TGF-β Pathways Stratify Colorectal Cancer into Two Subtypes with Distinct Cartilage Oligomeric Matrix Protein (COMP) Expression-Related Characteristics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Retrieval and Preprocessing
2.2. Consensus Clustering and Weighted Gene Co-Expression Network Analysis
2.3. Functional and Pathway Enrichment Analysis
2.4. Feature Selection with Machine Learning Algorithms
2.5. Association between COMP and Clinical Features
2.6. Tumor Immune Estimation Resource
2.7. Analysis of COMP Expression Correlation with Immunological Characteristics of TME
2.8. Interaction Network and Enrichment Analysis
2.9. Statistical Analysis
3. Results
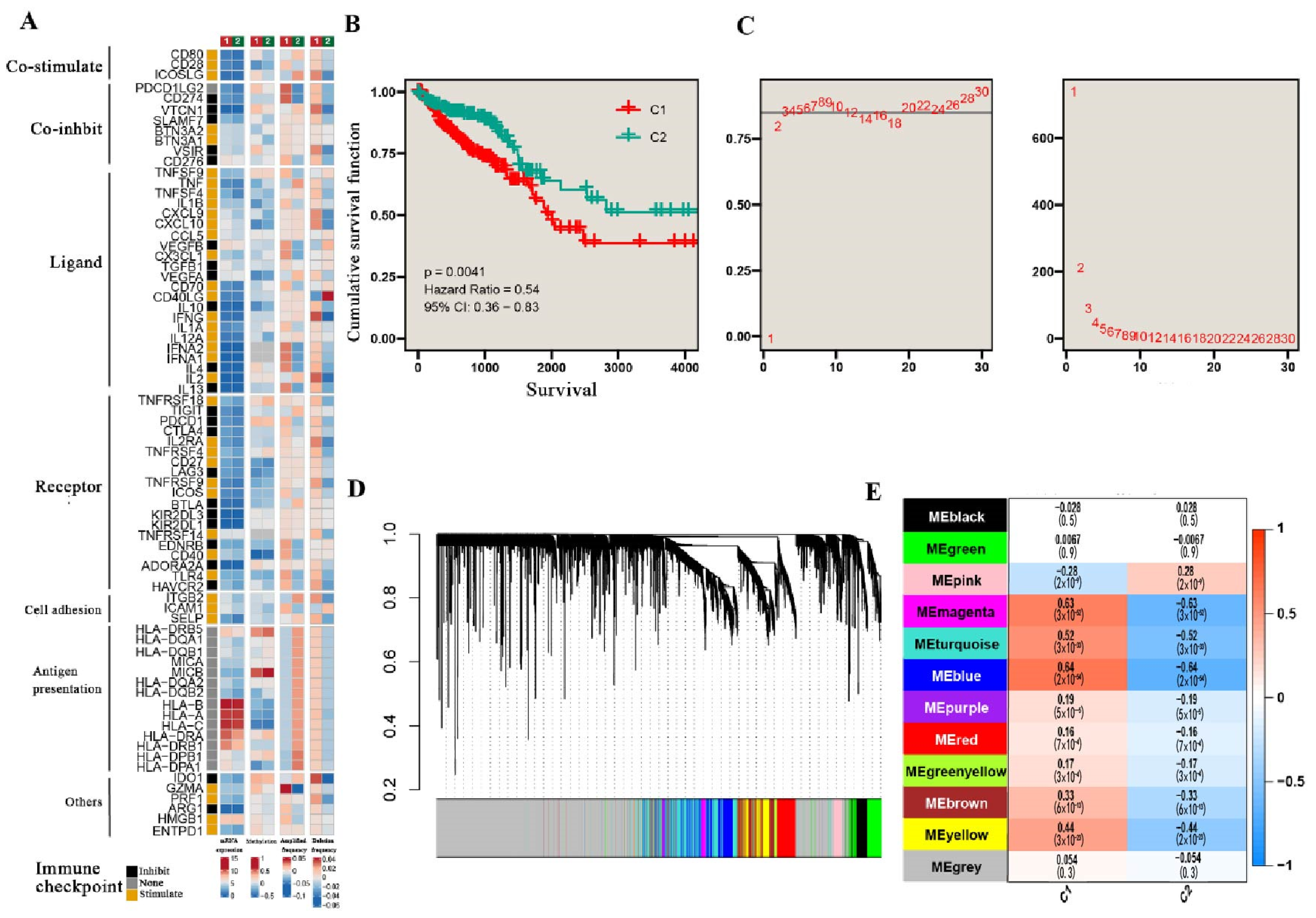
3.1. TGF-β Pathways Stratify CRC into Two Subtypes
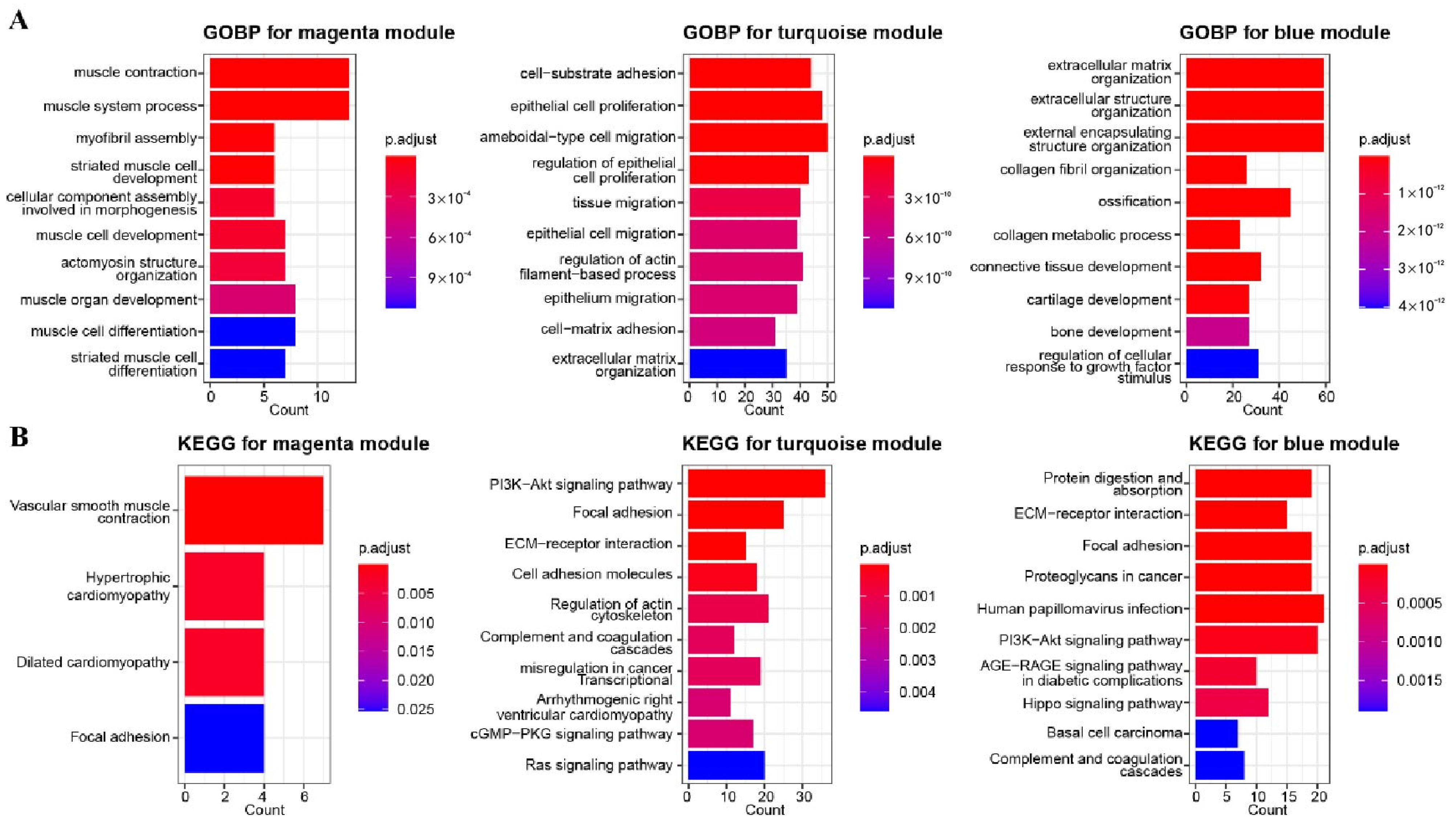
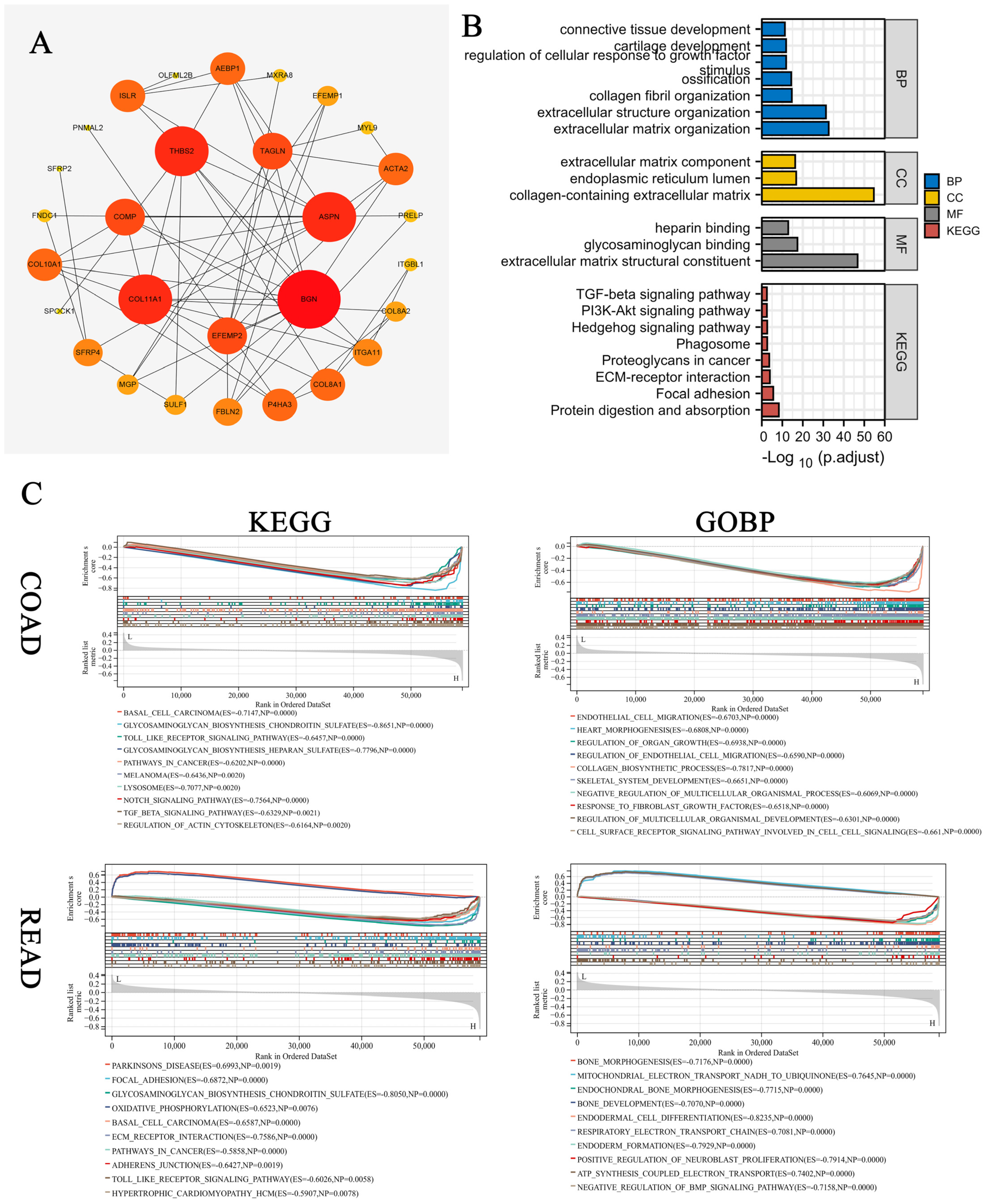
3.2. GO and KEGG Enrichment Analysis for Related Gene Modules
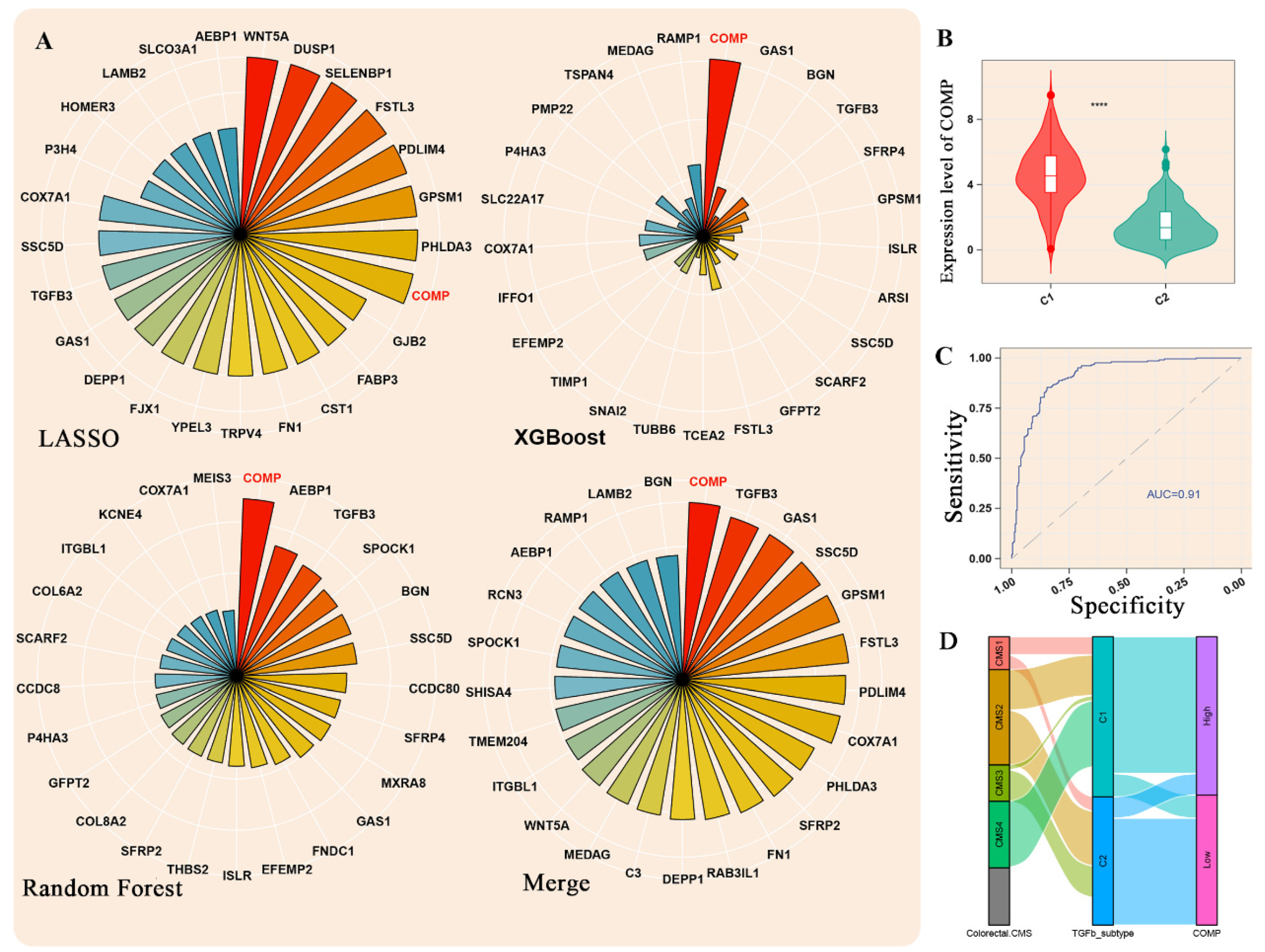
3.3. Machine Learning Methods Identified COMP as the Hub Gene
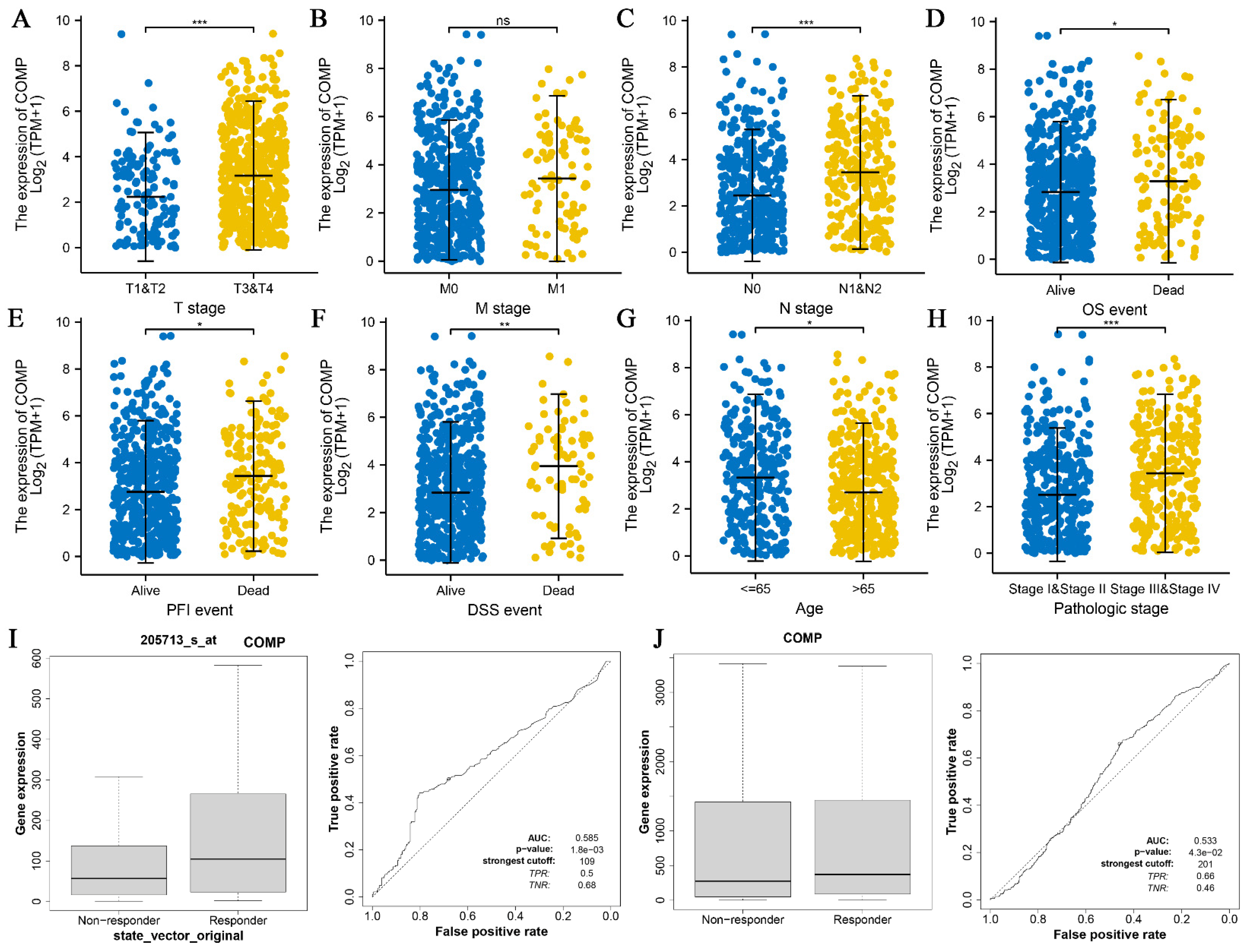
3.4. Association between COMP and CRC Clinical Features
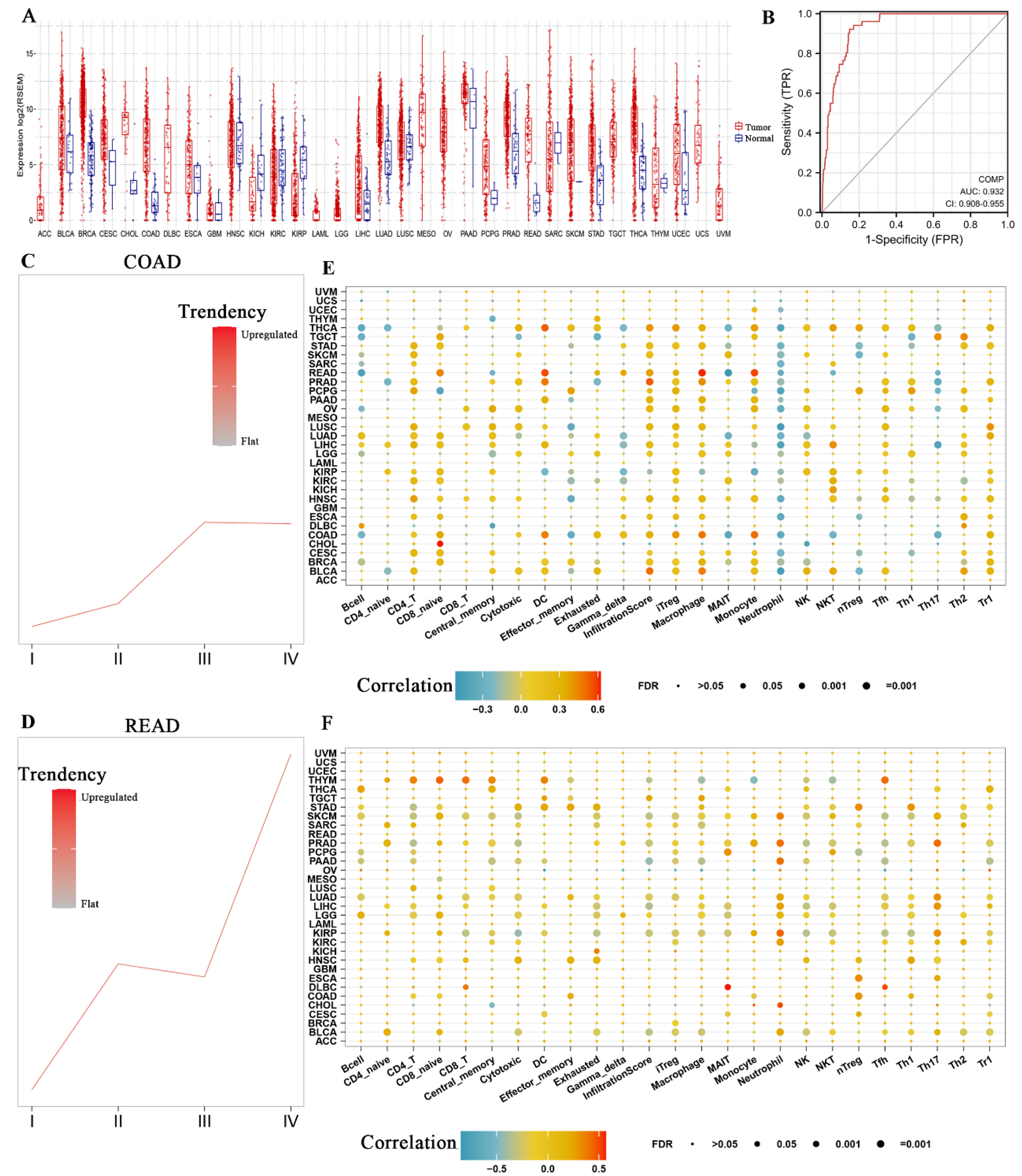
3.5. COMP Expression Level and Immune Infiltration Analysis in Pan Cancers
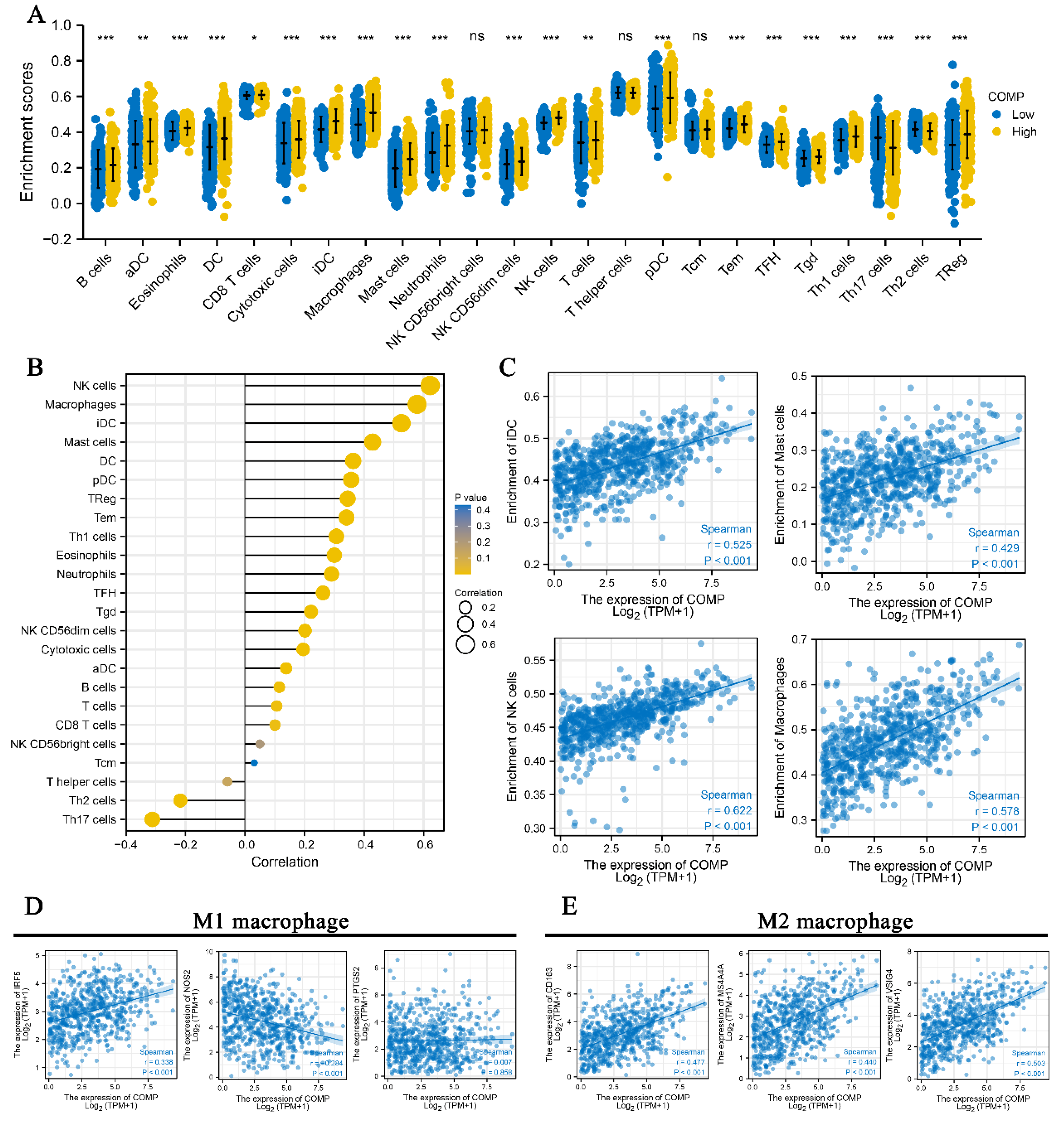
3.6. Correlation between COMP and Immune Infiltration in CRC
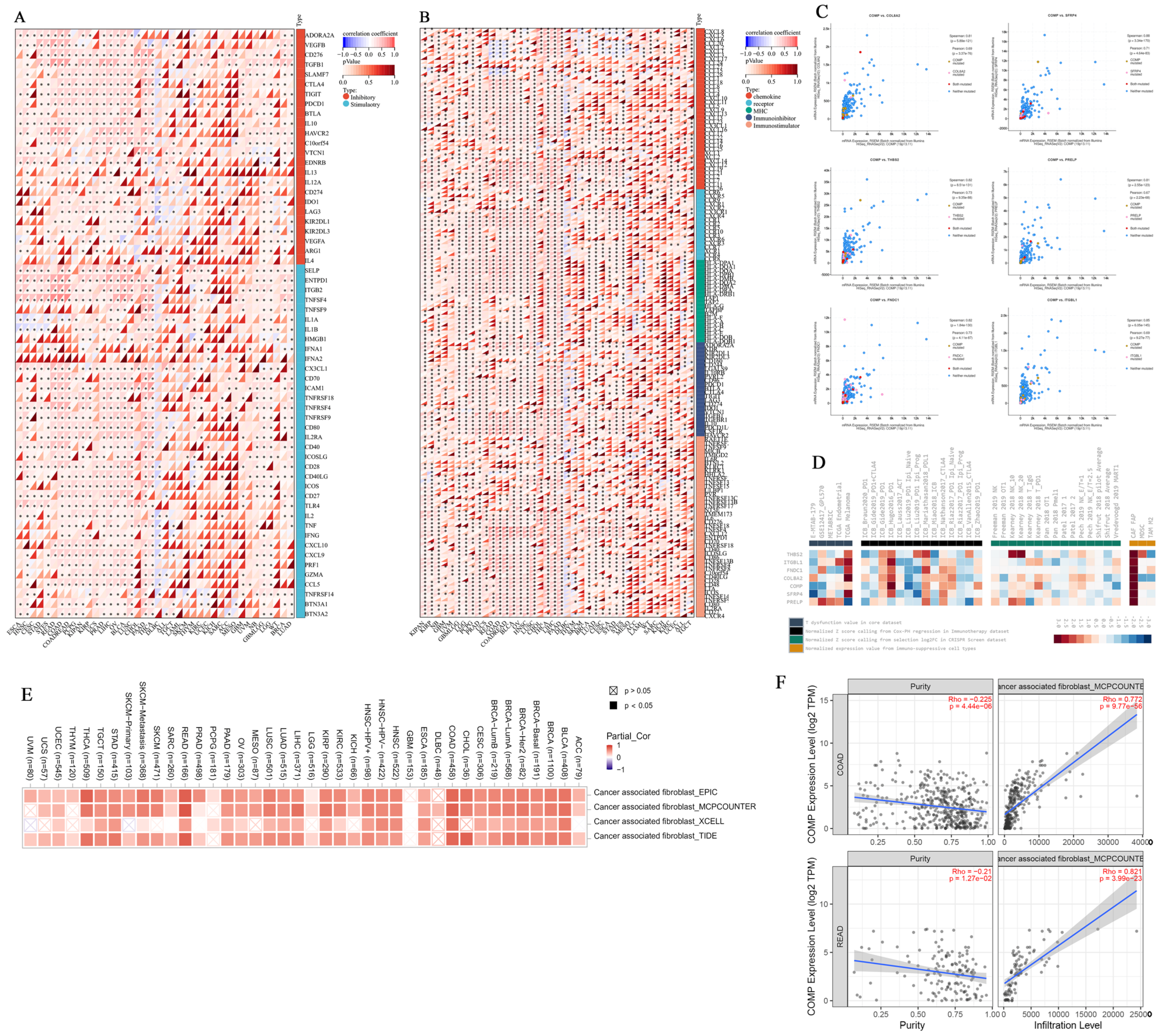
3.7. Immunomodulatory Relevance of COMP
3.8. Enrichment Analysis of COMP-Related Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Joanito, I.; Wirapati, P.; Zhao, N.; Nawaz, Z.; Yeo, G.; Lee, F.; Eng, C.L.P.; Macalinao, D.C.; Kahraman, M.; Srinivasan, H.; et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 2022, 54, 963–975. [Google Scholar] [CrossRef]
- Ashktorab, H.; Brim, H. Colorectal cancer subtyping. Nat. Rev. Cancer 2022, 22, 68–69. [Google Scholar] [CrossRef]
- Ubink, I.; Verhaar, E.R.; Kranenburg, O.; Goldschmeding, R. A potential role for CCN2/CTGF in aggressive colorectal cancer. J. Cell Commun. Signal. 2016, 10, 223–227. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, S.; Sharma, J.; Bukkapatnam, S.; Oweida, A.; Lennon, S.; Phan, A.; Milner, D.; Uyanga, N.; Jimeno, A.; Raben, D.; et al. Inhibition of EphB4-Ephrin-B2 Signaling Enhances Response to Cetuximab-Radiation Therapy in Head and Neck Cancers. Clin. Cancer Res. 2018, 24, 4539–4550. [Google Scholar] [CrossRef] [Green Version]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-beta Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-beta-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Ren, J.; Ten Dijke, P. Targeting TGFbeta signal transduction for cancer therapy. Signal Transduct. Target Ther. 2021, 6, 8. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Wang, Y.; Sun, L.; Liu, Z.; Wang, L.; Song, T.; Yao, Y.; Liu, Q.; Tu, K. HSCs-derived COMP drives hepatocellular carcinoma progression by activating MEK/ERK and PI3K/AKT signaling pathways. J. Exp. Clin. Cancer Res. 2018, 37, 231. [Google Scholar] [CrossRef]
- Nfonsam, V.N.; Jecius, H.C.; Janda, J.; Omesiete, P.N.; Elquza, E.; Scott, A.J.; Nfonsam, L.E.; Jandova, J. Cartilage oligomeric matrix protein (COMP) promotes cell proliferation in early-onset colon cancer tumorigenesis. Surg. Endosc. 2020, 34, 3992–3998. [Google Scholar] [CrossRef]
- Tran, V.; Karsai, A.; Fong, M.C.; Cai, W.; Fraley, J.G.; Yik, J.H.N.; Klineberg, E.; Haudenschild, D.R.; Liu, G.Y. Direct Visualization of the Binding of Transforming Growth Factor Beta 1 with Cartilage Oligomeric Matrix Protein via High-Resolution Atomic Force Microscopy. J. Phys. Chem. B 2020, 124, 9497–9504. [Google Scholar] [CrossRef]
- Wu, J.; Lubman, D.M.; Kugathasan, S.; Denson, L.A.; Hyams, J.S.; Dubinsky, M.C.; Griffiths, A.M.; Baldassano, R.N.; Noe, J.D.; Rabizadeh, S.; et al. Serum Protein Biomarkers of Fibrosis Aid in Risk Stratification of Future Stricturing Complications in Pediatric Crohn’s Disease. Am. J. Gastroenterol. 2019, 114, 777–785. [Google Scholar] [CrossRef]
- Melisi, D.; Garcia-Carbonero, R.; Macarulla, T.; Pezet, D.; Deplanque, G.; Fuchs, M.; Trojan, J.; Kozloff, M.; Simionato, F.; Cleverly, A.; et al. Fbeta receptor inhibitor galunisertib is linked to inflammation- and remodeling-related proteins in patients with pancreatic cancer. Cancer Chemother. Pharmacol. 2019, 83, 975–991. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Fekete, J.T.; Gyorffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Lawal, B.; Tseng, S.H.; Olugbodi, J.O.; Iamsaard, S.; Ilesanmi, O.B.; Mahmoud, M.H.; Ahmed, S.H.; Batiha, G.E.; Wu, A.T.H. Pan-Cancer Analysis of Immune Complement Signature C3/C5/C3AR1/C5AR1 in Association with Tumor Immune Evasion and Therapy Resistance. Cancers 2021, 13, 4124. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, A.J.; Elsawa, S.F. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic. Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Zhong, M.E.; Huang, Z.P.; Wang, X.; Cai, D.; Li, C.H.; Gao, F.; Wu, X.J.; Wang, W. A Transcription Factor Signature Can Identify the CMS4 Subtype and Stratify the Prognostic Risk of Colorectal Cancer. Front. Oncol. 2022, 12, 902974. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, J.; Di, Z.; Yuan, W.; Zhou, Z.; Liu, Z.; Han, S.; Liu, Y.; Ying, G.; Shu, X.; et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/beta-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 232. [Google Scholar] [CrossRef]
- Shang, A.; Gu, C.; Wang, W.; Wang, X.; Sun, J.; Zeng, B.; Chen, C.; Chang, W.; Ping, Y.; Ji, P.; et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol. Cancer 2020, 19, 117. [Google Scholar] [CrossRef]
- Ning, Z.K.; Hu, C.G.; Liu, J.; Tian, H.K.; Yu, Z.L.; Zhou, H.N.; Li, H.; Zong, Z. The Hypoxic Landscape Stratifies Gastric Cancer Into 3 Subtypes With Distinct M6a Methylation and Tumor Microenvironment Infiltration Characteristics. Front. Immunol. 2022, 13, 860041. [Google Scholar] [CrossRef]
- Yeh, H.W.; Lee, S.S.; Chang, C.Y.; Lang, Y.D.; Jou, Y.S. A New Switch for TGFbeta in Cancer. Cancer Res. 2019, 79, 3797–3805. [Google Scholar] [CrossRef] [Green Version]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef] [Green Version]
- Kai, F.; Drain, A.P.; Weaver, V.M. The Extracellular Matrix Modulates the Metastatic Journey. Dev. Cell. 2019, 49, 332–346. [Google Scholar] [CrossRef]
- Kim, M.S.; Ha, S.E.; Wu, M.; Zogg, H.; Ronkon, C.F.; Lee, M.Y.; Ro, S. Extracellular Matrix Biomarkers in Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 9185. [Google Scholar] [CrossRef]
- Karlsson, S.; Nystrom, H. The extracellular matrix in colorectal cancer and its metastatic settling—Alterations and biological implications. Crit. Rev. Oncol. Hematol. 2022, 175, 103712. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.; Farhood, B.; Mortezaee, K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell Biochem. 2019, 120, 2782–2790. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, S.; Novosad, V.; Engibaryan, N.; Ushkaryov, Y.; Nikulin, S.; Tonevitsky, A. ECM-Receptor Regulatory Network and Its Prognostic Role in Colorectal Cancer. Front. Genet. 2021, 12, 782699. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, N.N.; Kedrin, D.; Incio, J.; Liu, H.; Ho, W.W.; Nia, H.T.; Edrich, C.M.; Jung, K.; Daubriac, J.; Chen, I.; et al. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci. Transl. Med. 2016, 8, 360ra135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarthy, A.; Khan, L.; Bensler, N.P.; Bose, P.; De Carvalho, D.D. TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun. 2018, 9, 4692. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Young, C.D.; Zhou, H.; Wang, X. Transforming Growth Factor-beta Signaling in Fibrotic Diseases and Cancer-Associated Fibroblasts. Biomolecules 2020, 10, 1666. [Google Scholar] [CrossRef]
- Chen, J.; Ding, Z.Y.; Li, S.; Liu, S.; Xiao, C.; Li, Z.; Zhang, B.X.; Chen, X.P.; Yang, X. Targeting transforming growth factor-beta signaling for enhanced cancer chemotherapy. Theranostics 2021, 11, 1345–1363. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kawabata, K.; Kusaka-Kikushima, A.; Sugiyama, Y.; Mabuchi, T.; Takekoshi, S.; Miyasaka, M.; Ozawa, A.; Sakai, S. Cartilage Oligomeric Matrix Protein Increases in Photodamaged Skin. J. Investig. Dermatol. 2016, 136, 1143–1149. [Google Scholar] [CrossRef] [Green Version]
- Janša, V.; Klančič, T.; Pušić, M.; Klein, M.; Vrtačnik Bokal, E.; Ban Frangež, H.; Rižner, T.L. Proteomic analysis of peritoneal fluid identified COMP and TGFBI as new candidate biomarkers for endometriosis. Sci. Rep. 2021, 11, 20870. [Google Scholar] [CrossRef]
- Zhong, W.; Hou, H.; Liu, T.; Su, S.; Xi, X.; Liao, Y.; Xie, R.; Jin, G.; Liu, X.; Zhu, L.; et al. Cartilage Oligomeric Matrix Protein promotes epithelial-mesenchymal transition by interacting with Transgelin in Colorectal Cancer. Theranostics 2020, 10, 8790–8806. [Google Scholar] [CrossRef]
- Kather, J.N.; Halama, N. Harnessing the innate immune system and local immunological microenvironment to treat colorectal cancer. Br. J. Cancer 2019, 120, 871–882. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.; Wu, X.; Chen, X.; Yang, S.; Chen, W.; Wang, M.; Liu, Y.; Gu, D.; Zeng, G. A novel immune-related gene-based prognostic signature to predict biochemical recurrence in patients with prostate cancer after radical prostatectomy. Cancer Immunol. Immunother. 2021, 70, 3587–3602. [Google Scholar] [CrossRef]
- Väyrynen, J.P.; Haruki, K.; Lau, M.C.; Väyrynen, S.A.; Zhong, R.; Dias Costa, A.; Borowsky, J.; Zhao, M.; Fujiyoshi, K.; Arima, K.; et al. The Prognostic Role of Macrophage Polarization in the Colorectal Cancer Microenvironment. Cancer Immunol. Res. 2021, 9, 8–19. [Google Scholar] [CrossRef]
- Guerriero, J.L. Macrophages: The Road Less Traveled, Changing Anticancer Therapy. Trends. Mol. Med. 2018, 24, 472–489. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Y.; Wang, L.; Yao, B.; Chen, T.; Li, Q.; Liu, Z.; Liu, R.; Niu, Y.; Song, T.; et al. Resolvin D1 prevents epithelial-mesenchymal transition and reduces the stemness features of hepatocellular carcinoma by inhibiting paracrine of cancer-associated fibroblast-derived COMP. J. Exp. Clin. Cancer Res. 2019, 38, 170. [Google Scholar] [CrossRef]
- Ford, K.; Hanley, C.J.; Mellone, M.; Szyndralewiez, C.; Heitz, F.; Wiesel, P.; Wood, O.; Machado, M.; Lopez, M.A.; Ganesan, A.P.; et al. NOX4 Inhibition Potentiates Immunotherapy by Overcoming Cancer-Associated Fibroblast-Mediated CD8 T-cell Exclusion from Tumors. Cancer Res. 2020, 80, 1846–1860. [Google Scholar] [CrossRef] [Green Version]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [Green Version]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.L.; Li, F.; Yeo, J.Z.; Yong, K.J.; Bassal, M.A.; Ng, G.H.; Lee, M.Y.; Leong, C.Y.; Tan, H.K.; Wu, C.S.; et al. New High-Throughput Screening Identifies Compounds That Reduce Viability Specifically in Liver Cancer Cells That Express High Levels of SALL4 by Inhibiting Oxidative Phosphorylation. Gastroenterology 2019, 157, 1615–1629.e17. [Google Scholar] [CrossRef]
- Kim, H.C.; Chang, J.; Lee, H.S.; Kwon, H.J. Mitochondrial UQCRB as a new molecular prognostic biomarker of human colorectal cancer. Exp. Mol. Med. 2017, 49, e391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, W.; Ten Dijke, P.; Kostidis, S.; Giera, M.; Hornsveld, M. TGFbeta-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell Mol. Life Sci. 2020, 77, 2103–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pihl, E.; Hughes, E.S.; McDermott, F.T.; Johnson, W.R.; Katrivessis, H. Lung recurrence after curative surgery for colorectal cancer. Dis. Colon. Rectum. 1987, 30, 417–419. [Google Scholar] [CrossRef] [PubMed]
| Univariate Analysis | Multivariate Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Features | Number | HR 1 | 95% CI 1 | p-Value | q-Value 2 | HR 1 | 95% CI 1 | p-Value | q-Value 2 |
| Gender | 455 | 0.64 | 0.84 | ||||||
| Female | - | - | |||||||
| Male | 1.10 | 0.74, 1.64 | |||||||
| Age | 454 | 0.013 | 0.029 | ||||||
| <40 | - | - | - | - | |||||
| >40 | 0.54 | 0.12, 2.37 | 0.39 | 0.09, 1.75 | 0.2 | 0.3 | |||
| >60 | 1.13 | 0.28, 4.64 | 1.32 | 0.32, 5.42 | 0.7 | 0.7 | |||
| Molecular subtype | 455 | 0.84 | 0.84 | ||||||
| Unstable chromatin type | - | - | |||||||
| Stable genome type | 1.02 | 0.58, 1.80 | |||||||
| Super mutated single-nucleotide type | 0.46 | 0.06, 3.29 | |||||||
| Unstable microsatellite type | 1.03 | 0.59, 1.81 | |||||||
| geographic area | 360 | 0.79 | 0.84 | ||||||
| East Asia | - | - | |||||||
| Eastern Europe | 0.52 | 0.07, 4.05 | |||||||
| America | 0.54 | 0.07, 4.00 | |||||||
| Western Europe | 0.67 | 0.09, 5.05 | |||||||
| Organ | 455 | 0.028 | 0.050 | ||||||
| colon | - | - | - | - | |||||
| rectum | 0.55 | 0.31, 0.97 | 0.49 | 0.27, 0.89 | 0.019 | 0.043 | |||
| Stage | 443 | <0.001 | <0.001 | ||||||
| I | - | - | - | - | |||||
| II | 1.62 | 0.67, 3.92 | 1.44 | 0.59, 3.50 | 0.4 | 0.5 | |||
| III | 2.75 | 1.16, 6.57 | 2.71 | 1.13, 6.48 | 0.026 | 0.045 | |||
| IV | 6.10 | 2.50, 14.9 | 7.20 | 2.93, 17.7 | <0.001 | <0.001 | |||
| TGF-β subtype | 455 | 0.004 | 0.013 | ||||||
| C1 | - | - | - | - | |||||
| C2 | 0.54 | 0.36, 0.83 | 0.56 | 0.36, 0.88 | 0.011 | 0.038 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, J.-T.; Zhou, H.-N.; Huang, Y.-F.; Peng, J.; Huang, H.-Y.; Yi, H.; Zong, Z.; Ning, Z.-K. TGF-β Pathways Stratify Colorectal Cancer into Two Subtypes with Distinct Cartilage Oligomeric Matrix Protein (COMP) Expression-Related Characteristics. Biomolecules 2022, 12, 1877. https://doi.org/10.3390/biom12121877
Ding J-T, Zhou H-N, Huang Y-F, Peng J, Huang H-Y, Yi H, Zong Z, Ning Z-K. TGF-β Pathways Stratify Colorectal Cancer into Two Subtypes with Distinct Cartilage Oligomeric Matrix Protein (COMP) Expression-Related Characteristics. Biomolecules. 2022; 12(12):1877. https://doi.org/10.3390/biom12121877
Chicago/Turabian StyleDing, Jia-Tong, Hao-Nan Zhou, Ying-Feng Huang, Jie Peng, Hao-Yu Huang, Hao Yi, Zhen Zong, and Zhi-Kun Ning. 2022. "TGF-β Pathways Stratify Colorectal Cancer into Two Subtypes with Distinct Cartilage Oligomeric Matrix Protein (COMP) Expression-Related Characteristics" Biomolecules 12, no. 12: 1877. https://doi.org/10.3390/biom12121877







