Off-the-Vine Ripening of Tomato Fruit Causes Alteration in the Primary Metabolite Composition
Abstract
:1. Introduction
2. Results and Discussion
2.1. 1H NMR Spectra of Mature Tomato Fruits
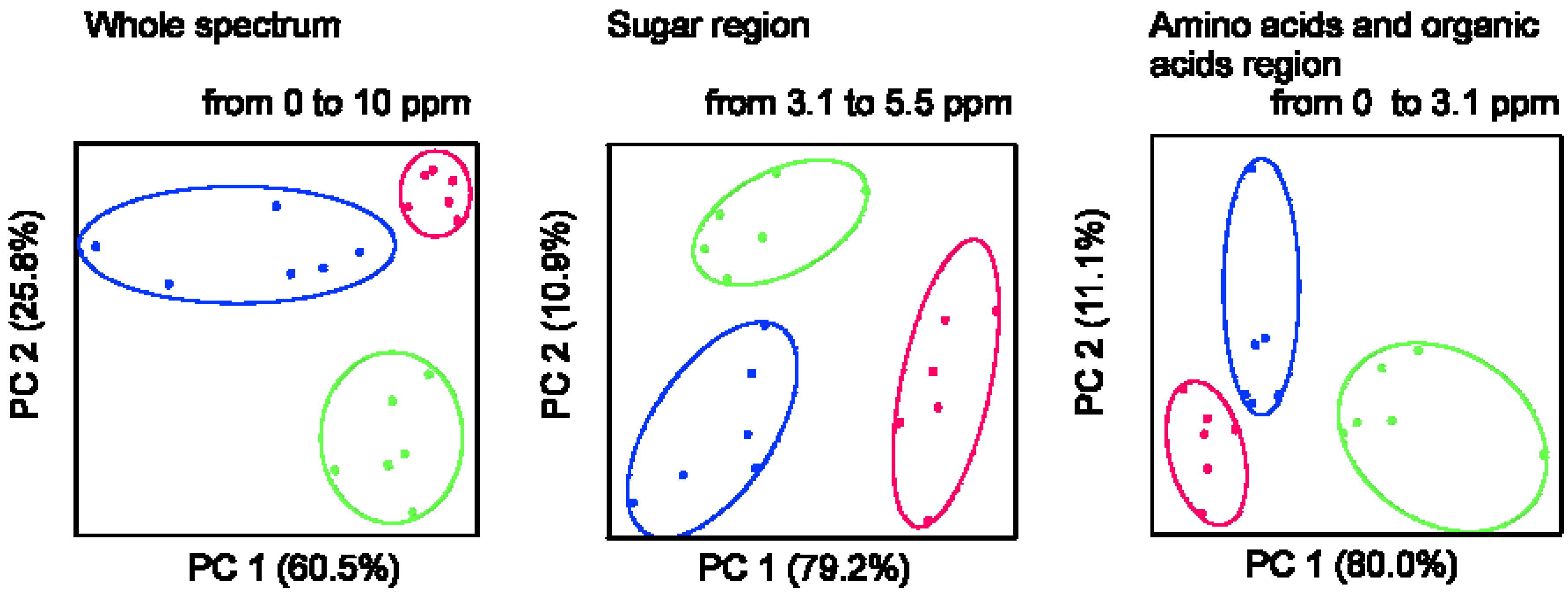
| Metabolite | Mature green | Red ripened on-the-vine | Red ripened off-the-vine |
|---|---|---|---|
| mol/ g FW | |||
| Sugars | |||
| Fructose | 64.16 ± 6.56 | 74.98 ± 6.71 | 50.76 ± 1.69* |
| Glucose | 25.98 ± 2.63 | 29.51 ± 2.83 | 17.69 ± 1.15* |
| Sucrose | 2.82 ± 0.63 | 0.15 ± 0.01 | 0.02 ± 0.01* |
| Total sugars | 92.96 ± 9.28 | 104.63 ± 9.54 | 68.47 ± 2.82* |
| Organic acids | |||
| 2-oxoglutarate | 5.11 ± 1.00 | 0.00 ± 0.00 | 0.99 ± 0.18* |
| Citrate | 28.17 ± 1.02 | 41.96 ± 0.95 | 41.32 ± 0.92 |
| Formate | 0.05 ± 0.00 | 0.10 ± 0.01 | 0.08 ± 0.00* |
| Fumarate | 0.02 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Malate | 19.57 ± 2.61 | 4.04 ± 0.45 | 3.49 ± 1.05 |
| Pyruvate | 0.55 ± 0.13 | 0.02 ± 0.01 | 0.01 ± 0.00 |
| Succinate | 0.52 ± 0.13 | 0.00 ± 0.00 | 0.02 ± 0.02 |
| Total org. acids | 52.98 ± 1.04 | 46.13 ± 0.95 | 45.91 ± 1.76 |
| Amino acids | |||
| Alanine | 0.61 ± 0.08 | 0.59 ± 0.05 | 0.42 ± 0.01* |
| Asparagine | 1.06 ± 0.04 | 0.88 ± 0.07 | 0.32 ± 0.08* |
| Aspartate | 0.51 ± 0.06 | 2.30 ± 0.04 | 1.36 ± 0.26* |
| GABA | 5.27 ± 0.32 | 2.52 ± 0.39 | 2.73 ± 0.92 |
| Glutamine | 4.68 ± 0.17 | 2.49 ± 0.29 | 2.10 ± 0.18 |
| Glutamate | 0.78 ± 0.09 | 5.19 ± 0.74 | 3.04 ± 0.14* |
| Isoleucine | 0.22 ± 0.04 | 0.09 ± 0.02 | 0.08 ± 0.01 |
| Phenylalanine | 0.20 ± 0.01 | 0.12 ± 0.02 | 0.05 ± 0.02* |
| Threonine | 0.23 ± 0.05 | 0.30 ± 0.01 | 0.33 ± 0.01 |
| Tryptophan | 0.10 ± 0.02 | 0.11 ± 0.01 | 0.06 ± 0.01* |
| Tyrosine | 0.19 ± 0.15 | 0.14 ± 0.01 | 0.07 ± 0.01* |
| Valine | 0.27 ± 0.04 | 0.07 ± 0.02 | 0.08 ± 0.01 |
| Total am. acids | 14.13 ± 0.93 | 15.32 ± 1.15 | 10.20 ± 0.89 |
| Others | |||
| Ethanol | 3.34 ± 0.53 | 2.30 ± 0.15 | 1.53 ± 0.06* |
| Methanol | 6.56 ± 0.60 | 5.94 ± 0.35 | 4.74 ± 0.07 |
| Trigonelline | 0.17 ± 0.01 | 0.15 ± 0.01 | 0.11 ± 0.01 |
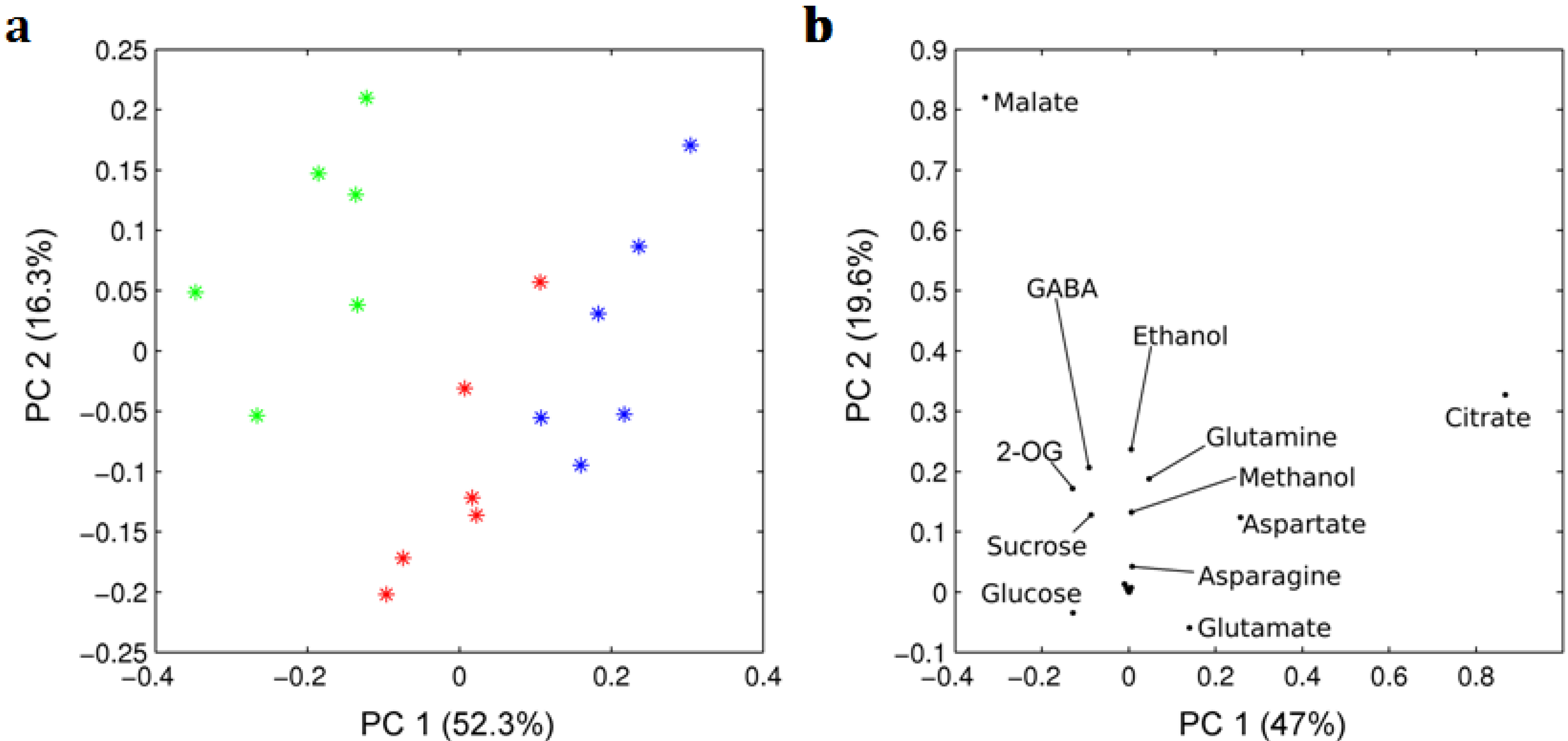
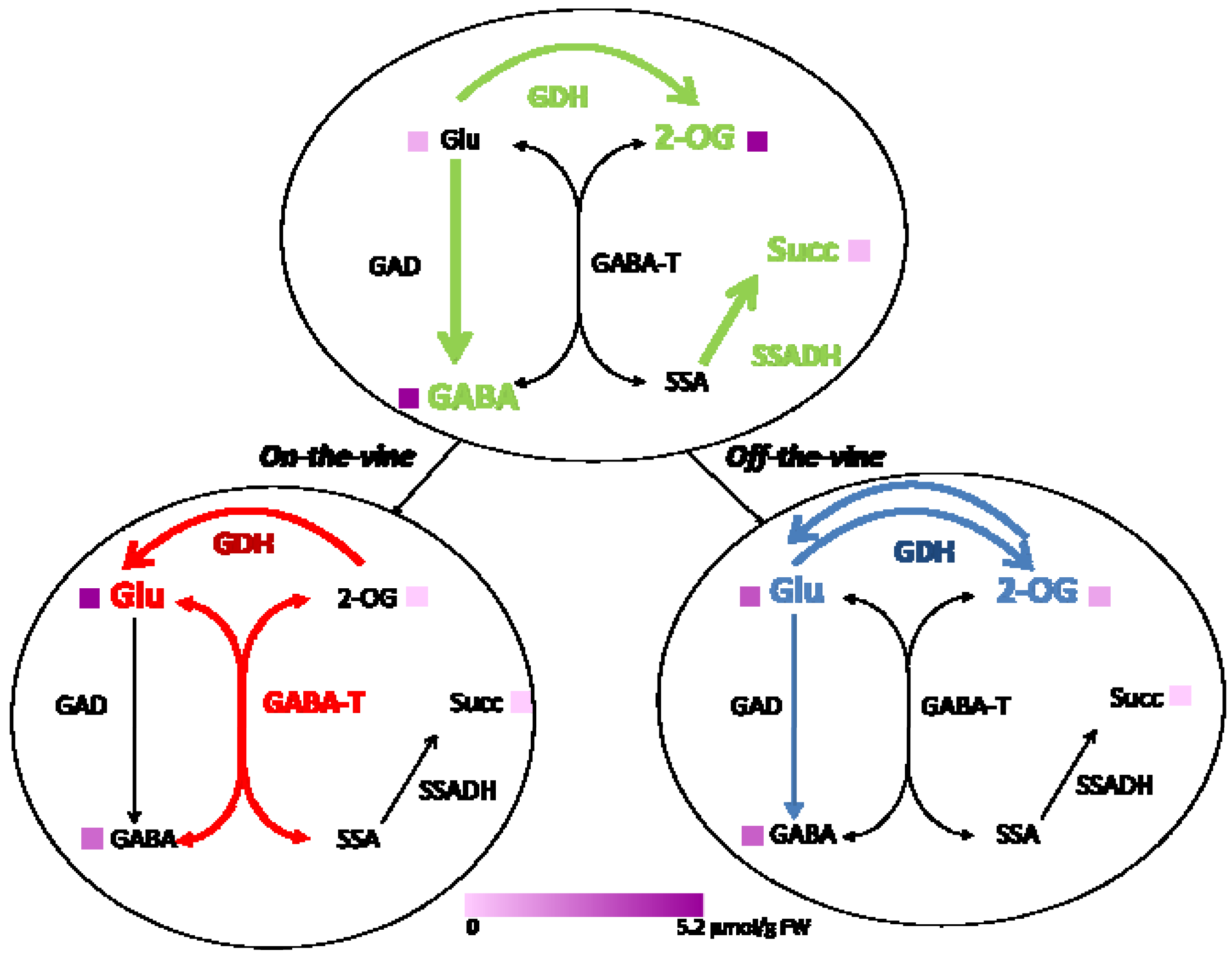
2.2. Enzymes of the GABA Shunt in Ripening Fruit
| Enzyme | Green | Red on-the-vine mU/mg protein | Red off-the-vine |
|---|---|---|---|
| Aminating GDH | 26.33 ± 5.56 | 25.04 ± 3.61 | 30.50 ± 1.71 |
| Deaminating GDH | 89.20 ± 11.71 | 7.22 ± 1.25 | 12.63 ± 2.55 |
| GDH aminating/deaminating | 0.29 ± 0.02 | 3.49 ± 0.10 | 2.49 ± 0.37 |
| GAD | 86.74 ± 15.27 | 5.11 ± 2.60 | 32.28 ± 7.72* |
| GABA-T | 8.30 ± 3.00 | 47.15 ± 1.25 | 13.19 ± 0.28* |
| SSADH | 9.88 ± 0.65 | 1.50 ± 0.47 | 2.25 ± 0.74 |
| Gene | Green | Red on-the-vine | Red off-the-vine |
|---|---|---|---|
| GAD1 | 1.025 ± 0.227 | 0.806 ± 0.253 | 0.087 ± 0.011* |
| GAD2 | 1.012 ± 0.153 | 0.130 ± 0.047 | 0.015 ± 0.008 |
| GAD3 | 1.036 ± 0.270 | 0.240 ± 0.006 | 0.198 ± 0.079 |
| GABA-T1 | 1.124 ± 0.512 | 0.089 ± 0.016 | 0.083 ± 0.010 |
| GABA-T2 | 1.000 ± 0.021 | 0.010 ± 0.004 | 0.004 ± 0.002 |
| GABA-T3 | 1.021 ± 0.206 | 0.680 ± 0.210 | 0.318 ± 0.004* |
| SSADH | 1.001 ± 0.069 | 2.028 ± 0.334 | 2.815 ± 0.596 |
| GDHA1 | 1.026 ± 0.209 | 0.625 ± 0.077 | 0.503 ± 0.103 |
| GADHB | 1.007 ± 0.115 | 0.359 ± 0.085 | 0.388 ± 0.020 |
3. Experimental Section
3.1. Plant Material and Ripening Conditions
3.2. NMR Spectroscopy
3.3. Enzyme Extraction and Analysis
3.4. Total RNA Extraction and Real-Time
3.5. Statistical Methods
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Gapper, N.E.; McQuinn, R.P.; Giovannoni, J.J. Molecular and genetic regulation of fruit ripening. Plant Mol. Biol. 2013, 82, 575–591. [Google Scholar] [CrossRef]
- Gillaspy, G.; Ben-David, H.; Gruissem, W. Fruits: A Developmental Perspective. Plant Cell. 1993, 5, 1439–1451. [Google Scholar]
- Carrari, F.; Fernie, A.R. Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 2006, 57, 1883–1897. [Google Scholar] [CrossRef]
- Bortolotti, S.; Boggio, S.B.; Delgado, L.; Orellano, E.G.; Valle, E.M. Different induction patterns of glutamate metabolising enzymes in ripening fruits of the tomato mutant green flesh. Physiol. Plant. 2003, 119, 384–391. [Google Scholar] [CrossRef]
- Oms-Oliu, G.; Hertog, M.L.A.T.M.; van de Poel, B.; Ampofo-Asiama, J.; Geeraerd, A.H.; Nicolaï, B.M. Metabolic characterization of tomato fruit during preharvest development, ripening, and postharvest shelf-life. Postharvest Biol. Technol. 2011, 62, 7–16. [Google Scholar] [CrossRef]
- Beckles, D.M. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biol. Technol. 2012, 63, 129–140. [Google Scholar] [CrossRef]
- Malacrida, C.; Valle, E.M.; Boggio, S.B. Postharvest chilling induces oxidative stress response in the dwarf tomato cultivar Micro-Tom. Physiol. Plant. 2006, 127, 10–18. [Google Scholar] [CrossRef]
- Scott, J.M.; Harbaugh, B.K. Micro-Tom: a miniature dwarf tomato. Florida Agr. Expt. Sta. Circ. 1989, 371, 1–6. [Google Scholar]
- Sobolev, A.P.; Segre, A.; Lamanna, R. Proton high-field NMR study of tomato juice. Magn. Reson. Chem. 2003, 41, 237–245. [Google Scholar] [CrossRef]
- Mounet, F.; Lemaire-Chamley, M.; Maucourt, M.; Cabasson, C.; Giraudel, J.L.; Deborde, C.; Lessire, R.; Gallusci, P.; Bertrand, A.; Gaudillere, M.; et al. Quantitative metabolic profiles of tomato flesh and seeds during fruit development: complementary analysis with ANN and PCA. Metabolomics 2007, 3, 273–288. [Google Scholar] [CrossRef]
- Deborde, C.; Maucourt, M.; Baldet, P.; Bernillon, S.; Biais, B.; Talon, G.; Ferrand, C.; Jacob, D.; Ferry-Dumazet, H.; de Daruvar, A.; et al. Proton NMR quantitative profiling for quality assessment of greenhouse-grown tomato fruit. Metabolomics 2009, 5, 183–198. [Google Scholar] [CrossRef]
- Le Gall, G.; Colquhoun, I.J.; Davis, A.L.; Collins, G.J.; Verhoeyen, M.E. Metabolite profiling of tomato (Lycopersicon esculentum) using 1H NMR spectroscopy as a tool to detect potential unintended effects following a genetic modification. J. Agric. Food Chem. 2003, 51, 2447–2456. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Sobolev, A.P.; Neelam, A.; Goyal, R.K.; Handa, A.K.; Segre, A.L. Nuclear magnetic resonance spectroscopy-based metabolite profiling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiol. 2006, 142, 1759–1770. [Google Scholar]
- Sorrequieta, A.; Ferraro, G.; Boggio, S.B.; Valle, E.M. Free amino acid production during tomato fruit ripening: a focus on L-glutamate. Amino Acids 2010, 38, 1523–1532. [Google Scholar] [CrossRef]
- Clark, S.M.; Di Leo, R.; van Cauwenberghe, O.R.; Mullen, R.T.; Shelp, B.J. Subcellular localization and expression of multiple tomato gamma-aminobutyrate transaminases that utilize both pyruvate and glyoxylate. J. Exp. Bot. 2009, 60, 3255–3267. [Google Scholar] [CrossRef]
- Fait, A.; Fromm, H.; Walter, D.; Galili, G.; Fernie, A.R. Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci. 2008, 13, 14–19. [Google Scholar]
- Forde, B.G.; Lea, P.J. Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Akihiro, T.; Koike, S.; Tani, R.; Tominaga, T.; Watanabe, S.; Iijima, Y.; Aoki, K.; Shibata, D.; Ashihara, H.; Matsukura, C.; et al. Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol. 2008, 49, 1378–1389. [Google Scholar] [CrossRef]
- Abriata, L.A. Utilization of NMR spectroscopy to study biological fluids and metabolic processes: Two introductory activities. Conc. Magn. Reson. Part. A 2012, 40A, 171–178. [Google Scholar]
- Zanor, M.I.; Lopez, M.; Boggio, S.; Sorrequieta, A.; Pratta, G.; Rodriguez, G.R.; Zorzoli, R.; Picardi, L.A.; Carrari, F.; Valle, E.M. Comparison of 1H-NMR and GC-MS metabolic profile of tomato fruits from a breeding program progeny. Biocell 2010, 34, 140. [Google Scholar]
- Yin, Y.G.; Tominaga, T.; Iijima, Y.; Aoki, K.; Shibata, D.; Ashihara, H.; Nishimura, S.; Ezura, H.; Matsukura, C. Metabolic alterations in organic acids and gamma-aminobutyric acid in developing tomato (Solanum lycopersicum L.) fruits. Plant Cell Physiol. 2010, 51, 1300–1314. [Google Scholar] [CrossRef]
- Steinhauser, M.C.; Steinhauser, D.; Koehl, K.; Carrari, F.; Gibon, Y.; Fernie, A.R.; Stitt, M. Enzyme activity profiles during fruit development in tomato cultivars and Solanum. pennellii. Plant Physiol. 2010, 153, 80–98. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Marro, M.L.; Bortolotti, S.; Boggio, S.B.; Valle, E.M. Plant nutritional status modulates glutamine synthetase levels in ripe tomatoes (Solanum lycopersicum cv. Micro-Tom). Plant Physiol. 2007, 164, 137–145. [Google Scholar] [CrossRef]
- Breitkreuz, K.E.; Shelp, B.J. Subcellular compartmentation of the 4-aminobutyrate shunt in protoplasts from developing soybean cotyledons. Plant Physiol. 1995, 108, 99–103. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ferraro, G.; Bortolotti, S.; Mortera, P.; Schlereth, A.; Stitt, M.; Carrari, F.; Kamenetzky, L.; Valle, E.M. Novel glutamate dehydrogenase genes show increased transcript and protein abundances in mature tomato fruits. Plant Physiol. 2012, 169, 899–907. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Neily, M.H.; Matsukuraa, C.; Maucourt, M.; Bernillon, S.; Deborde, C.; Moing, A.; Yin, Y.; Saito, T.; Mori, K.; Asamizu, E.; et al. Enhanced polyamine accumulation alters carotenoid metabolism at the transcriptional level in tomato fruit over-expressing spermidine synthase. J. Plant Physiol. 2011, 168, 242–252. [Google Scholar] [CrossRef] [Green Version]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sorrequieta, A.; Abriata, L.A.; Boggio, S.B.; Valle, E.M. Off-the-Vine Ripening of Tomato Fruit Causes Alteration in the Primary Metabolite Composition. Metabolites 2013, 3, 967-978. https://doi.org/10.3390/metabo3040967
Sorrequieta A, Abriata LA, Boggio SB, Valle EM. Off-the-Vine Ripening of Tomato Fruit Causes Alteration in the Primary Metabolite Composition. Metabolites. 2013; 3(4):967-978. https://doi.org/10.3390/metabo3040967
Chicago/Turabian StyleSorrequieta, Augusto, Luciano A. Abriata, Silvana B. Boggio, and Estela M. Valle. 2013. "Off-the-Vine Ripening of Tomato Fruit Causes Alteration in the Primary Metabolite Composition" Metabolites 3, no. 4: 967-978. https://doi.org/10.3390/metabo3040967
APA StyleSorrequieta, A., Abriata, L. A., Boggio, S. B., & Valle, E. M. (2013). Off-the-Vine Ripening of Tomato Fruit Causes Alteration in the Primary Metabolite Composition. Metabolites, 3(4), 967-978. https://doi.org/10.3390/metabo3040967







