2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis
Abstract
:1. Introduction
2. Results
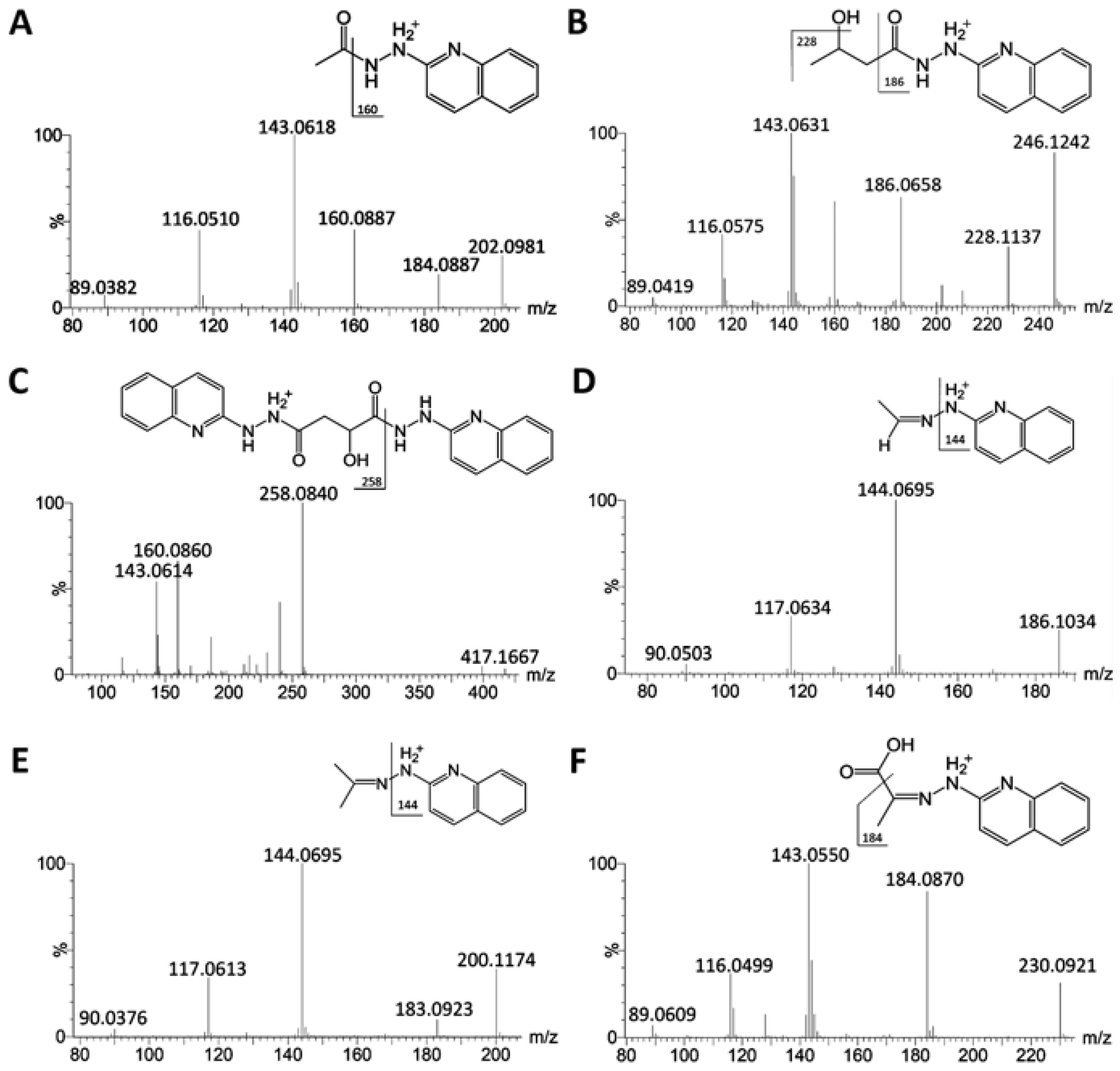
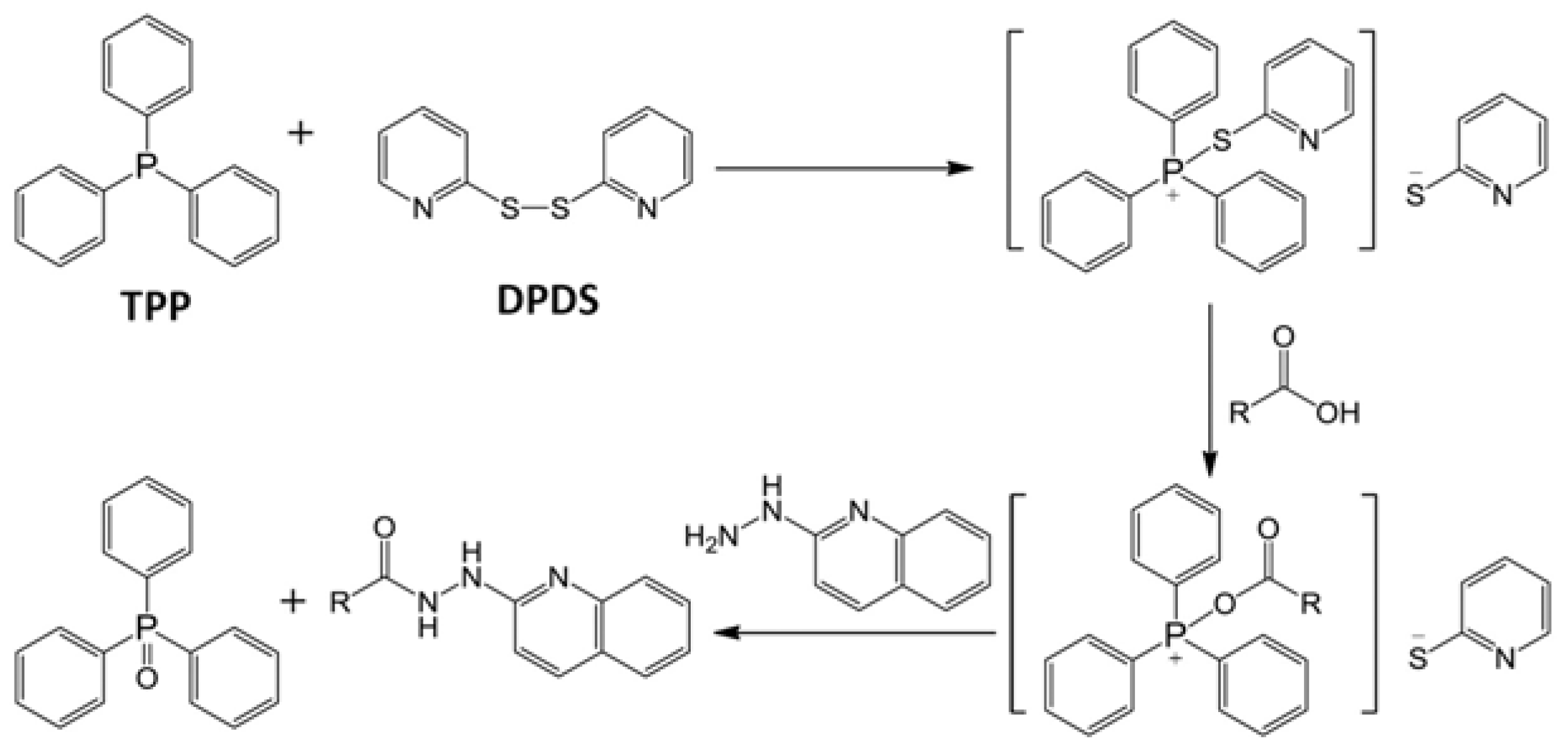
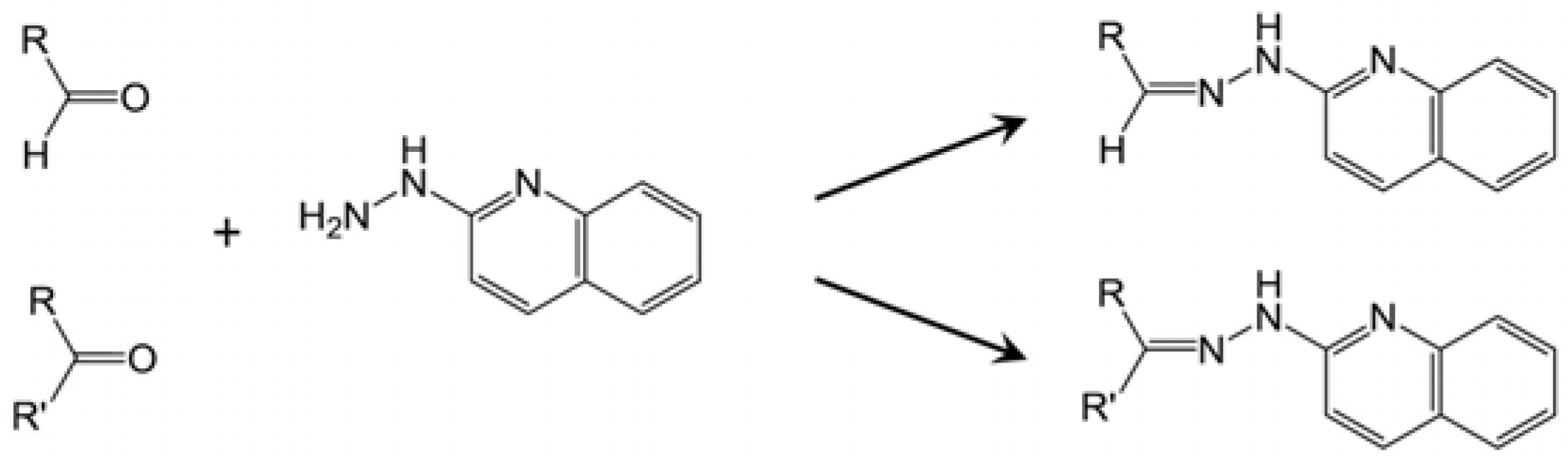
2.1. Identification of HQ As an Effective Derivatization Agent for LC-MS Detection of Short-Chain Carboxylic Acids, Aldehydes and Ketones
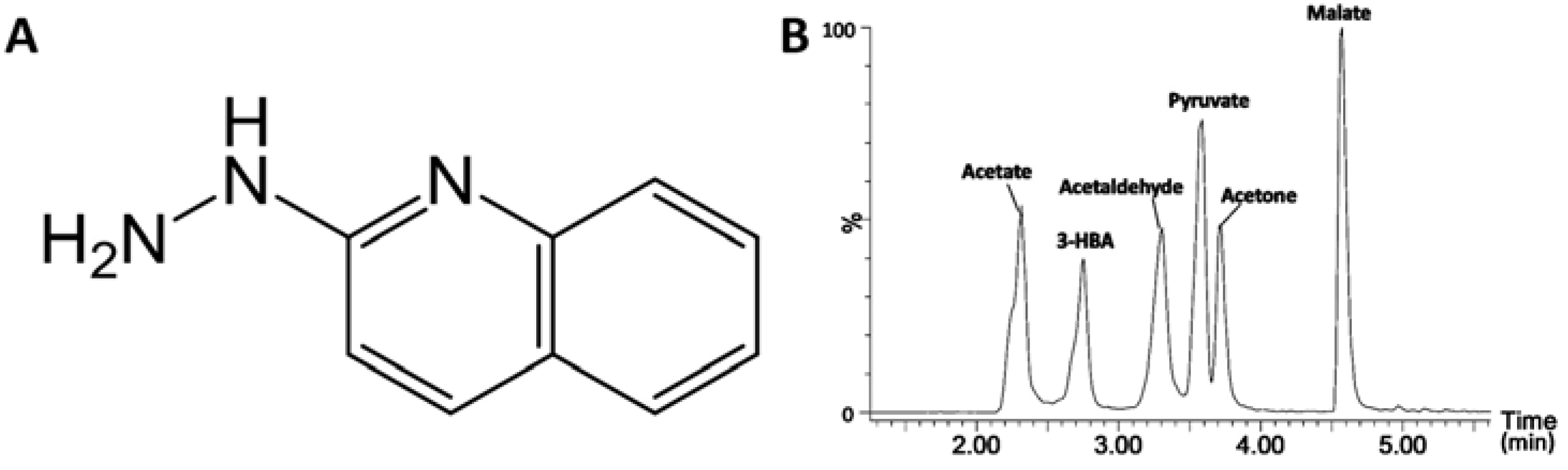
| Derivatization reactions | Acetate  | HBA  | Malate  | Acetaldehyde  | Acetone  | Pyruvate  |
|---|---|---|---|---|---|---|
 | Forming amide C8H10N2O | Forming amide C10H14N2O2 | Forming di-amide C16H18N4O3 | N.D. | N.D. | Forming amide C9H10N2O2 |
 | Forming hydrazide C7H9N3O | Forming hydrazide C9H13N3O2 | Forming di-hydrazide C14H16N6O3 | N.D. | N.D. | Forming hydrazide C8H9N3O2 |
 | Forming hydrazide C11H11N3O | Forming hydrazide C13H15N3O2 | Forming di-hydrazide C22H20N6O3 | Forming hydrazine C11H11N3 | Forming hydrazine C12H13N3 | Forming hydrazine C12H11N3O2 |
 | N.D. | N.D. | N.D. | Forming hydrazine C14H17N3O2S | Forming hydrazine C15H19N3O2S | Forming hydrazine C15H17N3O4S |
2.2. Optimization of HQ-Mediated Derivatization Reaction Conditions

2.3. Compatibility of HQ Derivatization with Biological Samples
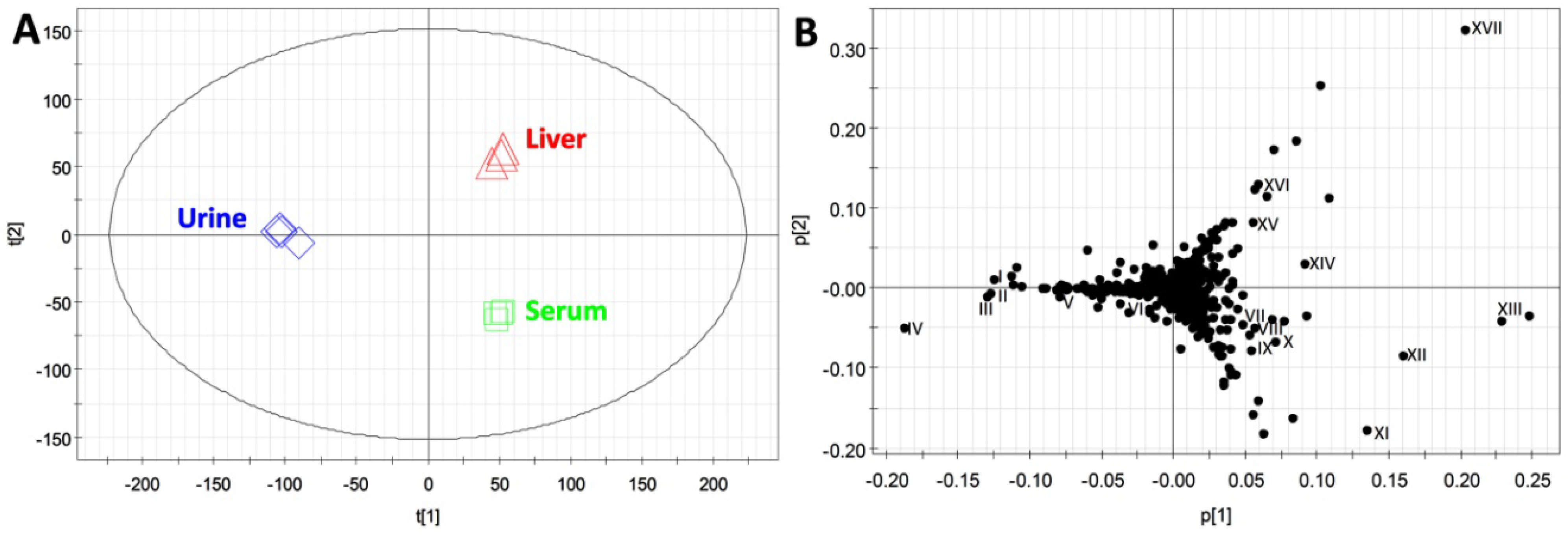
| Compounds | ID | Formula | Derivative Formula (No. of HQ moiety) | Exact mass of [M+H]+ | Distribution |
|---|---|---|---|---|---|
| Formaldehyde | XIV | CH2O | C10H9N3 (1) | 172.0875 | L |
| Acetaldehyde | XVII | C2H4O | C11H11N3 (1) | 186.1031 | L |
| Acetic acid | VIII | C2H4O2 | C11H11N3O (1) | 202.0980 | S |
| Acetone | XIII | C3H6O | C12H13N3 (1) | 200.1188 | U/S/L |
| Propionic acid | XV | C3H6O2 | C12H13N3O (1) | 216.1137 | U/L |
| Pyruvic acid | VII | C3H4O3 | C12H11N3O2 (1) | 230.0930 | S/L |
| Lactic acid | XII | C3H6O3 | C12H13N3O2 (1) | 232.1086 | S/L |
| Acetoin | V | C4H8O2 | C13H15N3O (1) | 230.1288 | U |
| Butyric acid | - | C4H8O2 | C13H15N3O (1) | 230.1288 | - |
| Acetoacetic acid | - | C4H6O3 | C13H13N3O2 (1) | 244.1086 | U |
| HBA | XI | C4H8O3 | C13H15N3O2 (1) | 246.1243 | S |
| Fumaric acid | - | C4H4O4 | C13H11N3O3 (1) | 258.0873 | |
| Succinic acid | - | C4H6O4 | C13H13N3O3 (1) | 260.1030 | U |
| Malic acid | - | C4H6O5 | C22H20N6O3 (2) | 417.1675 | - |
| α-Ketoisovaleric acid | I | C5H8O3 | C14H15N3O2 (1) | 258.1243 | U |
| α-Ketoglutaric acid | IV | C5H6O5 | C14H13N3O4 (1) | 288.0984 | U |
| Dehydroascorbic acid | XVI | C6H6O6 | C15H13N3O5 (1) | 316.0928 | U/L |
| α-Ketoisocaproic acid | III | C6H10O3 | C15H17N3O2 (1) | 272.1399 | U |
| Glucose | IX | C6H12O6 | C15H19N3O5 (1) | 322.1397 | S/L |
| Mannose | X | C6H12O6 | C15H19N3O5 (1) | 322.1397 | S/L |
| Citric acid | - | C6H8O7 | C13H11N3O3 (1) | 258.0873 | U |
| Formiminoglutamic acid | II | C6H10N2O4 | C15H17N5O3 (1) | 316.1404 | U |
| 4-Hydroxyphenylacetic acid | VI | C8H8O3 | C17H15N3O2 (1) | 294.1237 | U |
| 4-Hydroxyphenylpyruvic acid | - | C9H8O4 | C18H15N3O3 (1) | 322.1192 | U |
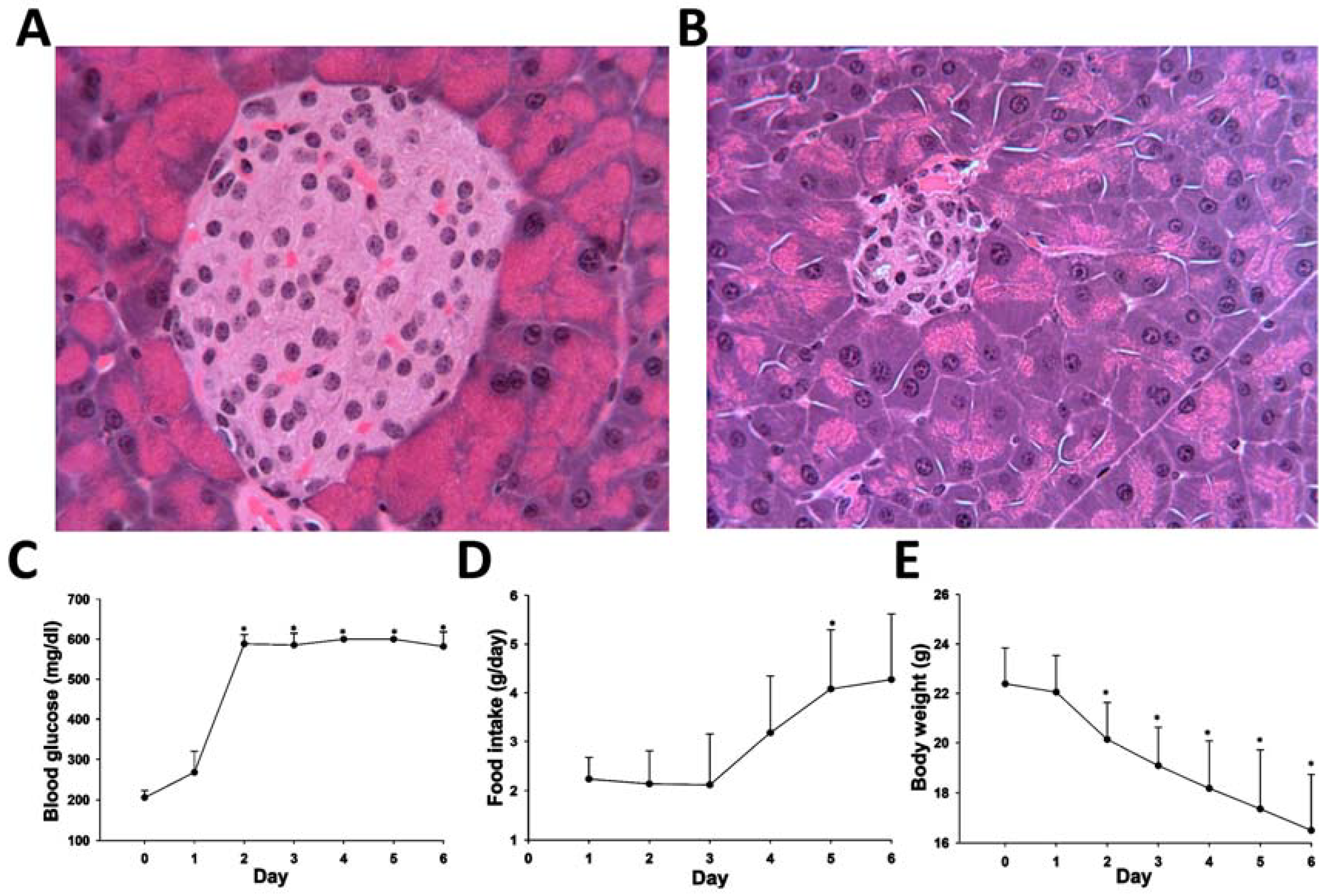
2.4. Application of HQ Derivatization in LC-MS-based Metabolomic Analysis of Diabetes-Induced Ketoacidosis
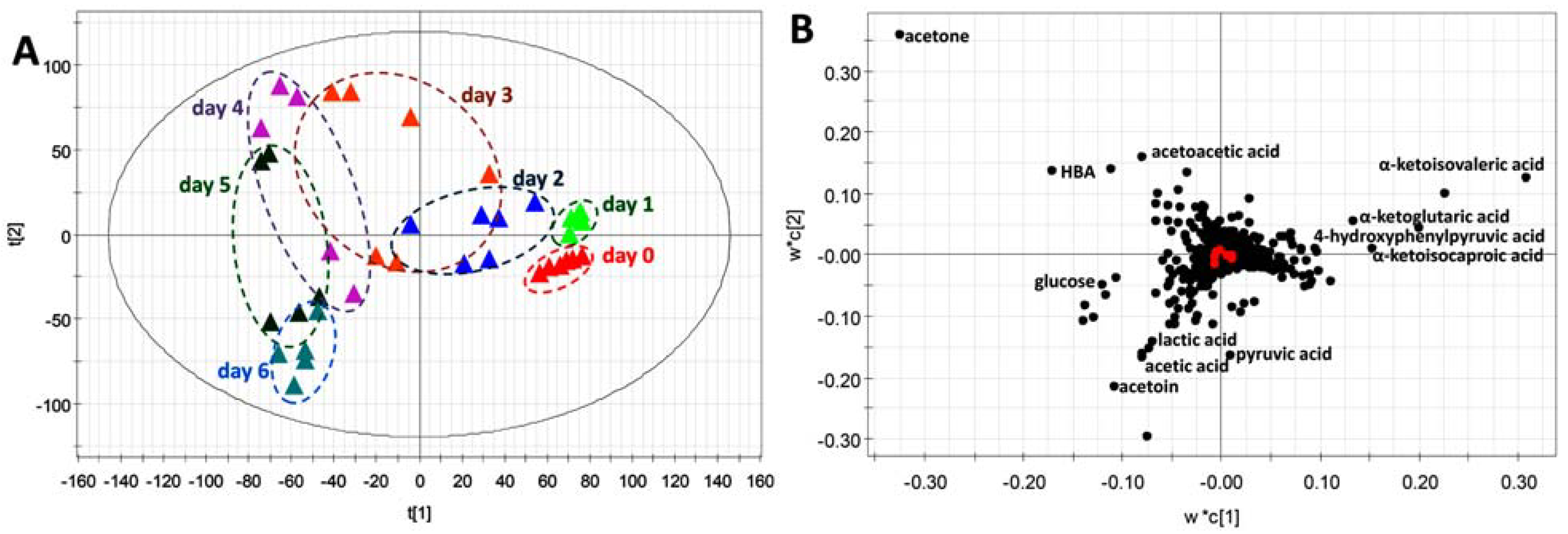
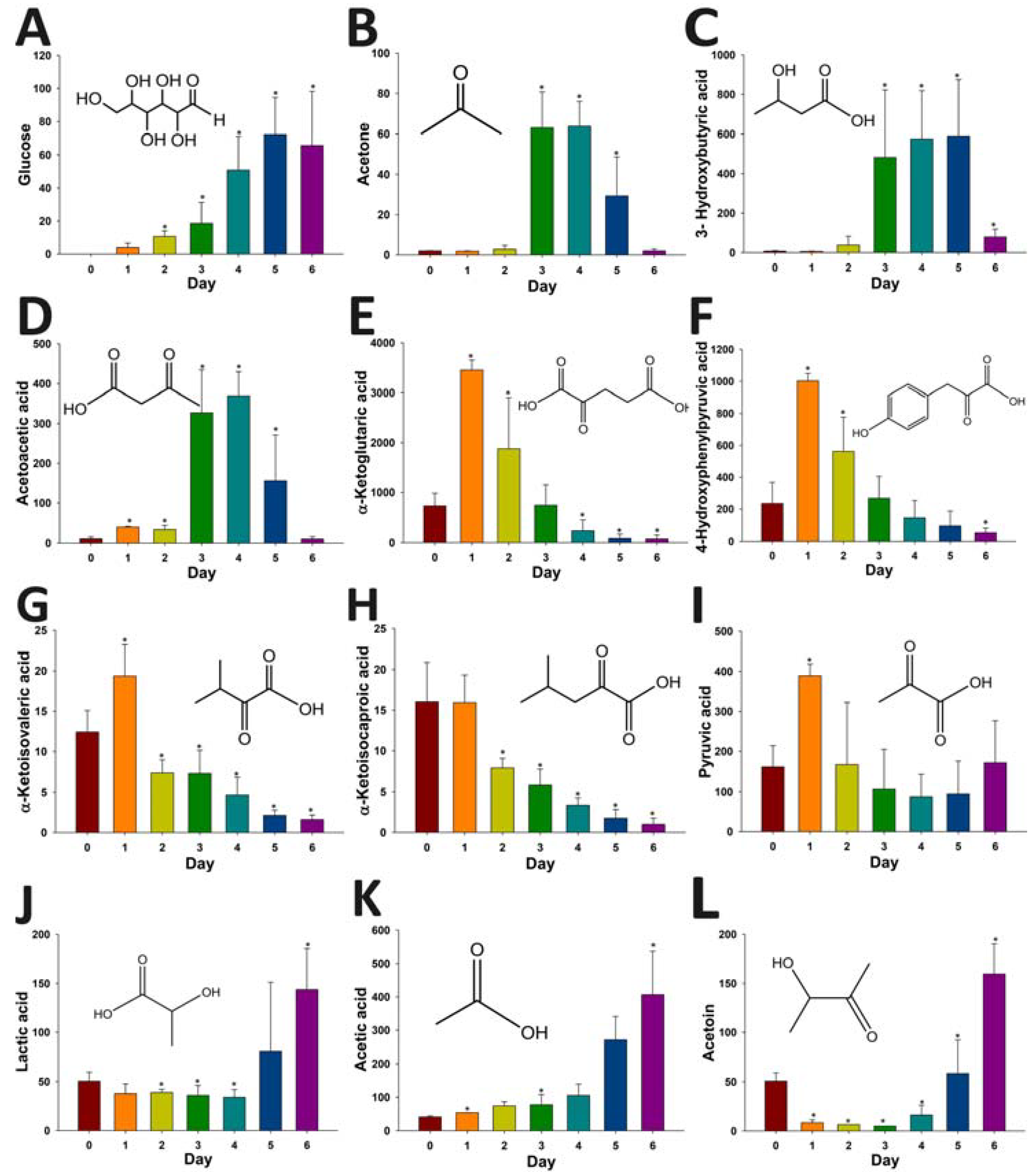
3. Discussion
4. Experimental Section
4.1. Reagents
4.2. Animal Treatment and Sample Collection
4.3. Biochemical and Histological Analysis of STZ-Induced Diabetes
4.4. Preparation of Aqueous Liver Extraction
4.5. Derivatization of Six Test Compounds with PA, HP, DH and HQ
4.6. Optimization of HQ Derivatization Reactions
4.7. HQ Derivatization of Biological Samples
4.8. LC-MS Analysis of Derivatization Products
4.9. Chemometric Analysis of LC-MS Data
4.10. Statistics
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Kumps, A.; Duez, P.; Mardens, Y. Metabolic, nutritional, iatrogenic, and artifactual sources of urinary organic acids: A comprehensive table. Clin. Chem. 2002, 48, 708–717. [Google Scholar]
- Laffel, L. Ketone bodies: A review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab. Res. Rev. 1999, 15, 412–426. [Google Scholar] [CrossRef]
- Draper, H.H.; Csallany, A.S.; Hadley, M. Urinary aldehydes as indicators of lipid peroxidation in vivo. Free Radical Bio. Med. 2000, 29, 1071–1077. [Google Scholar] [CrossRef]
- Jellum, E.; Stokke, O.; Eldjarn, L. Application of gas chromatography, mass spectrometry, and computer methods in clinical biochemistry. Anal. Chem. 1973, 46, 1099–1106. [Google Scholar]
- Knapp, D.R. Handbook of Analytical Derivatization Reactions; Wiley: New York, NY, USA, 1979; pp. 342–346. [Google Scholar]
- Hong, Z.; Lin, Z.; Liu, Y.; Tan, G.; Lou, Z.; Zhu, Z.; Chai, Y.; Fan, G.; Zhang, J.; Zhang, L. Innovative microwave-assisted oximation and silylation procedures for metabolomic analysis of plasma samples using gas chromatography-mass spectrometry. J. Chromatogr. A 2012, 1254, 14–22. [Google Scholar]
- Pierce, A.E. Silylation of Organic Compounds: a Technique for Gas-Phase Analysis; Pierce Chemical Co.: Rockford, IL, USA, 1968; pp. 1–487. [Google Scholar]
- Grob, K.; Li, Z.W. Introduction of water and water-containing solvent mixtures in capillary gas-chromatography: 1. Failure to produce water-wettable precolumns (retention gaps). J. Chromatogr. A 1989, 473, 381–390. [Google Scholar]
- Kind, T.; Tolstikov, V.; Fiehn, O.; Weiss, R.H. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal. Biochem. 2007, 363, 185–195. [Google Scholar]
- Lanz, C.; Patterson, A.D.; Slavik, J.; Krausz, K.W.; Ledermann, M.; Gonzalez, F.J.; Idle, J.R. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat. Res. 2009, 172, 198–212. [Google Scholar]
- Zhang, Q.; Wang, G.; Du, Y.; Zhu, L.; Jiye, A. GC/MS analysis of the rat urine for metabonomic research. J. Chromatogr. B 2007, 854, 20–25. [Google Scholar]
- Jian, W.; Edom, R.W.; Xu, Y.; Weng, N. Recent advances in application of hydrophilic interaction chromatography for quantitative bioanalysis. J. Sep. Sci. 2010, 33, 681–697. [Google Scholar] [CrossRef]
- Bajad, S.U.; Lu, W.; Kimball, E.H.; Yuan, J.; Peterson, C.; Rabinowitz, J.D. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 2006, 1125, 76–88. [Google Scholar]
- Xu, F.; Zou, L.; Liu, Y.; Zhang, Z.; Ong, C.N. Enhancement of the capabilities of liquid chromatography-mass spectrometry with derivatization: General principles and applications. Mass Spectrom. Rev. 2011, 30, 1143–1172. [Google Scholar]
- Santa, T. Derivatization reagents in liquid chromatography/electrospray ionization tandem mass spectrometry. Biomed. Chromatogr. 2011, 25, 1–10. [Google Scholar] [CrossRef]
- Higashi, T.; Ichikawa, T.; Inagaki, S.; Min, J.Z.; Fukushima, T.; Toyo’oka, T. Simple and practical derivatization procedure for enhanced detection of carboxylic acids in liquid chromatography-electrospray ionization-tandem mass spectrometry. J. Pharmaceut. Biomed. 2010, 52, 809–818. [Google Scholar]
- Zhao, L.; Liu, X.; Xie, L.; Gao, H.; Lin, D. 1H NMR-based metabonomic analysis of metabolic changes in streptozotocin-induced diabetic rats. Anal. Sci. 2010, 26, 1277–1282. [Google Scholar] [CrossRef]
- Li, X.; Xu, Z.; Lu, X.; Yang, X.; Yin, P.; Kong, H.; Yu, Y.; Xu, G. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal. Chim. Acta 2009, 633, 257–262. [Google Scholar]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- Huffman, K.M.; Shah, S.H.; Stevens, R.D.; Bain, J.R.; Muehlbauer, M.; Slentz, C.A.; Tanner, C.J.; Kuchibhatla, M.; Houmard, J.A.; Newgard, C.B.; et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care 2009, 32, 1678–1683. [Google Scholar] [CrossRef]
- Maestri, L.; Ghittori, S.; Imbriani, M.; Capodaglio, E. Determination of 2,5-hexandione by high-performance liquid chromatography after derivatization with dansylhydrazine. J. Chromatogr. B 1994, 657, 111–117. [Google Scholar]
- Matsueda, R.; Maruyama, H.; Ueki, M.; Mukaiyama, T. Peptide synthesis by oxidation-reduction condensation. Ii. The use of disulfide as an oxidant. Bull. Chem. Soc. Jpn. 1971, 44, 1373–1378. [Google Scholar]
- Tanaka, K.; Hine, D.G.; West-Dull, A.; Lynn, T.B. Gas-chromatographic method of analysis for urinary organic acids. I. Retention indices of 155 metabolically important compounds. Clin. Chem. 1980, 26, 1839–1846. [Google Scholar]
- Binding, N.; Klaning, H.; Karst, U.; Potter, W.; Czeschinski, P.A.; Witting, U. Analytical reliability of carbonyl compound determination using 1,5-dansylhydrazine-derivatization. Fresen. J. Anal. Chem. 1998, 362, 270–273. [Google Scholar]
- Kitabchi, A.E.; Umpierrez, G.E.; Murphy, M.B.; Barrett, E.J.; Kreisberg, R.A.; Malone, J.I.; Wall, B.M. Hyperglycemic crises in patients with diabetes mellitus. Diabetes Care 2003, 26, S109–S117. [Google Scholar] [CrossRef]
- Felig, P.; Wahren, J.; Sherwin, R.; Palaiologos, G. Amino acid and protein metabolism in diabetes mellitus. Arch. Intern. Med. 1977, 137, 507–513. [Google Scholar]
- Felig, P.; Marliss, E.; Ohman, J.L.; Cahill, C.F., Jr. Plasma amino acid levels in diabetic ketoacidosis. Diabetes 1970, 19, 727–728. [Google Scholar]
- Aftring, R.P.; Miller, W.J.; Buse, M.G. Effects of diabetes and starvation on skeletal muscle branched-chain alpha-keto acid dehydrogenase activity. Am. J. Physiol. 1988, 254, E292–E300. [Google Scholar]
- Stanley, J.C.; Fisher, M.J.; Pogson, C.I. The metabolism of l-phenylalanine and l-tyrosine by liver cells isolated from adrenalectomized rats and from streptozotocin-diabetic rats. Biochem. J. 1985, 228, 249–255. [Google Scholar]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar]
- Hassinen, I. Acetoin as a metabolite of ethanol. Acta Physiol. Scand. 1963, 57, 135–143. [Google Scholar] [CrossRef]
- Casazza, J.P.; Song, B.J.; Veech, R.L. Short chain diol metabolism in human disease states. Trends Biochem. Sci. 1990, 15, 26–30. [Google Scholar] [CrossRef]
- Tesch, G.H.; Allen, T.J. Rodent models of streptozotocin-induced diabetic nephropathy. Nephrology 2007, 12, 261–266. [Google Scholar] [CrossRef]
- Clee, S.M.; Attie, A.D. The genetic landscape of type 2 diabetes in mice. Endocr. Rev. 2007, 28, 48–83. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Chen, C.; Kim, S. LC-MS-based metabolomics of xenobiotic-induced toxicities. Comput. Struct. Biotechnol. J. 2013, 4, e20130108. [Google Scholar]
Supplementary Materials
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lu, Y.; Yao, D.; Chen, C. 2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis. Metabolites 2013, 3, 993-1010. https://doi.org/10.3390/metabo3040993
Lu Y, Yao D, Chen C. 2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis. Metabolites. 2013; 3(4):993-1010. https://doi.org/10.3390/metabo3040993
Chicago/Turabian StyleLu, Yuwei, Dan Yao, and Chi Chen. 2013. "2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis" Metabolites 3, no. 4: 993-1010. https://doi.org/10.3390/metabo3040993





