NK Cells Can Preferentially Target Prostate Cancer Stem-like Cells via the TRAIL/DR5 Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture and Preparation of Prostate Cancer Stem-like Cells
2.3. Cell Viability Assay
2.4. Crystal Violet Staining
2.5. Isolation of mRNA and Real-Time Quantitative PCR (qPCR)
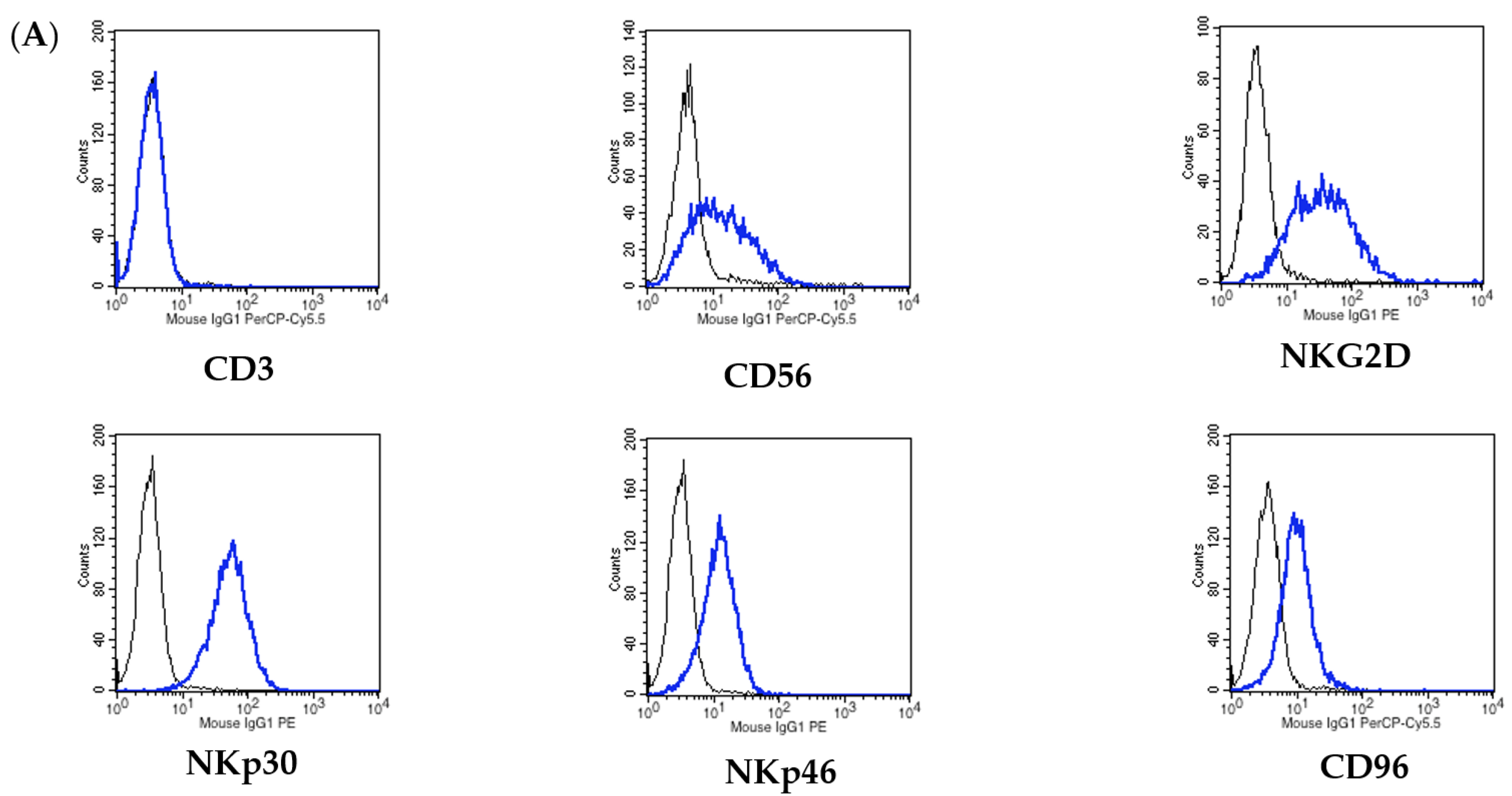
2.6. Flow Cytometry Analysis
2.7. Statistical Analysis
3. Results
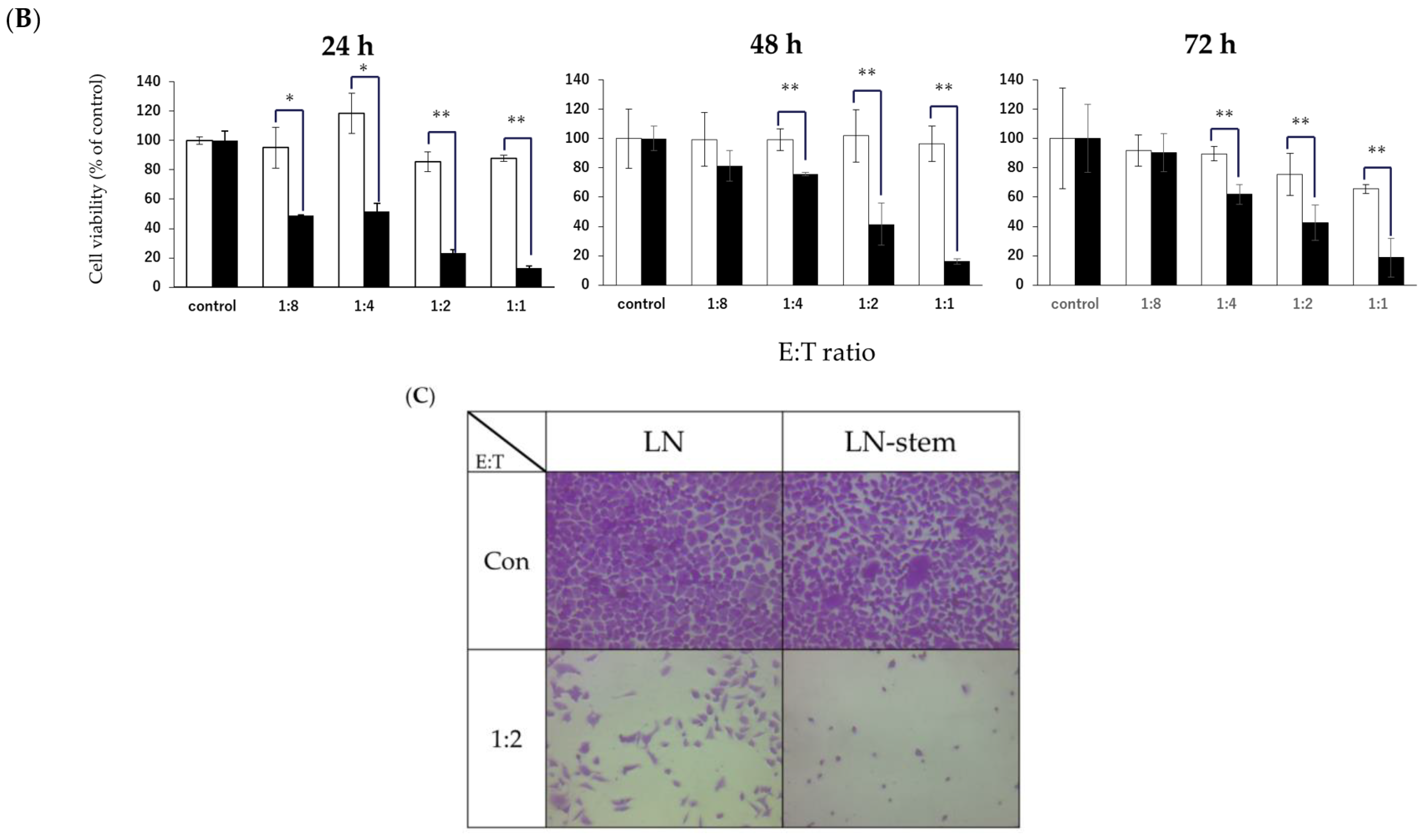
3.1. Comparison between the Sensitivity of LN-Stem Cells and LN Cells to NK Cell-Mediated Cytotoxicity
3.2. Relationship between KHYG-1 Cell-Mediated Cytotoxicity against LN-Stem Cells and Activation Receptor NKG2D and Its Ligands
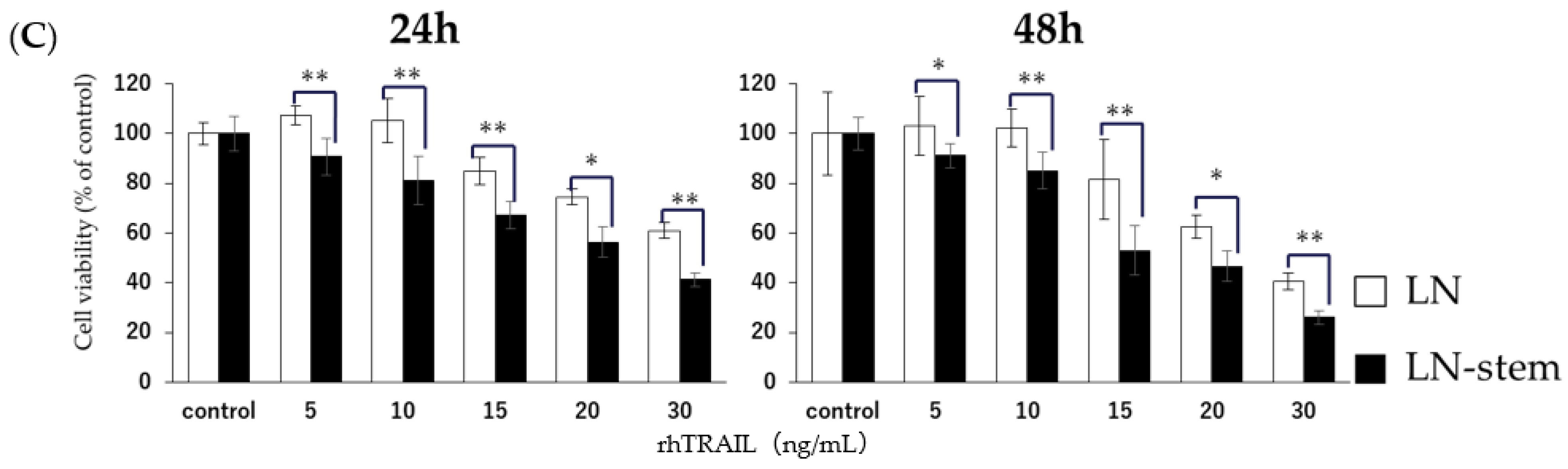
3.3. Relationship between KHYG-1 Cell-Mediated Cytotoxicity against LN-Stem Cells and the Death Receptor Pathway
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics 2018. CA. Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Bolla, M.; de Reijke, T.M.; Van Tienhoven, G.; Van den Bergh, A.C.; Oddens, J.; Poortmans, P.M.; Gez, E.; Kil, P.; Akdas, A.; Soete, G.; et al. Duration of androgen suppression in the treatment of prostate cancer. N. Engl. J. Med. 2009, 360, 2516–2527. [Google Scholar] [CrossRef] [PubMed]
- Goktas, E.D. Optimal hormonal therapy for advanced prostatic carcinoma. Semin. Oncol. 1999, 26, 162–173. [Google Scholar] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
- Giordano, A.; Fucito, A.; Romano, G.; Marino, I.R. Carcinogenesis and environment: The cancer stem cell hypothesis and implications for the development of novel therapeutics and diagnostics. Front. Biosci. 2007, 12, 3475–3482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skvortsov, S.; Skvortsova, I.I.; Tang, D.G.; Dubrovska, A. Concise Review: Prostate Cancer Stem Cells: Current Understanding. Stem Cells 2018, 36, 1457–1474. [Google Scholar] [CrossRef] [Green Version]
- O’Reilly, D.; Johnson, P.; Buchanan, P.J. Hypoxia induced cancer stem cell enrichment promotes resistance to androgen deprivation therapy in prostate cancer. Steroids 2019, 152, 108497. [Google Scholar] [CrossRef]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef]
- Imai, K.; Matsuyama, S.; Miyake, S.; Suga, K.; Nakachi, K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: An 11-year follow-up study of a general population. Lancet 2000, 356, 1795–1799. [Google Scholar] [CrossRef]
- Ames, E.; Canter, R.J.; Grossenbacher, S.K.; Mac, S.; Chen, M.; Smith, R.C.; Hagino, T.; Perez-Cunningham, J.; Sckisel, G.D.; Urayama, S.; et al. NK Cells Preferentially Target Tumor Cells with a Cancer Stem Cell Phenotype. J. Immunol. 2015, 195, 4010–4019. [Google Scholar] [CrossRef] [Green Version]
- van Steenbrugge, G.J.; van Uffelen, C.J.; Bolt, J.; Schröder, F.H. The human prostatic cancer cell line LNCaP and its derived sublines: An in vitro model for the study of androgen sensitivity. J. Steroid Biochem. Mol. Biol. 1991, 40, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Suck, G.; Tan, S.M.; Chu, S.; Niam, M.; Vararattanavech, A.; Lim, T.J.; Koh, M.B. KHYG-1 and NK-92 represent different subtypes of LFA-1-mediated NK cell adhesiveness. Front. Biosci. 2011, 3, 166–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, D.; Kaneko, S.; Ishii, K.; Kohno, N.; Sato, A.; Virgona, N.; Yano, T. The effect of Bowman-Birk inhibitor from Soybeans on the sensitivity of prostate cancer stem-like cells to anti-androgen. Food Sci. Technol. Res. 2020, 26, 553–559. [Google Scholar] [CrossRef]
- Suck, G.; Branch, D.R.; Smyth, M.J.; Miller, R.G.; Vergidis, J.; Fahim, S.; Keating, A. KHYG-1, a model for the study of enhanced natural killer cell cytotoxicity. Exp. Hematol. 2005, 33, 1160–1171. [Google Scholar] [CrossRef]
- Lee, S.H.; Hyun, S.K.; Kim, H.B.; Kang, C.D.; Kim, S.H. Potential Role of CD133 Expression in the Susceptibility of Human Liver Cancer Stem-Like Cells to TRAIL. Oncol. Res. 2016, 24, 495–509. [Google Scholar] [CrossRef]
- Eun, K.; Ham, S.W.; Kim, H. Cancer stem cell heterogeneity: Origin and new perspectives on CSC targeting. BMB. Rep. 2017, 50, 117–125. [Google Scholar] [CrossRef]
- Medema, J.P. Cancer stem cells: The challenges ahead. Nat. Cell Biol. 2013, 15, 338–344. [Google Scholar] [CrossRef]
- Sharifi, N.; Kawasaki, B.T.; Hurt, E.M.; Farrar, W.L. Stem cells in prostate cancer: Resolving the castrate-resistant conundrum and implications for hormonal therapy. Cancer Biol. Ther. 2006, 5, 901–906. [Google Scholar] [CrossRef]
- Mimeault, M.; Batra, S.K. Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J. Cell Mol. Med. 2013, 17, 30–54. [Google Scholar] [CrossRef] [PubMed]
- Leão, R.; Domingos, C.; Figueiredo, A.; Hamilton, R.; Tabori, U.; Castelo-Branco, P. Cancer Stem Cells in Prostate Cancer: Implications for Targeted Therapy. Urol. Int. 2017, 99, 125–136. [Google Scholar] [CrossRef]
- Leão, R.; Domingos, C.; Figueiredo, A.; Ames, R.E.; Canter, R.J.; Grossenbacher, S.K.; Mac, S.; Smith, R.C.; Monjazeb, A.M.; Chen, M.; et al. Enhanced targeting of stem-like solid tumor cells with radiation and natural killer cells. Oncoimmunology 2015, 4, e1036212. [Google Scholar] [CrossRef] [Green Version]
- Kaur, K.; Nanut, M.P.; Ko, M.W.; Safaie, T.; Kos, J.; Jewett, A. Natural killer cells target and differentiate cancer stem-like cells_undifferentiated tumors_strategies to optimize their growth and expansion for effective cancer immunotherapy. Curr. Opin. Immunol. 2018, 51, 170–180. [Google Scholar] [CrossRef]
- Frazao, A.; Rethacker, L.; Messaoudene, M.; Avril, M.F.; Toubert, A.; Dulphy, N.; Caignard, A. NKG2D/NKG2-Ligand Pathway Offers New Opportunities in Cancer Treatment. Front. Immunol. 2019, 10, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.R.; Ha, G.H.; Bae, J.H.; Oh, S.O.; Kim, S.H.; Kang, C.D. Metastatic colon cancer cell populations contain more cancer stem-like cells with a higher susceptibility to natural killer cell-mediated lysis compared with primary colon cancer cells. Oncol. Lett. 2015, 9, 1641–1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.C.; Ping, Y.F.; Yang, J.; Xu, S.L.; Ye, X.Z.; Xu, C.; et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.S. TRAIL as a target in anti-cancer therapy. Cancer Lett. 2009, 285, 1–5. [Google Scholar] [CrossRef]
- Yuan, X.; Gajan, A.; Chu, Q.; Xiong, H.; Wu, K.; Wu, G.S. Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 2018, 37, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Crowder, R.N.; El-Deiry, W.S. Caspase-8 regulation of TRAIL-mediated cell death. Exp. Oncol. 2012, 34, 160–164. [Google Scholar]
- Prager, I.; Liesche, C.; van Ooijen, H.; Urlaub, D.; Verron, Q.; Sandstrom, N.; Fasbender, F.; Claus, M.; Eils, R.; Beaudouin, J.; et al. NK cells switch from granzyme B to death receptor-mediated cytotoxicity during serial killing. J. Exp. Med. 2019, 216, 2113–2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobie, J.J.; Shah, P.R.; Yang, L.; Rebhahn, J.A.; Fowell, D.J.; Mosmann, T.R. T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5’-adenosine monophosphate to adenosine. J. Immunol. 2006, 177, 6780–6786. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Xu, W.; Huang, Y.; Zhang, Z.; Huang, Q.; Xin, K.Z.; Ma, Y.; Han, L. Methotrexate restores the function of peripheral blood regulatory T cells in psoriasis vulgaris via the CD73/AMPK/mTOR pathway. Br. J. Dermatol. 2018, 179, 896–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, R.; Vishnoi, K.; Viswakarma, N.; Santha, S.; Das, S.; Rana, A.; Rana, B. Involvement of AMP-activated protein kinase and Death Receptor 5 in TRAIL-Berberine-induced apoptosis of cancer cells. Sci. Rep. 2018, 8, 5521. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| NKG2D | Forward primer | TGAGAGTAAAAACTGGTATGAGAGCCA |
| Reverse primer | TGCATGCAGATGTATGTATTTGGAG | |
| MICA | Forward primer | AGACTTGACAGGGAACGGAAAG |
| Reverse primer | TCCAGGTTTTGGGAGAGGAA | |
| MICB | Forward primer | ACCTTGGCTATGAACGTCACA |
| Reverse primer | CCCTCTGAGACCTCGC | |
| DR5 | Forward primer | GTCGTTGTGAGCTTCTGTCC |
| Reverse primer | GCCTCTCCCTGTTCTCTCTC | |
| RPL32 | Forward primer | AACCCTGTTGTCAATGCCTC |
| Reverse primer | CATCTCCTTCTCGGCATCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seki, T.; Shimizu, Y.; Ishii, K.; Takahama, Y.; Kato, K.; Yano, T. NK Cells Can Preferentially Target Prostate Cancer Stem-like Cells via the TRAIL/DR5 Signaling Pathway. Biomolecules 2021, 11, 1702. https://doi.org/10.3390/biom11111702
Seki T, Shimizu Y, Ishii K, Takahama Y, Kato K, Yano T. NK Cells Can Preferentially Target Prostate Cancer Stem-like Cells via the TRAIL/DR5 Signaling Pathway. Biomolecules. 2021; 11(11):1702. https://doi.org/10.3390/biom11111702
Chicago/Turabian StyleSeki, Taiga, Yui Shimizu, Kyota Ishii, Yuzuki Takahama, Kazunori Kato, and Tomohiro Yano. 2021. "NK Cells Can Preferentially Target Prostate Cancer Stem-like Cells via the TRAIL/DR5 Signaling Pathway" Biomolecules 11, no. 11: 1702. https://doi.org/10.3390/biom11111702





