Galectin-1 Cooperates with Yersinia Outer Protein (Yop) P to Thwart Protective Immunity by Repressing Nitric Oxide Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Culture and Purification of Yops
2.2. Mice and Infection
2.3. Stimulation of Peritoneal Macrophages
2.4. NO and Urea Determination
2.5. Apoptosis Assays
2.6. Preparation and Purification of RGal1
2.7. ELISA Assays
2.8. Oxidative Burst Assay
2.9. In Vivo and In Vitro Supplementation of RGal1
2.10. Analysis of Yops-Gal1 Interactions by SDS-PAGE and Western Blot
2.11. Lectin Blotting
2.12. Yops Proteolysis Using Trypsin Digestion
2.13. Schiff Staining
2.14. Flow Cytometry
2.15. Mass Spectrometry
2.16. Bioinformatics Analysis
2.17. Western Blot Analysis of INOS Expression
2.18. Statistical Analysis
3. Results
3.1. Galectin-1 Binds to YopP in Y. enterocolitica
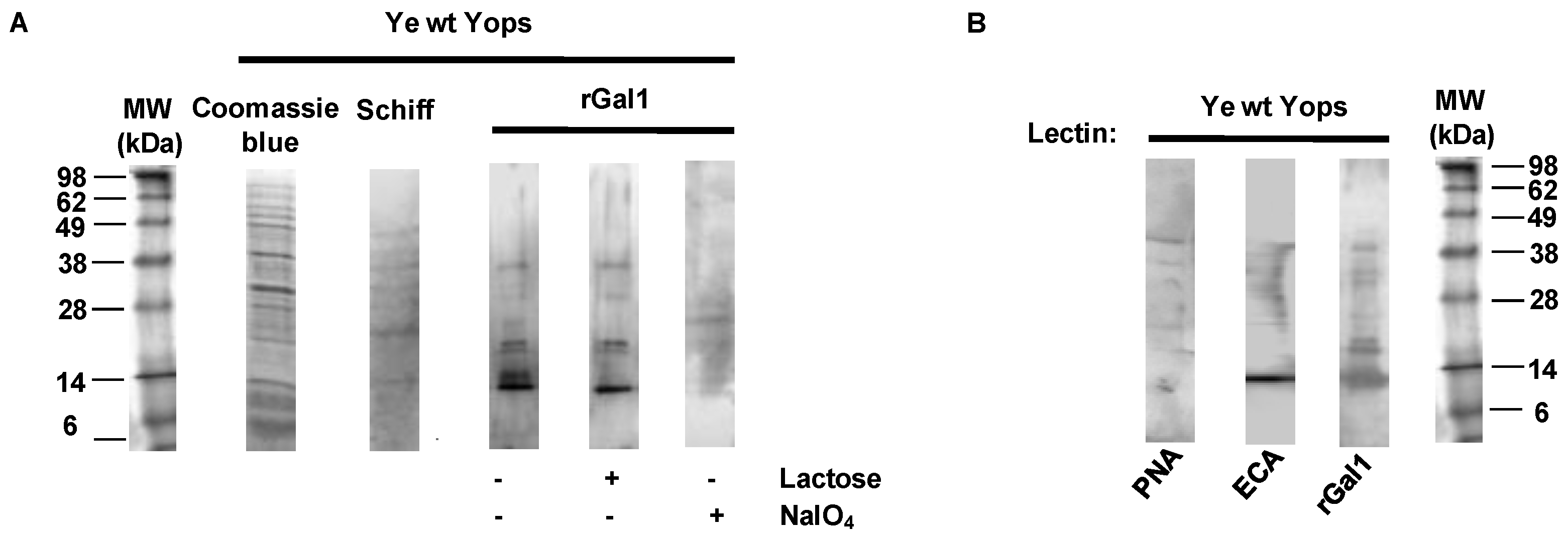
3.2. Galectin-1 Recognizes Yops in a Carbohydrate-Dependent Manner
3.3. Mass Spectrometry-Based Proteomics Analysis of Yops
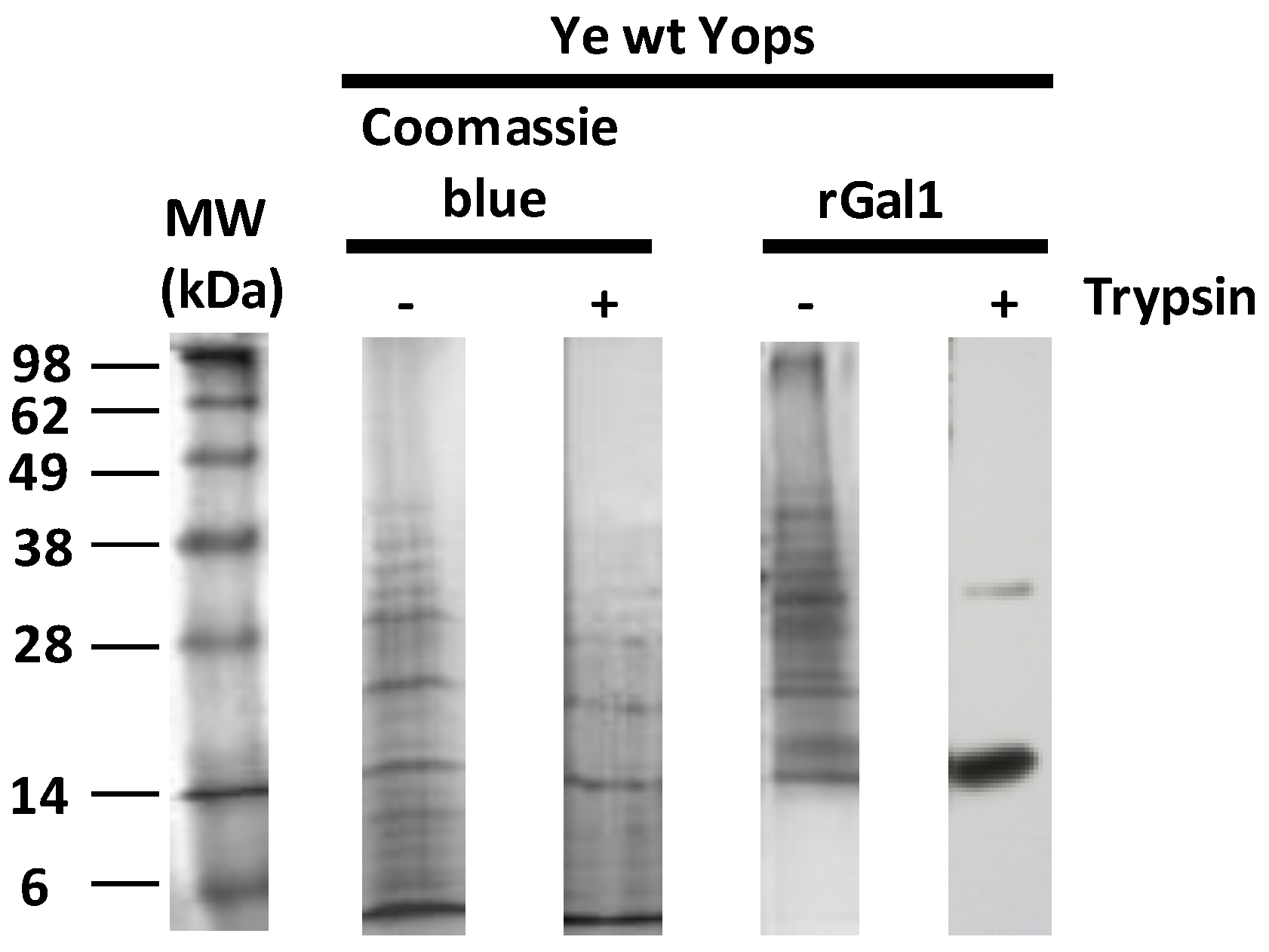
3.4. Gal1 Protects Yops from Protease Degradation
3.5. Gal1 and YopP Control Y. enterocolitica Infection by Decreasing NO Production
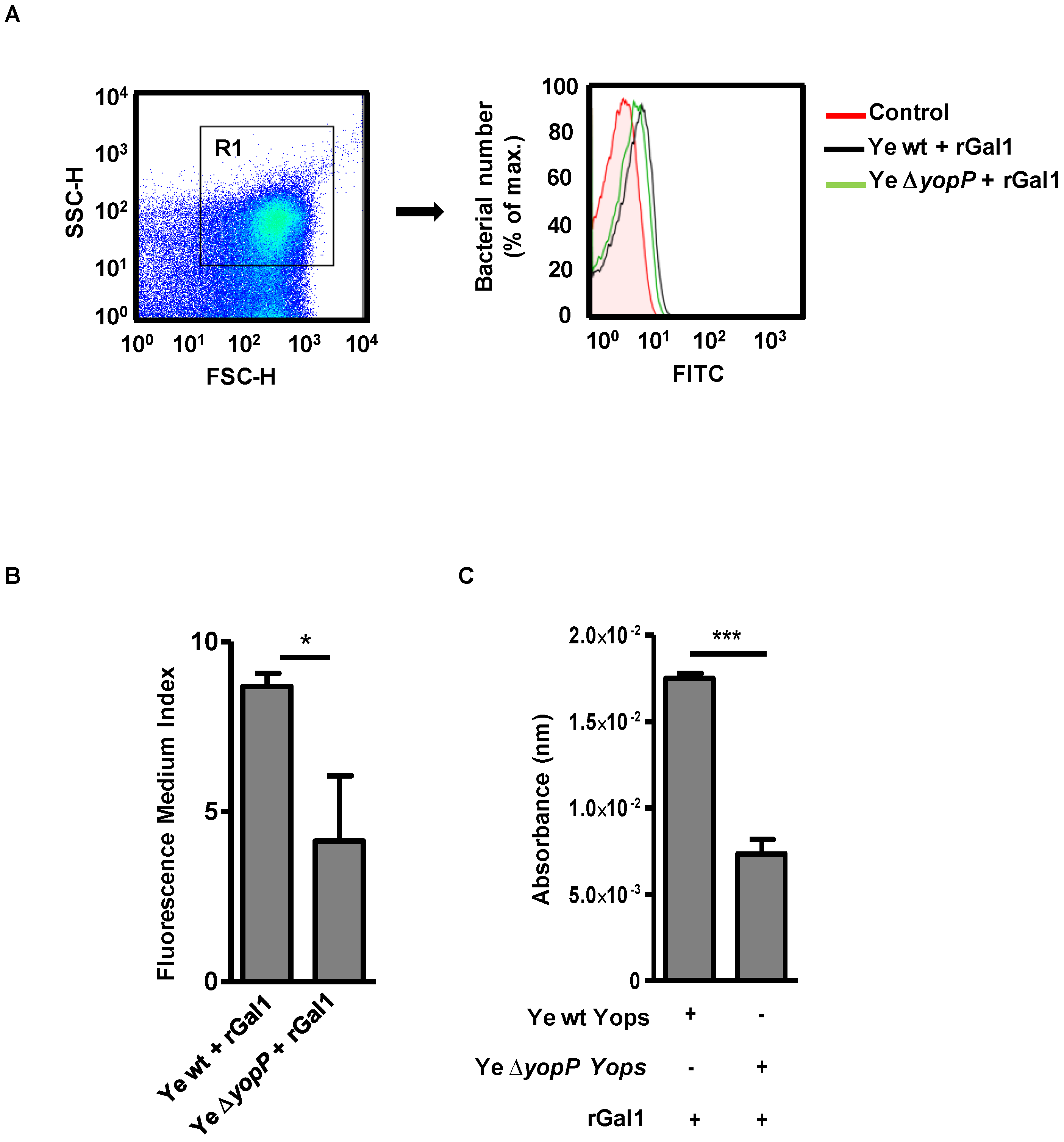
3.6. Exogenous Supplementation of rGal1 Does Not Influence the Protective Anti-Y. enterocolitica Response Observed in the Absence of YopP
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wren, B.W. The Yersiniae—A model Genus to Study the Rapid Evolution of Bacterial Pathogens. Nat. Rev. Microbiol. 2003, 1, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Autenrieth, I.B.; Firsching, R. Penetration of M Cells and Destruction of Peyer’s Patches by Yersinia Enterocolitica: An ultrastructural and Histological Study. J. Med. Microbiol. 1996, 44, 285–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottone, E.J. Yersinia Enterocolitica: Overview and Epidemiologic Correlates. Microbes Infect. 1999, 1, 323–333. [Google Scholar] [CrossRef]
- Cornelis, G.R.; Boland, A.; Boyd, A.P.; Geuijen, C.; Iriarte, M.; Neyt, C.; Sory, M.-P.; Stainier, I. The Virulence Plasmid of Yersinia, an Antihost Genome. Microbiol. Mol. Biol. Rev. 1998, 62, 1315–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, A.; Bottini, N.; Bruckner, S.; Rahmouni, S.; Williams, S.; Schoenberger, S.P.; Mustelin, T. Lck Dephosphorylation AT Tyr-394 and Inhibition of T Cell Antigen Receptor Signaling by Yersinia phosphatase yopH. J. Biol. Chem. 2004, 279, 4922–4928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosdent, N.; Maridonneau-Parini, I.; Sory, M.P.; Cornelis, G.R. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immunity 2002, 70, 4165–4176. [Google Scholar] [CrossRef] [Green Version]
- Viboud, G.I.; Bliska, J.B. Yersinia outer Proteins: Role in Modulation of Host Cell Signaling Responses and Pathogenesis. Annu. Rev. Microbiol. 2005, 59, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, B.; Schmidt, M.A.; Rüter, C. Immunomodulatory Yersinia Outer Proteins (Yops)—Useful Tools for Bacteria and Humans Alike. Virulence 2017, 8, 1124–1147. [Google Scholar] [CrossRef] [Green Version]
- Ruckdeschel, K.; Mannel, O.; Richter, K.; Jacobi, C.A.; Trülzsch, K.; Rouot, B.; Heesemann, J. Yersinia Outer Protein P of Yersinia Enterocolitica Simultaneously Blocks the nuclear factor-κb pathway and exploits lipopolysaccharide Signaling to Trigger Apoptosis in Macrophages. J. Immunol. 2001, 166, 1823–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erfurth, S.E.; Gröbner, S.; Kramer, U.; Gunst, D.S.; Soldanova, I.; Schaller, M.; Autenrieth, I.B.; Borgmann, S. Yersinia enterocolitica Induces Apoptosis and Inhibits Surface Molecule Expression and Cytokine Production in Murine Dendritic Cells. Infect. Immun. 2004, 72, 7045–7054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiefes, A.; Wolf, A.; Doerrie, A.; Grassl, G.A.; Matsumoto, K.; Autenrieth, I.; Bohn, E.; Sakurai, H.; Niedenthal, R.; Resch, K.; et al. The Yersinia Enterocolitica Effector YopP Inhibits Host Cell Signalling by Inactivating the Protein Kinase TAK1 in the IL-1 Signalling Pathway. EMBO Rep. 2006, 7, 838–844. [Google Scholar] [CrossRef] [Green Version]
- Mittal, R.; Peak-Chew, S.-Y.; McMahon, H.T. Acetylation of MEK2 and I B kinase (IKK)Activation Loop Residues by YopJ Inhibits Signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 18574–18579. [Google Scholar] [CrossRef] [Green Version]
- Autenrieth, S.E.; Soldanova, I.; Rösemann, R.; Gunst, D.; Zahir, N.; Kracht, M.; Ruckdeschel, K.; Wagner, H.; Borgmann, S.; Autenrieth, I.B. Yersinia Enterocolitica Yopp Inhibits Map Kinase-Mediated Antigen Uptake in Dendritic Cells. Cell. Microbiol. 2007, 9, 425–437. [Google Scholar] [CrossRef]
- Boland, A.; Cornelis, G.R. Role of Yopp in Suppression of Tumor Necrosis Factor Alpha Release by Macrophages during Yersinia Infection. Infect. Immun. 1998, 66, 1878–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruckdeschel, K.; Harb, S.; Roggenkamp, A.; Hornef, M.; Zumbihl, R.; Köhler, S.; Heesemann, J.; Rouot, B. Yersinia Enterocolitica Impairs Activation of Transcription Factor Nf-Κb: Involvement in the Induction of Programmed Cell Death and in the Suppression of the Macrophage Tumor Necrosis Factor α Production. J. Exp. Med. 1998, 187, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Orth, K. Inhibition of the Mitogen-Activated Protein Kinase Kinase Superfamily by a Yersinia Effector. Science 1999, 285, 1920–1923. [Google Scholar] [CrossRef] [Green Version]
- Ma, K.W.; Ma, W. YopJ Family Effectors Promote Bacterial Infection through a Unique Acetyltransferase Activity. Microbiol. Mol. Biol. Rev. 2016, 80, 1011–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.S.; Filbert, E.L. Scaffold proteins and immune-cell signaling. Nat. Rev. Immunol. 2009, 9, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Peak-Chew, S.Y.; Sade, R.S.; Vallis, Y.; McMahon, H.T. The Acetyltransferase Activity of the Bacterial Toxin YopJ of Yersinia Is Activated by Eukaryotic Host Cell Inositol Hexakisphosphate. J. Biol. Chem. 2010, 285, 19927–19934. [Google Scholar] [CrossRef] [Green Version]
- Fink, S.L.; Cookson, B.T. Apoptosis, Pyroptosis, and Necrosis: Mechanistic Description of Dead and Dying Eukaryotic Cells. Infect. Hnmun. 2005, 73, 1907–1916. [Google Scholar] [CrossRef] [Green Version]
- Bergsbaken, T.; Cookson, B.T. Innate immune response during Yersinia infection: Critical modulation of cell death mechanisms through phagocyte activation. Leukoc. Biol. 2009, 86, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Philip, N.H.; Brodsky, I.E. Cell death programs in Yersinia immunity and pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 149. [Google Scholar] [CrossRef] [Green Version]
- Monack, D.M.; Mecsas, J.; Bouley, D.; Falkow, S. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J. Expr. Med. 1998, 188, 2127–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergsbaken, T.; Cookson, B.T. Macrophage Activation Redirects Yersinia-Infected Host Cell Death from Apoptosis to Caspase-l-Dependent Pyroptosis. PLoS Pathog. 2007, 3, e161. [Google Scholar] [CrossRef]
- Hanski, C.; Naumann, M.; Grützkau, A.; Pluschke, G.; Friedrich, B.; Hahn, H.; Riecken, E.O. Humoral and Cellular Defense against Intestinal Murine Infection with Yersinia Enterocolitica. Infect. Immun. 1991, 59, 1106–1111. [Google Scholar] [CrossRef] [Green Version]
- Autenrieth, I.B.; Hantschmann, P.; Heymer, B.; Heesemann, J. Immunohistological Characterization of the Cellular Immune Response against Yersinia Enterocolitica in Mice: Evidence for the Involvement of t Lymphocytes. Immunobiology 1993, 187, 1–16. [Google Scholar] [CrossRef]
- Olson, R.M.; Dhariwala, M.O.; Mitchell, W.J.; Anderson, D.M. Yersinia Pestis Exploits Early Activation of myd88 for Growth in the Lungs during Pneumonic Plague. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Tufano, M.A.; Rossano, F.; Catalanotti, P.; Liguori, G.; Marinelli, A.; Baroni, A.; Marinelli, P. Properties of Yersinia Enterocolitica Porins: Interference with Biological Functions of Phagocytes, Nitric Oxide Production and Selective Cytokine Release. Res. Microbiol. 1994, 145, 297–307. [Google Scholar] [CrossRef]
- Carlos, I.Z.; Silva Monnazzi, L.G.; Falcão, D.P.; de Medeiros, B.M.M. TNF-α, H2O2 and No Response of Peritoneal Macrophages to Yersinia Enterocolitica O:3 Derivatives. Microbes Infect. 2004, 6, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Eliçabe, R.J.; Arias, J.L.; Rabinovich, G.A.; Di Genaro, M.S. TNFRp55 Modulates Il-6 and Nitric oxide Responses Following Yersinia Lipopolysaccharide Stimulation in Peritoneal Macrophages. Immunobiology 2011, 216, 1322–1330. [Google Scholar] [CrossRef]
- Davicino, R.C.; Méndez-Huergo, S.P.; Eliçabe, R.J.; Stupirski, J.C.; Autenrieth, I.; Di Genaro, M.S.; Rabinovich, G.A. Galectin-1–Driven Tolerogenic Programs Aggravate Yersinia enterocolitica infection by Repressing antibacterial immunity. J. Immunol. 2017, 199, 1382–1392. [Google Scholar] [CrossRef] [Green Version]
- Baum, L.G.; Garner, O.B.; Schaefer, K.; Lee, B. Microbe-Host Interactions Are Positively and Negatively Regulated BY Galectin-Glycan Interactions. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerliani, J.P.; Blidner, A.G.; Toscano, M.A.; Croci, D.O.; Rabinovich, G.A. Translating the ‘Sugar Code’ into Immune and Vascular Signaling Programs. Trends Biochem. Sci. 2017, 42, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Tharakaraman, K.; Sasisekharan, V.; Sasisekharan, R. Glycan–Protein Interactions in viral pathogenesis. Curr. Opin. Struct. Biol. 2016, 40, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Davicino, R.C.; Eliçabe, R.J.; Di Genaro, M.S.; Rabinovich, G.A. Coupling Pathogen Recognition to Innate Immunity through Glycan-Dependent Mechanisms. Int. Immunopharmacol. 2011, 11, 1457–1463. [Google Scholar] [CrossRef]
- Rabinovich, G.A.; Gruppi, A. Galectins as immunoregulators during infectious Processes: From Microbial Invasion to the Resolution of the Disease. Parasite Immunol. 2005, 27, 103–114. [Google Scholar] [CrossRef]
- Vasta, G.R. Roles of Galectins in Infection. Nat. Rev. Microbiol. 2009, 7, 424–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.-T.; Rabinovich, G.A. Galectins: Regulators of Acute and Chronic Inflammation. Ann. N. Y. Acad. Sci. 2010, 1183, 158–182. [Google Scholar] [CrossRef]
- Salatino, M.; Rabinovich, G.A. Fine-Tuning Antitumor Responses through the Control of Galectin–Glycan Interactions: An Overview. Methods Mol. Biol. 2010, 355–374. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Weng, I.-C.; Hong, M.-H.; Liu, F.-T. Galectins as Bacterial Sensors in the HOST Innate Response. Curr. Opin. Microbiol. 2014, 17, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Huergo, S.P.; Blidner, A.G.; Rabinovich, G.A. Galectins: Emerging Regulatory Checkpoints Linking tumor Immunity and angiogenesis. Curr. Opin. Immunol. 2017, 45, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Lujan, A.L.; Croci, D.O.; Tudela, J.A.G.; Losinno, A.D.; Cagnoni, A.J.; Mariño, K.V.; Damiani, M.T.; Rabinovich, G.A. Glycosylation-Dependent Galectin–Receptor Interactions Promotechlamydia Trachomatisinfection. Proc. Natl. Acad. Sci. USA 2018, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adkins, I.; Köberle, M.; Gröbner, S.; Bohn, E.; Autenrieth, I.B.; Borgmann, S. Yersinia Outer Proteins E, H, P, and T Differentially Target the Cytoskeleton and Inhibit Phagocytic Capacity of Dendritic Cells. Int. J. Med. Microbiol. 2007, 297, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Eliçabe, R.J.; Cargnelutti, E.; Serer, M.I.; Stege, P.W.; Valdez, S.R.; Toscano, M.A.; Rabinovich, G.A.; Di Genaro, M.S. Lack of Tnfr P55 Results in Heightened Expression of Ifn-γ and IL-17 during the Development of Reactive Arthritis. J. Immunol. 2010, 185, 4485–4495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trülzsch, K.; Sporleder, T.; Igwe, E.I.; Rüssmann, H.; Heesemann, J. Contribution of the Major Secreted Yops of Yersinia enterocolitica O:8 to Pathogenicity in the mouse Infection Model. Infect. Immun. 2004, 72, 5227–5234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autenrieth, I.B.; Beer, M.; Bohn, E.; Kaufmann, S.H.; Heesemann, J. Immune Responses to Yersinia enterocolitica in Susceptible BALB/c and Resistant C57BL/6 Mice: An Essential Role for gamma interferon. Infect. Immun. 1994, 62, 2590–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáez, J.M.P.; Hockl, P.F.; Cagnoni, A.J.; Huergo, S.P.M.; García, P.A.; Gatto, S.G.; Cerliani, J.P.; Croci, D.O.; Rabinovich, G.A. Characterization of a neutralizing anti-human galectin-1 monoclonal antibody with angioregulatory and immunomodulatory activities. Angiogenesis 2021, 24, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chatzipanagiotou, S.; Legakis, J.N.; Boufidou, F.; Petroyianni, V.; Nicolaou, C. Prevalence of Yersinia Plasmid-Encoded Outer protein (Yop) Class-Specific Antibodies in Patients with Hashimoto’s Thyroiditis. Clin. Microbiol. Infect. 2001, 7, 138–143. [Google Scholar] [CrossRef] [Green Version]
- Schopf, R.E.; Mattar, J.; Meyenburg, W.; Scheiner, O.; Hammann, K.P.; Lemmel, E.-M. Measurement of the Respiratory Burst in Human Monocytes and Polymorphonuclear Leukocytes by Nitro Blue Tetrazolium Reduction and Chemiluminescence. J. Immunol. Methods 1984, 67, 109–117. [Google Scholar] [CrossRef]
- Correa, S.G. Opposite Effects of Galectin-1 on Alternative Metabolic Pathways of l-Arginine in Resident, Inflammatory, and Activated Macrophages. Glycobiology 2002, 13, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabinovich, G.A.; Iglesias, M.M.; Modesti, N.M.; Castagna, L.F.; Wolfenstein-Todel, C.; Riera, C.M.; Sotomayor, C.E. Activated rat macrophages produce a galectin-1-like protein that induces apoptosis of T cells: Biochemical and functional characterization. J. Immunol. 1998, 160, 4831–4840. [Google Scholar]
- Reinhold, B.B.; Hauer, C.R.; Plummer, T.H.; Reinhold, V.N. Detailed Structural Analysis of a novel, Specific O-Linked glycan from the prokaryote Flavobacterium Meningosepticum. J. Biol. Chem. 1995, 270, 13197–13203. [Google Scholar] [CrossRef] [Green Version]
- Croci, D.O.; Cerliani, J.P.; Dalotto-Moreno, T.; Méndez-Huergo, S.P.; Mascanfroni, I.D.; Dergan-Dylon, S.; Toscano, M.A.; Caramelo, J.J.; García-Vallejo, J.J.; Ouyang, J.; et al. Glycosylation-Dependent Lectin-Receptor Interactions preserve Angiogenesis IN Anti-VEGF Refractory Tumors. Cell 2014, 156, 744–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.; Srinivas, V.R.; Adhikari, P.; Vijayan, M.; Surolia, A. Molecular Basis of Recognition by Gal/Galnac Specific Legume Lectins: Influence of Glu 129 on the Specificity of Peanut agglutinin (pna) towards c2-substituents of Galactose. Glycobiology 1998, 8, 1007–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debray, H.; Montreuil, J.; Lis, H.; Sharon, N. Affinity of Four Immobilized Erythrina lectins toward Various N-linked Glycopeptides and Related Oligosaccharides. Carbohydr. Res. 1986, 151, 359–370. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Kiyoshi, M.; Anraku, M.; Hashii, N.; Oda-Ueda, N.; Ueda, T.; Ohkuri, T. Glycosylation Decreases Aggregation and Immunogenicity of Adalimumab Fab Secreted from Pichia Pastoris. J. Biochem. 2020, 169, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Andre, S.; Pei, Z.; Siebert, H.; Ramstrom, O.; Gabius, H. Glycosyldisulfides from Dynamic Combinatorial Libraries as o-Glycoside Mimetics for Plant and Endogenous Lectins: Their Reactivities in Solid-Phase and Cell Assays and Conformational Analysis by Molecular Dynamics Simulations. Bioorganic Med. Chem. 2006, 14, 6314–6326. [Google Scholar] [CrossRef] [PubMed]
- Findlow, J.; Taylor, S.; Aase, A.; Horton, R.; Heyderman, R.; Southern, J.; Andrews, N.; Barchha, R.; Harrison, E.; Lowe, A.; et al. Comparison and Correlation of Neisseria meningitidis serogroup b immunologic assay results and human antibody responses following three Doses of the Norwegian Meningococcal Outer Membrane Vesicle Vaccine MenBvac. Infect. Immun. 2006, 74, 4557–4565. [Google Scholar] [CrossRef] [Green Version]
- Pizarro-Guajardo, M.; Ravanal, M.C.; Paez, M.D.; Callegari, E.; Paredes-Sabja, D. Identification of Clostridium Difficile Immunoreactive Spore Proteins of the Epidemic Strain r20291. Proteom. Clin. Appl. 2018, 12, 1700182. [Google Scholar] [CrossRef] [PubMed]
- Wilharm, G.; Lehmann, V.; Krauss, K.; Lehnert, B.; Richter, S.; Ruckdeschel, K.; Heesemann, J.; Trülzsch, K. Yersinia Enterocolitica Type III Secretion Depends on the Proton Motive Force but Not on the flagellar motor Components MotA and MotB. Infect. Immun. 2004, 72, 4004–4009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, F.; Fan, Y.-Y.; Schubert, M.; Aebi, M. Cytoplasmic n-Glycosyltransferase of Actinobacillus pleuropneumoniae Is an Inverting Enzyme and Recognizes the NX(S/T) Consensus Sequence. J. Biol. Chem. 2011, 286, 35267–35274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.-J.; Grass, S.; Paek, S.; St. Geme, J.W.; Yeo, H.-J. The Actinobacillus Pleuropneumoniae HMW1C-Like Glycosyltransferase Mediates N-linked Glycosylation of the Haemophilus Influenzae Hmw1 Adhesin. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sing, A.; Roggenkamp, A.; Geiger, A.M.; Heesemann, J. Yersinia enterocolitica evasion of the Host Innate Immune Response by v Antigen-Induced Il-10 Production of Macrophages Is Abrogated in Il-10-Deficient Mice. J. Immunol. 2002, 168, 1315–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Z.Q.; Guo, X.L. The role of galectin-4 in physiology and diseases. Protein Cell 2016, 7, 314–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, J.T.R.; Miriyala, S.; Noel, T.; Moscow, J.A.; St. Clair, D.K.; Butterfield, D.A. Superoxide Induces Protein Oxidation in Plasma and Tnf-α Elevation in Macrophage Culture: Insights into Mechanisms of Neurotoxicity Following Doxorubicin Chemotherapy. Cancer Lett. 2015, 367, 157–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toscano, M.A.; Campagna, L.; Molinero, L.L.; Cerliani, J.P.; Croci, D.O.; Ilarregui, J.M.; Fuertes, M.B.; Nojek, I.M.; Fededa, J.P.; Zwirner, N.W.; et al. Nuclear Factor (NF)-ΚB controls expression of the immunoregulatory glycan-binding protein Galectin-1. Mol. Immunol. 2011, 48, 1940–1949. [Google Scholar] [CrossRef]
- Zuñiga, E.; Rabinovich, G.A.; Iglesias, M.M.; Gruppi, A. Regulated expression of galectin-1 during B-cell activation and implications for T-cell apoptosis. J. Leukoc. Biol. 2001, 70, 73–79. [Google Scholar]
- Ilaregui, J.M.; Croci, D.O.; Bianco, G.A.; Toscano, M.A.; Salatino, M.; Vermeulen, M.E.; Geffner, J.R.; Rabinovich, G.A. Tolerogenic Signals Delivered by Dendritic Cells to T Cells through a Galectin-1-driven immunoregulatory Circuit Involving Interleukin 27 and Interleukin 10. Nat. Immunol. 2009, 10, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Toscano, M.A.; Bianco, G.A.; Ilarregui, J.M.; Croci, D.O.; Correale, J.; Hernandez, J.D.; Zwirner, N.W.; Poirier, F.; Riley, E.M.; Baum, L.G.; et al. Differential Glycosylation OF Th1, Th2 and Th-17 Effector Cells selectively Regulates Susceptibility to Cell Death. Nat. Immunol. 2007, 8, 825–834. [Google Scholar] [CrossRef]
- Toscano, M.A.; Commodaro, A.G.; Ilarregui, J.M.; Bianco, G.A.; Liberman, A.; Serra, H.M.; Hirabayashi, J.; Rizzo, L.V.; Rabinovich, G.A. Galectin-1 Suppresses Autoimmune retinal disease by promoting Concomitant th2- and T Regulatory-Mediated Anti-Inflammatory Responses. J. Immunol. 2006, 176, 6323–6332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poncini, C.V.; Ilarregui, J.M.; Batalla, E.I.; Engels, S.; Cerliani, J.P.; Cucher, M.A.; van Kooyk, Y.; González-Cappa, S.M.; Rabinovich, G.A. Trypanosoma Cruzi infection Imparts a Regulatory Program in Dendritic Cells and T Cells via Galectin-1–Dependent Mechanisms. J. Immunol. 2015, 195, 3311–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cedeno-Laurent, F.; Opperman, M.; Barthel, S.R.; Kuchroo, V.K.; Dimitroff, C.J. Galectin-1 Triggers an Immunoregulatory Signature in Th Cells Functionally Defined BY IL-10 Expression. J. Immunol. 2012, 188, 3127–3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalotto-Moreno, T.; Croci, D.O.; Cerliani, J.P.; Martinez-Allo, V.C.; Dergan-Dylon, S.; Méndez-Huergo, S.P.; Stupirski, J.C.; Mazal, D.; Osinaga, E.; Toscano, M.A.; et al. Targeting Galectin-1 Overcomes Breast Cancer-Associated immunosuppression and prevents Metastatic Disease. Cancer Res. 2012, 73, 1107–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starossom, S.C.; Mascanfroni, I.D.; Imitola, J.; Cao, L.; Raddassi, K.; Hernandez, S.F.; Bassil, R.; Croci, D.O.; Cerliani, J.P.; Delacour, D.; et al. Galectin-1 Deactivates Classically Activated Microglia and Protects from Inflammation-Induced Neurodegeneration. Immunity 2012, 37, 249–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okumura, C.Y.; Baum, L.G.; Johnson, P.J. Galectin-1 on Cervical Epithelial Cells Is a Receptor for the Sexually Transmitted Human parasite Trichomonas vaginalis. Cell. Microbiol. 2008, 10, 2078–2090. [Google Scholar] [CrossRef] [Green Version]
- Nita-Lazar, M.; Banerjee, A.; Feng, C.; Amin, M.N.; Frieman, M.B.; Chen, W.H.; Cross, A.S.; Wang, L.-X.; Vasta, G.R. Desialylation of Airway Epithelial Cells during Influenza Virus Infection Enhances Pneumococcal Adhesion via Galectin Binding. Mol. Immunol. 2015, 65, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mahdavi, A.; Szychowski, J.; Ngo, J.T.; Sweredoski, M.J.; Graham, R.L.; Hess, S.; Schneewind, O.; Mazmanian, S.K.; Tirrell, D.A. Identification of Secreted Bacterial Proteins by Noncanonical Amino Acid Tagging. Proc. Natl. Acad. Sci. USA 2013, 111, 433–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heesemann, J.; Gross, U.; Schmidt, N.; Laufs, R. Immunochemical Analysis of Plasmid-Encoded Proteins Released by Enteropathogenic Yersinia Sp. Grown in Calcium-Deficient Media. Infect. Immun. 1986, 54, 561–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plowman, S.J.; Hancock, J.F. Ras Signaling from Plasma Membrane AND Endomembrane Microdomains. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2005, 1746, 274–283. [Google Scholar] [CrossRef] [Green Version]
- Belanis, L.; Plowman, S.J.; Rotblat, B.; Hancock, J.F.; Kloog, Y. Galectin-1 Is a Novel Structural Component and a Major Regulator of H-Ras Nanoclusters. Mol. Biol. Cell 2008, 19, 1404–1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Behloul, N.; Baha, S.; Liu, Z.; Aslam, M.S.; Meng, J. Dimerization: A Structural Feature for the Protection of Hepatitis e Virus Capsid Protein against Trypsinization. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Tumitan, A.R.; Monnazzi, L.G.; Ghiraldi, F.R.; Cilli, E.M.; de Medeiros, B.M. Pattern of Macrophage Activation Inyersinia-Resistant Andyersinia-Susceptible Strains of Mice. Microbiol. Immunol. 2007, 51, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Liew, F.Y.; Li, Y.; Moss, D.; Parkinson, C.; Rogers, M.V.; Moncada, S. Resistance Toleishmania Major Infection Correlates with the Induction of Nitric Oxide Synthase in Murine Macrophages. Eur. J. Immunol. 1991, 21, 3009–3014. [Google Scholar] [CrossRef] [PubMed]
- Monnazzi, L.G.; Carlos, I.Z.; deMedeiros, B.M. Influence of Yersinia pseudotuberculosis Outer Proteins (YOPS) on Interleukin-12, Tumor Necrosis Factor Alpha and Nitric Oxide Production BY Peritoneal Macrophages. Immunol. Lett. 2004, 94, 91–98. [Google Scholar] [CrossRef]
- Tansini, A.; de Medeiros, B.M. Susceptibility to Yersinia Pseudotuberculosis infection Is Linked to the Pattern of Macrophage Activation. Scand. J. Immunol. 2009, 69, 310–318. [Google Scholar] [CrossRef] [Green Version]
- Virág, L.; Jaén, R.I.; Regdon, Z.; Boscá, L.; Prieto, P. Self-Defense of Macrophages against Oxidative Injury: Fighting for Their Own Survival. Redox Biol. 2019, 26, 101261. [Google Scholar] [CrossRef]
- Kim, P.K.M.; Zamora, R.; Petrosko, P.; Billiar, T.R. The Regulatory Role of Nitric Oxide in Apoptosis. Int. Immunopharmacol. 2001, 1, 1421–1441. [Google Scholar] [CrossRef]
- Giordano, D.; Li, C.; Suthar, M.S.; Draves, K.E.; Ma, D.Y.; Gale, M., Jr.; Clark, E.A. Nitric oxide controls an inflammatory-like Ly6C(hi)PDCA1+DC subset that regulates Th1 immune responses. J. Leukoc. Biol. 2011, 89, 443–455. [Google Scholar] [CrossRef] [Green Version]
- Xue, Q.; Yan, Y.; Zhang, R.; Xiong, H. Regulation of iNOS on Immune Cells and Its Role in Diseases. Int. J. Mol. Sci. 2018, 19, 3805. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Su, Y.; Kang, B.H.; Fan, Z.; Dong, T.; Brown, D.R.; Cheah, J.; Wittrup, K.D.; Chen, J. High-throughput phenotypic screen and transcriptional analysis identify new compounds and targets for macrophage reprogramming. Nat. Commun. 2021, 12, 773. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lilo, S.; Brodsky, I.E.; Zhang, Y.; Medzhitov, R.; Marcu, K.B.; Bliska, J.B. A Yersinia effector with Enhanced Inhibitory Activity on the Nf-Κb PATHWAY Activates the nlrp3/Asc/Caspase-1 Inflammasome in Macrophages. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, M.B.; Molinero, L.L.; Toscano, M.A.; Ilarregui, J.M.; Rubinstein, N.; Fainboim, L.; Zwirner, N.W.; Rabinovich, G.A. Regulated Expression of Galectin-1 during T-Cell Activation Involves lck and Fyn Kinases and Signaling through mek1/Erk, P38 MAP Kinase And p70S6kinase. Mol. Cell. Biochem. 2004, 267, 177–185. [Google Scholar] [CrossRef] [PubMed]


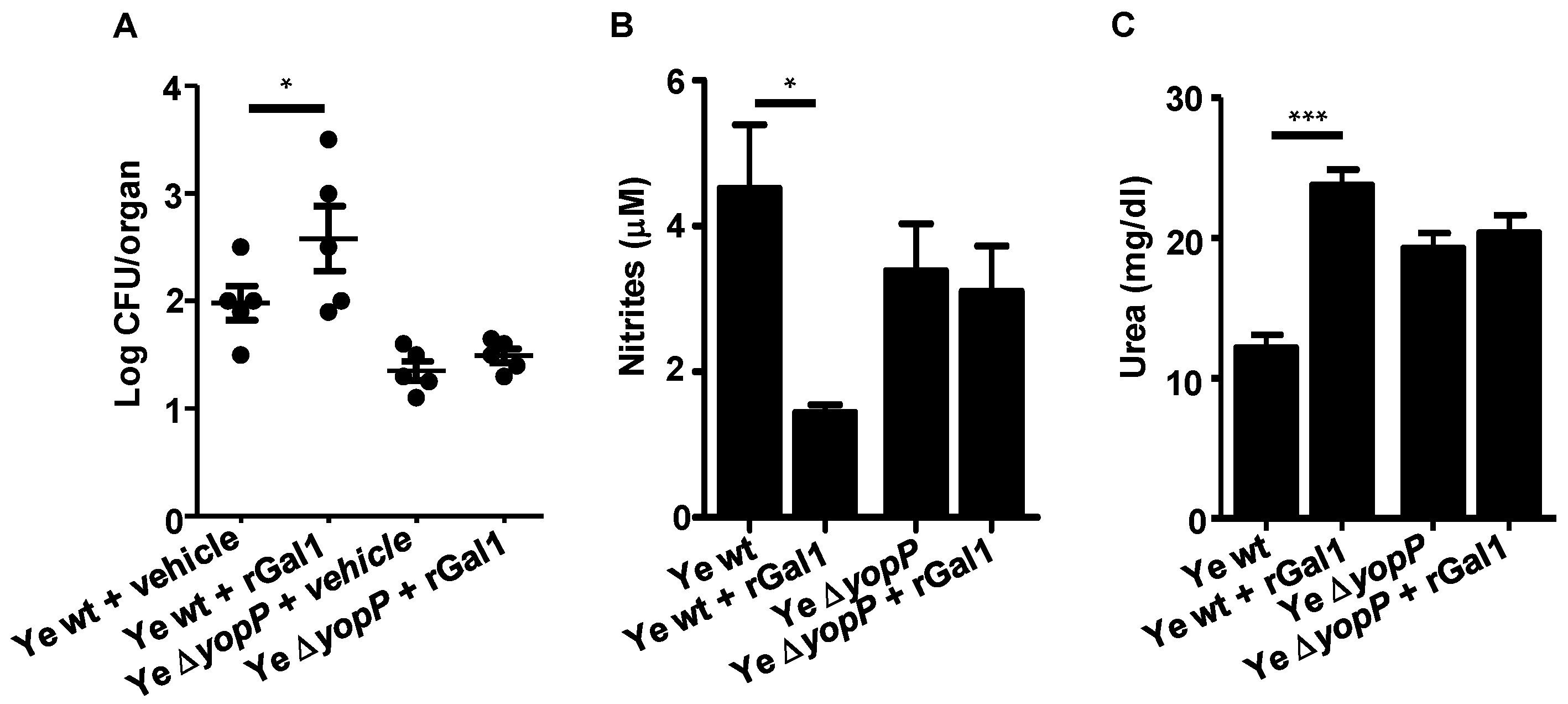
| Lectin | Ligands Described | Reference |
|---|---|---|
| Arachis Hypogaea (Peanut agglutinin) (PNA) | Galβ(1–3)GalNAc Galβ(1–3)GlcNAc Galβ(1–4)GlcNAc Lactose Galactose | [55] |
| Erythrina crystagalli (ECA) | Galβ(1–4)GlcNAc Lactose > GalNAc > Gal | [56] |
| Observed a | Mass Expt b | Mass (Theor) c | Delta Error (Da) d | Pep_exp_z e | Start f | End g | Peptide Sequence h | Modifications i |
|---|---|---|---|---|---|---|---|---|
| 707.816 | 2120.4261 | 2120.0208 | 0.4053 | +3 | 232 | 250 | LNEYLNTNPQGVGTVVNKK | Deamidated (NQ); Lys->CamCys (K) |
| 1123.8687 | 3368.5843 | 3367.5 | 1.0843 | +3 | 100 | 122 | FIINMGEGGIHFSVIDYKHINGK | Deamidated (NQ); Lys->CamCys (K); Hex(1)HexNAc(1) (N); Hex(1)HexNAc(1) (ST); Oxidation (M) |
| 1344.4664 | 2686.9182 | 2686.2353 | 0.6829 | +2 | 48 | 67 | NYSRLDIEVMPALVIQANNK | Deamidated (NQ); Hex(1)HexNAc(1) (N); Lys->CamCys (K) |
| 954.9102 | 2861.7088 | 2861.3441 | 0.3647 | +3 | 258 | 276 | FDNNKSIIDGKELSVSVHK | Deamidated (NQ); 2 Hex(1)HexNAc(1) (ST) |
| 470.2974 | 938.5802 | 937.5219 | 1.0583 | +2 | 284 | 288 | TLLKV | Hex(1)HexNAc(1) (ST) |
| 604.0027 | 1205.9908 | 1205.5122 | 0.4787 | +2 | 182 | 187 | KLYTER | Hex(1)HexNAc(1) (ST); Lys->CamCys (K) |
| 559.2594 | 1674.7563 | 1673.6991 | 1.0572 | +3 | 250 | 262 | KNETIFNRFDNNK | Deamidated (NQ); Lys->CamCys (K) |
| 599.2961 | 1794.8664 | 1793.7499 | 1.1165 | +3 | 21 | 29 | SLISNEELK | Hex(1)HexNAc(1) (N); Hex(1)HexNAc(1) (ST); Lys->CamCys (K) |
| 761.4185 | 1520.8224 | 1520.7471 | 0.0754 | +2 | 200 | 213 | GILSDSENPLPHNK | Deamidated (NQ) |
| 515.7982 | 1544.3728 | 1544.6201 | −0.2473 | +3 | 251 | 262 | NETIFNRFDNNK | Deamidated (NQ); Lys->CamCys (K) |
| 1092.9822 | 3275.9248 | 3275.4749 | 0.4499 | +3 | 1 | 20 | MIGPISQINSFGGLSEKETR | Deamidated (NQ); Hex(1)HexNAc(1) (N); 2 Hex(1)HexNAc(1) (ST); Oxidation (M) |
| 525.9216 | 1574.7429 | 1574.7069 | 0.036 | +3 | 263 | 276 | SIIDGKELSVSVHK | Lys->CamCys (K) |
| 525.2371 | 2621.1492 | 2620.2921 | 0.8571 | +5 | 30 | 51 | NIIIQLETDIADGSWFHKNYSR | Deamidated (NQ) |
| 890.7302 | 3558.8918 | 3558.7498 | 0.142 | +4 | 1 | 29 | MIGPISQINSFGGLSEKETRSLISNEELK | Deamidated (NQ); Hex(1)HexNAc(1) (N); Oxidation (M) |
| 532.2823 | 1062.55 | 1062.5193 | 0.0307 | +2 | 225 | 230 | HTQGKK | Hex(1)HexNAc(1) (ST) |
| 533.9495 | 1598.8266 | 1598.8767 | −0.05 | +1 | 278 | 288 | RIAEYKTLLK | Hex(1)HexNAc(1) (ST) |
| 793.7171 | 2378.1295 | 2378.0999 | 0.0296 | +2 | 2 | 17 | IGPISQINSFGGLSEK | Deamidated (NQ); 2 Hex(1)HexNAc(1) (ST) |
| 642.8429 | 1283.6713 | 1282.4918 | 1.1795 | +3 | 225 | 230 | HTQGKKR | Hex(1)HexNAc(1) (ST); 2 Lys->CamCys (K) |
| 793.4021 | 2377.1845 | 2376.9625 | 0.2219 | +3 | 258 | 268 | FDNNKSIIDGK | Hex(1)HexNAc(1) (N); Hex(1)HexNAc(1) (ST); Lys->CamCys (K) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jofre, B.L.; Eliçabe, R.J.; Silva, J.E.; Pérez Sáez, J.M.; Paez, M.D.; Callegari, E.; Mariño, K.V.; Di Genaro, M.S.; Rabinovich, G.A.; Davicino, R.C. Galectin-1 Cooperates with Yersinia Outer Protein (Yop) P to Thwart Protective Immunity by Repressing Nitric Oxide Production. Biomolecules 2021, 11, 1636. https://doi.org/10.3390/biom11111636
Jofre BL, Eliçabe RJ, Silva JE, Pérez Sáez JM, Paez MD, Callegari E, Mariño KV, Di Genaro MS, Rabinovich GA, Davicino RC. Galectin-1 Cooperates with Yersinia Outer Protein (Yop) P to Thwart Protective Immunity by Repressing Nitric Oxide Production. Biomolecules. 2021; 11(11):1636. https://doi.org/10.3390/biom11111636
Chicago/Turabian StyleJofre, Brenda Lucila, Ricardo Javier Eliçabe, Juan Eduardo Silva, Juan Manuel Pérez Sáez, Maria Daniela Paez, Eduardo Callegari, Karina Valeria Mariño, María Silvia Di Genaro, Gabriel Adrián Rabinovich, and Roberto Carlos Davicino. 2021. "Galectin-1 Cooperates with Yersinia Outer Protein (Yop) P to Thwart Protective Immunity by Repressing Nitric Oxide Production" Biomolecules 11, no. 11: 1636. https://doi.org/10.3390/biom11111636
APA StyleJofre, B. L., Eliçabe, R. J., Silva, J. E., Pérez Sáez, J. M., Paez, M. D., Callegari, E., Mariño, K. V., Di Genaro, M. S., Rabinovich, G. A., & Davicino, R. C. (2021). Galectin-1 Cooperates with Yersinia Outer Protein (Yop) P to Thwart Protective Immunity by Repressing Nitric Oxide Production. Biomolecules, 11(11), 1636. https://doi.org/10.3390/biom11111636






