. Immunomodulatory Potential of Fungal Extracellular Vesicles: Insights for Therapeutic Applications. Biomolecules. 2023; 13(10):1487. https://doi.org/10.3390/biom13101487
Chicago/Turabian StyleNenciarini, Stefano, and Duccio Cavalieri. 2023. "Immunomodulatory Potential of Fungal Extracellular Vesicles: Insights for Therapeutic Applications" Biomolecules 13, no. 10: 1487. https://doi.org/10.3390/biom13101487