Branched-Chain Fatty Acids as Mediators of the Activation of Hepatic Peroxisome Proliferator-Activated Receptor Alpha by a Fungal Lipid Extract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Submerged Cultivation of Conidiobolus heterosporus, Fungal Lipid Extraction and Preparation of Fatty Acid Methyl Esters
2.2. Analysis of Fatty Acid Composition
2.3. Chemical Reagents
2.4. Cell Culture
2.5. Cell Treatments
2.6. Cell Viability Assay
2.7. Transient Transfection and Dual Luciferase Reporter Assay
2.8. Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction (qPCR)
2.9. Statistics
3. Results
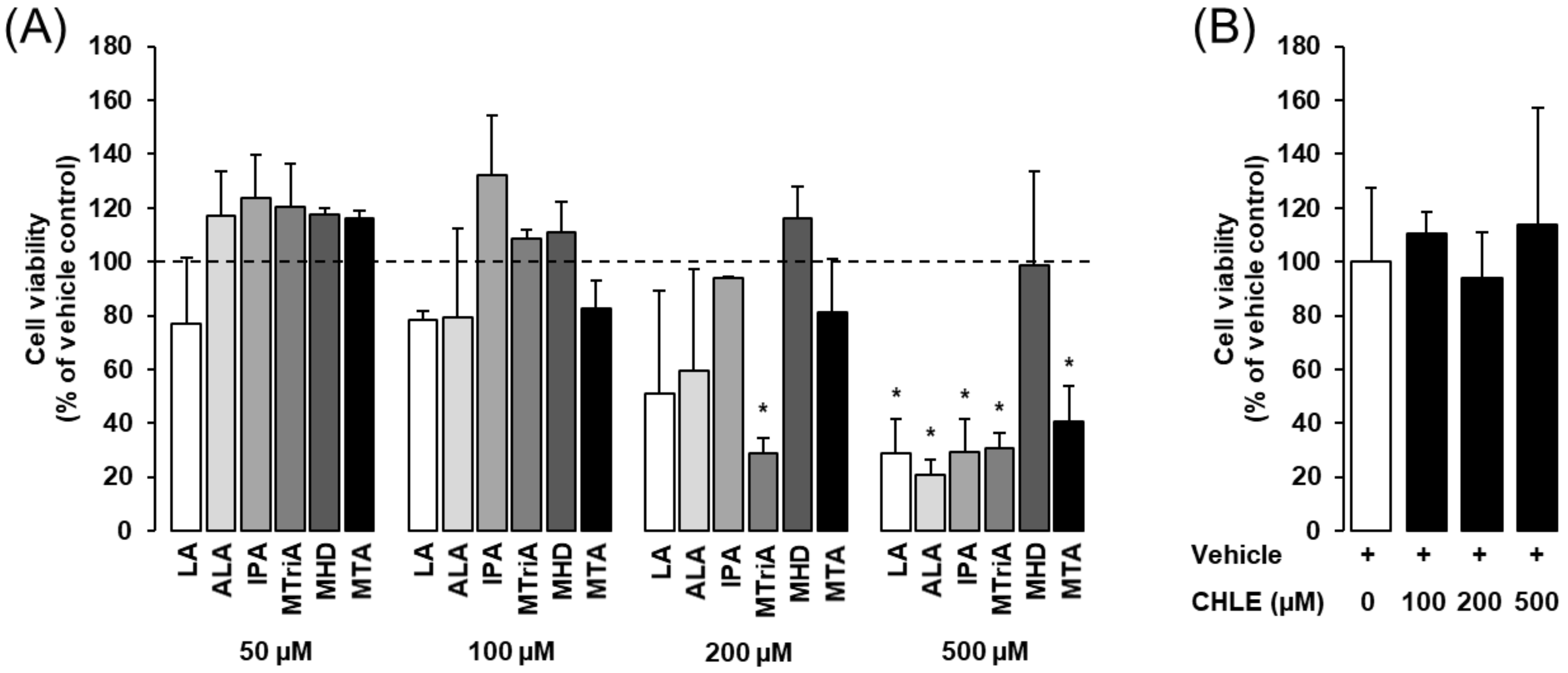
3.1. Effects of Isolated Fatty Acids and CHLE on Cell Viability of Rat Fao Cells
3.2. Effects of Isolated Fatty Acids and CHLE on PPARalpha Transactivation
3.3. Effect of CHLE on the mRNA Concentration of PPARalpha Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krey, O.; Braissant, F.; Lhorset, E.; Kalkhoven, M.; Perroud, M.; Parker, M.G.; Wahli, W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997, 11, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S.; Wahli, W. Peroxisome proliferator activated receptor agonists. EXS 2000, 89, 141–151. [Google Scholar] [PubMed]
- Mandard, S.; Müller, M.; Kersten, S. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 2004, 61, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Marx, N.; Duez, H.; Fruchart, J.C.; Staels, B. Peroxisome proliferator-activated receptors and atherogenesis: Regulators of gene expression in vascular cells. Circ. Res. 2004, 94, 1168–1178. [Google Scholar] [CrossRef]
- Loomba, R.S.; Arora, R. Prevention of cardiovascular disease utilizing fibrates—A pooled meta-analysis. Am. J. Ther. 2010, 17, 182–188. [Google Scholar] [CrossRef]
- Ellinghaus, P.; Wolfrum, C.; Assmann, G.; Spener, F.; Seedorf, U. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein 2-/sterol carrier protein x-deficient mice. J. Biol. Chem. 1999, 274, 2766–2772. [Google Scholar] [CrossRef] [Green Version]
- Wolfrum, C.; Ellinghaus, P.; Fobker, M.; Seedorf, U.; Assmann, G.; Börchers, T.; Spener, F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J. Lipid. Res. 1999, 40, 708–714. [Google Scholar]
- Zomer, A.W.; van Der Burg, B.; Jansen, G.A.; Wanders, R.J.; Poll-The, B.T.; van Der Saag, P.T. Pristanic acid and phytanic acid: Naturally occurring ligands for the nuclear receptor peroxisome proliferator-activated receptor alpha. J. Lipid. Res. 2000, 41, 1801–1807. [Google Scholar]
- Hostetler, H.A.; Kier, A.B.; Schroeder, F. Very-long-chain and branched-chain fatty acyl-CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha). Biochemistry 2006, 45, 7669–7681. [Google Scholar] [CrossRef] [Green Version]
- Gloerich, J.N.; van Vlies, G.A.; Jansen, S.; Denis, J.P.; Ruiter, J.P.N.; van Werkhoven, M.A.; Duran, M.; Vaz, F.M.; Wanders, R.J.A.; Ferdinandusse, S. A phytol-enriched diet induces changes in fatty acid metabolism in mice both via PPARalpha-dependent and—Independent pathways. J. Lipid. Res. 2005, 46, 716–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo-Lamas, L.; Barron, L.J.R.; Farmer, L.; Aldai, N. Fatty acid composition of intramuscular fat and odour-active compounds of lamb commercialized in northern Spain. Meat Sci. 2018, 139, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Ran-Ressler, R.R.; Bae, S.; Lawrence, P.; Wang, D.H.; Brenna, J.T. Branched-chain fatty acid content of foods and estimated intake in the USA. Br. J. Nutr. 2014, 112, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.H.; Yang, Y.; Wang, Z.; Lawrence, P.; Worobo, R.W.; Brenna, J.T. High levels of branched chain fatty acids in nātto and other Asian fermented foods. Food Chem. 2019, 286, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Ratnayakem, W.M.; Olsson, B.; Ackman, R.G. Novel branched-chain fatty acids in certain fish oils. Lipids 1989, 24, 630–637. [Google Scholar] [CrossRef]
- Kates, M. Bacterial lipids. Adv. Lipid. Res. 1964, 2, 17–90. [Google Scholar]
- Tyrrell, D. The fatty acid compositions of 17 Entomophthora isolates. Can. J. Microbiol. 1967, 13, 755–760. [Google Scholar] [CrossRef]
- Tyrrell, D. The fatty acid composition of some Entomophthoraceae. Can. J. Microbiol. 1971, 17, 1115–1118. [Google Scholar] [CrossRef]
- Fraatz, M.A.; Goldmann, M.; Geissler, T.; Gross, E.; Backes, M.; Hilmer, J.M.; Ley, J.; Rost, J.; Francke, A.; Zorn, H. Biotechnological production of methyl-branched aldehydes. J. Agric. Food Chem. 2016, 66, 2387–2392. [Google Scholar] [CrossRef]
- Deschatrette, J.; Weiss, M.C. Characterization of differentiated clones from a rat hepatoma. Biochimie 1974, 56, 1603–1611. [Google Scholar] [CrossRef]
- König, B.; Eder, K. Differential action of 13-HPODE on PPARalpha downstream genes in rat Fao and human HepG2 hepatoma cell lines. J. Nutr. Biochem. 2006, 17, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Ringseis, R.; Rosenbaum, S.; Gessner, D.K.; Herges, L.; Kubens, J.F.; Mooren, F.C.; Krüger, K.; Eder, K. Supplementing obese Zucker rats with niacin induces the transition of glycolytic to oxidative skeletal muscle fibers. J. Nutr. 2013, 143, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanhoff, T.; Wolfrum, C.; Ellinghaus, P.; Seedorf, U.; Spener, F. Pristanic acid is activator of peroxisome proliferator activated receptor α. Eur. J. Lipid. Sci. Technol. 2001, 103, 75–80. [Google Scholar] [CrossRef]
- Lampen, A.; Carlberg, C.; Nau, H. Peroxisome proliferator-activated receptor delta is a specific sensor for teratogenic valproic acid derivatives. Eur. J. Pharmacol. 2001, 431, 25–33. [Google Scholar] [CrossRef]
- Shirouchi, B.; Nagao, K.; Furuya, K.; Nagai, T.; Ichioka, K.; Tokairin, S.; Iida, Y.; Yanagita, T. Physiological functions of iso-type short-chain fatty acid and omega 3 polyunsaturated fatty acids containing oil in obese OLETF rats. J. Oleo Sci. 2010, 59, 299–305. [Google Scholar] [CrossRef] [Green Version]
- Castillero, E.; Martín, A.I.; Nieto-Bona, M.P.; Fernández-Galaz, C.; López-Menduiña, M.; Villanúa, M.Á.; López-Calderón, A. Fenofibrate administration to arthritic rats increases adiponectin and leptin and prevents oxidative muscle wasting. Endocr. Connect. 2012, 1, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, J.; Ganapathy, V.; Longo, N. Mutations in the organic cation/carnitine transporter OCTN2 in primary carnitine deficiency. Proc. Natl. Acad. Sci. USA 1999, 96, 2356–2360. [Google Scholar] [CrossRef] [Green Version]
- Ran-Ressler, R.R.; Khailova, L.; Arganbright, K.M.; Adkins-Rieck, C.K.; Jouni, Z.E.; Koren, O.; Ley, R.E.; Brenna, J.T.; Dvorak, B. Branched chain fatty acids reduce the incidence of necrotizing enterocolitis and alter gastrointestinal microbial ecology in a neonatal rat model. PLoS ONE 2011, 6, e29032. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, J.; Shiraishi, T.; Iwasaki, A.; Maekawa, S.; Higuchi, T.; Hiratuka, M.; Tanaka, T.; Shibaguchi, H.; Kuroki, M.; Shirakusa, T. PPARalpha ligand WY14643 reduced acute rejection after rat lung transplantation with the upregulation of IL-4, IL-10 and TGFbeta mRNA expression. J. Heart Lung Transplant. 2009, 28, 1172–1179. [Google Scholar] [CrossRef]
- Deetz, L.E.; Richardson, C.R.; Pritchard, R.H.; Preston, R.L. Feedlot performance and carcass characteristics of steers fed diets containing ammonium salts of the branched-chain fatty acids and valeric acid. J. Anim. Sci. 1985, 61, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Cline, T.R.; Garrigus, U.S.; Hatfield, E.E. Addition of branched- and straight-chain volatile fatty acids to purified lamb diets and effects on utilization of certain dietary components. J. Anim. Sci. 1966, 25, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Lough, A.K.; Earl, C.R. Fatty acid composition of tissue lipids of rats given dietary branched-chain fatty acids. Proc. Nutr. Soc. 1978, 37, 76. [Google Scholar]
- Chateauvieux, S.; Morceau, F.; Dicato, M.; Diederich, M. Molecular and therapeutic potential and toxicity of valproic acid. J. Biomed. Biotechnol. 2010, 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driever, P.H.; Knüpfer, M.M.; Cinatl, J.; Wolff, J.E. Valproic acid for the treatment of pediatric malignant glioma. Klin. Padiatr. 1999, 211, 323–328. [Google Scholar] [CrossRef]
- Reddy, J.K.; Azarnoff, D.L.; Hignite, C.E. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature 1980, 283, 397–398. [Google Scholar] [CrossRef]
- Peters, J.M.; Cheung, C.; Gonzalez, F.J. Peroxisome proliferator-activated receptor-α and liver cancer: Where do we stand? J. Mol. Med. 2005, 83, 774–785. [Google Scholar] [CrossRef]
- Fan, C.Y.; Pan, J.; Chu, R.; Lee, D.; Kluckman, K.D.; Usuda, N.; Singh, I.; Yeldandi, A.V.; Rao, M.S.; Maeda, N.; et al. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J. Biol. Chem. 1996, 271, 24698–24710. [Google Scholar] [CrossRef] [Green Version]
- Idel, S.; Ellinghaus, P.; Wolfrum, C.; Nofer, J.R.; Gloerich, J.; Assmann, G.; Spener, F.; Seedorf, U. Branched chain fatty acids induce nitric oxide-dependent apoptosis in vascular smooth muscle cells. J. Biol. Chem. 2002, 277, 49319–49325. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Collin, P.; Madden, T.; Chan, D.; Sweeney-Gotsch, B.; McConkey, D.; Newman, R.A. Inhibition of proliferation of PC3 cells by the branched-chain fatty acid, 12-methyltetradecanoic acid, is associated with inhibition of 5-lipoxygenase. Prostate 2003, 55, 281–291. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, S.; Chen, X.; Chen, H.; Huang, M.; Zheng, J. Induction of apoptotic cell death and in vivo growth inhibition of human cancer cells by a saturated branched-chain fatty acid, 13-methyltetradecanoic acid. Cancer. Res. 2000, 60, 505–509. [Google Scholar] [PubMed]


| Fatty acids 1 | Area (%) |
|---|---|
| Branched-chain fatty acids (BCFAs) | 52.7 |
| iso-C14:0 (MTriA) | 33.0 |
| anteiso-C15:0 (MTA) | 13.1 |
| iso-C16:0 (IPA) | 5.9 |
| anteiso-C17:0 (MHD) | 0.7 |
| Straight-chain fatty acids | 39.5 |
| C12:0 | 0.3 |
| C13:0 | 1.4 |
| C14:0 | 5.4 |
| C14:1 | 0.1 |
| C15:0 | 4.8 |
| C16:0 | 7.8 |
| C16:1 | 0.6 |
| C17:0 | 0.4 |
| C18:0 | 1.3 |
| C18:1 | 1.7 |
| C18:2 | 0.7 |
| C18:3 (γ-linolenic acid) | 0.8 |
| C20:0 | 0.2 |
| C20:1 | 0.2 |
| C20:2 | 0.3 |
| C20:3 | 1.1 |
| C20:4 | 9.4 |
| C20:5 | 0.4 |
| C22:0 | 0.2 |
| C22:1 | 0.3 |
| C22:2 | 0.3 |
| C23:0 | 0.1 |
| C24:0 | 1.4 |
| C24:1 | 0.3 |
| Sum of saturated straight-chain fatty acids | 23.3 |
| Sum of monounsaturated straight-chain fatty acids | 3.2 |
| Sum of polyunsaturated straight-chain fatty acids | 13.0 |
| Total others | 7.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maheshwari, G.; Ringseis, R.; Wen, G.; Gessner, D.K.; Rost, J.; Fraatz, M.A.; Zorn, H.; Eder, K. Branched-Chain Fatty Acids as Mediators of the Activation of Hepatic Peroxisome Proliferator-Activated Receptor Alpha by a Fungal Lipid Extract. Biomolecules 2020, 10, 1259. https://doi.org/10.3390/biom10091259
Maheshwari G, Ringseis R, Wen G, Gessner DK, Rost J, Fraatz MA, Zorn H, Eder K. Branched-Chain Fatty Acids as Mediators of the Activation of Hepatic Peroxisome Proliferator-Activated Receptor Alpha by a Fungal Lipid Extract. Biomolecules. 2020; 10(9):1259. https://doi.org/10.3390/biom10091259
Chicago/Turabian StyleMaheshwari, Garima, Robert Ringseis, Gaiping Wen, Denise K. Gessner, Johanna Rost, Marco A. Fraatz, Holger Zorn, and Klaus Eder. 2020. "Branched-Chain Fatty Acids as Mediators of the Activation of Hepatic Peroxisome Proliferator-Activated Receptor Alpha by a Fungal Lipid Extract" Biomolecules 10, no. 9: 1259. https://doi.org/10.3390/biom10091259





