An NMR-Based Model to Investigate the Metabolic Phenoreversion of COVID-19 Patients throughout a Longitudinal Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Recruitment and Sample Collection
2.2. Sample Preparation, Instrumentation, and Sample Acquisition with IVDr NMR Metabolomics
2.3. NMR Quantifications
2.4. Cytokine and Chemokine Quantification
2.5. Filtering of Samples
2.6. Nomenclature for Days since COVID
2.7. Statistical Analysis
2.8. COVID-19 Model
2.9. Estimation of Recovery Days
3. Results
3.1. Cohorts under Consideration
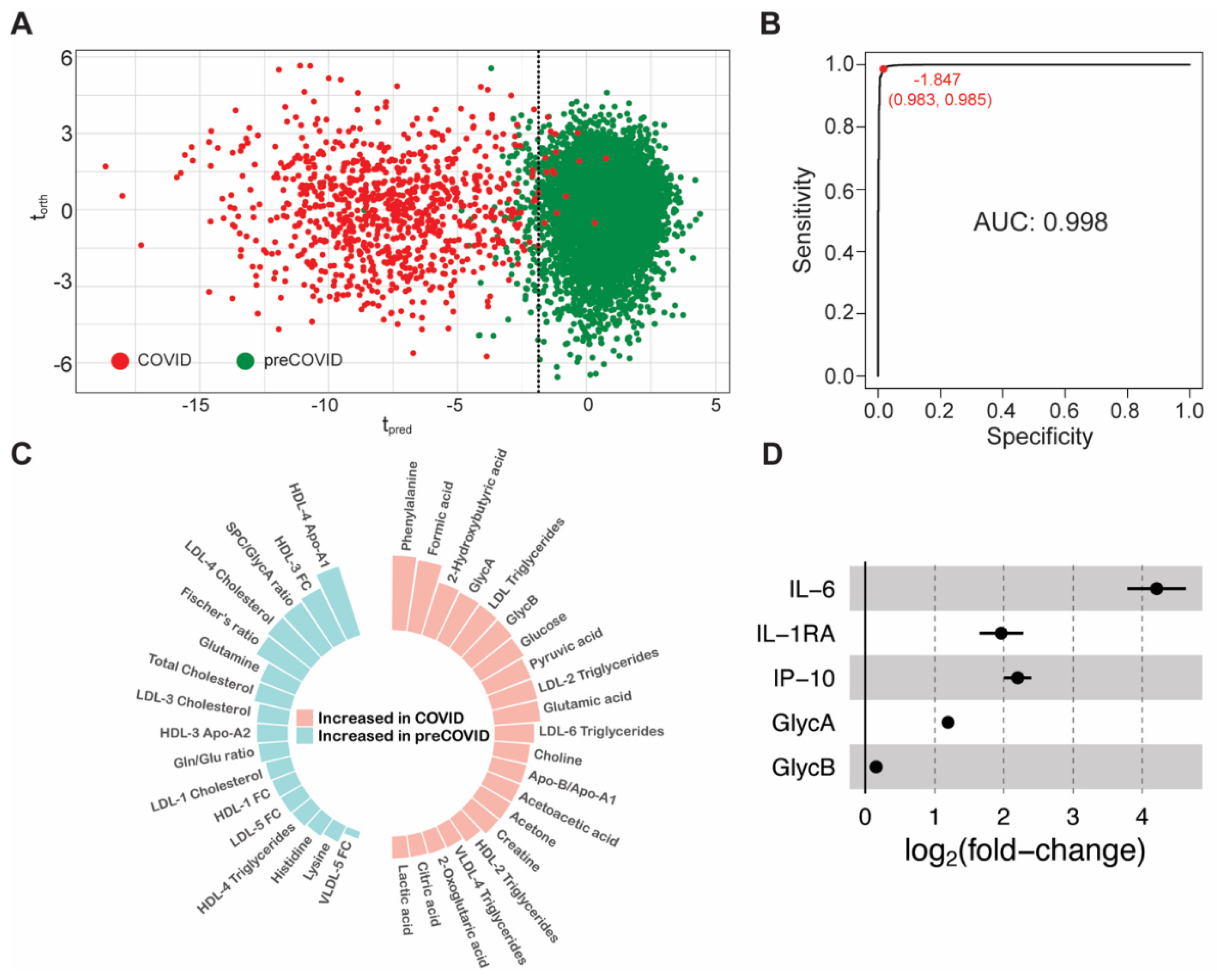
3.2. Early Metabolic Alterations in Acute and Mild COVID-19 Patients
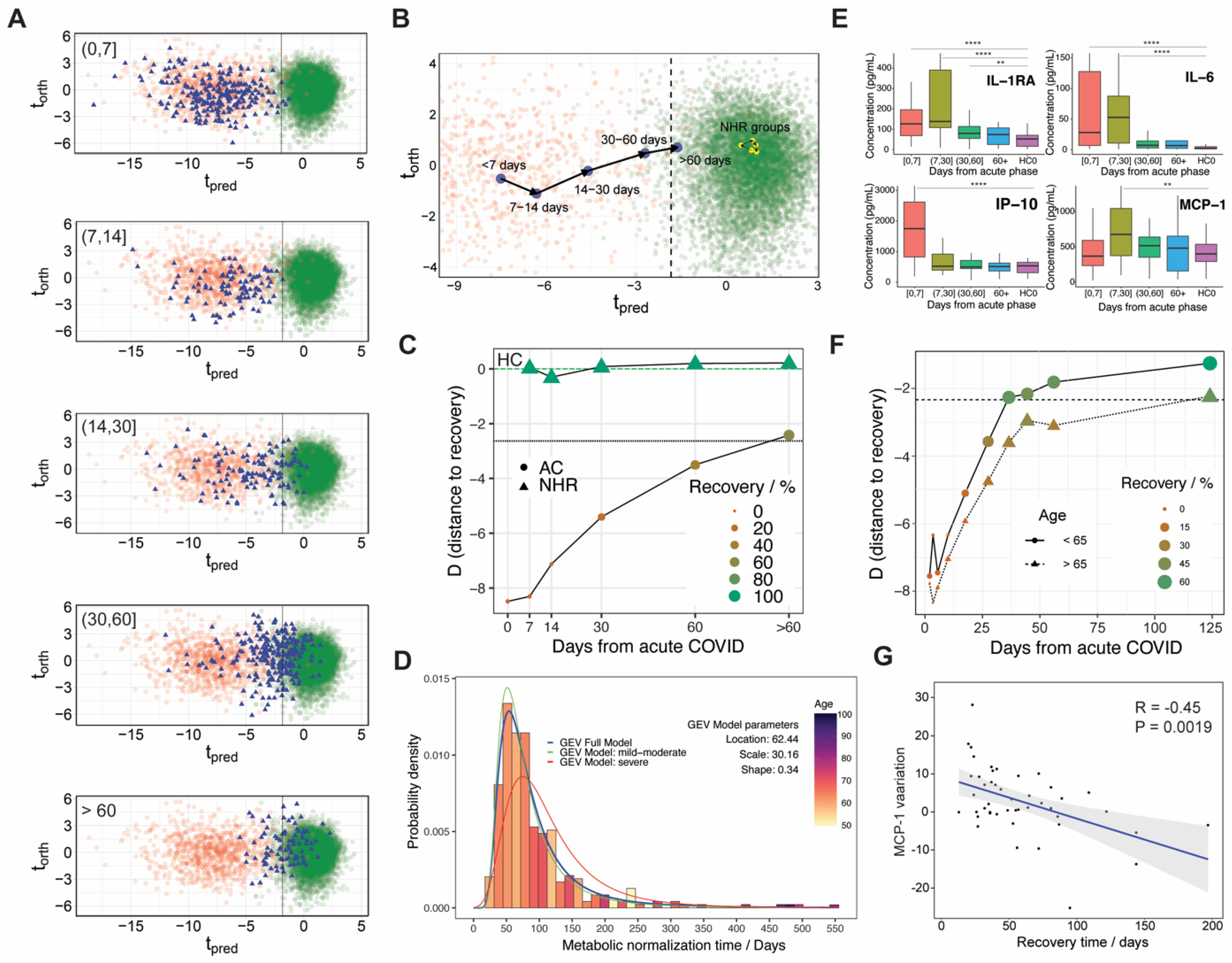
3.3. Metabolic Phenoreversion over Time for Hospitalized and Non-Hospitalized Recovered Patients
3.4. On the Lineage-Specific Metabolic Response of SARS-CoV-2
3.5. Potential Confounding Factors and Limitations of the Study
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marín-Corral, J.; Rodríguez-Morató, J.; Gomez-Gomez, A.; Pascual-Guardia, S.; Muñoz-Bermúdez, R.; Salazar-Degracia, A.; Pérez-Terán, P.; Restrepo, M.I.; Khymenets, O.; Haro, N.; et al. Metabolic Signatures Associated with Severity in Hospitalized COVID-19 Patients. Int. J. Mol. Sci. 2021, 22, 4794. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; García de Vicuña, A.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.D.; Slessarev, M.; Martin, C.M.; Daley, M.; Patel, M.A.; Miller, M.R.; Patterson, E.K.; O’Gorman, D.B.; Gill, S.E.; Wishart, D.S.; et al. Metabolomics Profiling of Critically Ill Coronavirus Disease 2019 Patients: Identification of Diagnostic and Prognostic Biomarkers. Crit. Care Explor. 2020, 2, e0272. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, F.; Föh, B.; Mallagaray, A.; Rahmöller, J.; Ehlers, M.; Lehrian, S.; von Kopylow, V.; Künsting, I.; Lixenfeld, A.S.; Martin, E.; et al. Metabolic and Lipidomic Markers Differentiate COVID-19 from Non-Hospitalized and Other Intensive Care Patients. Front. Mol. Biosci. 2021, 8, 1091. [Google Scholar] [CrossRef] [PubMed]
- Kimhofer, T.; Lodge, S.; Whiley, L.; Gray, N.; Loo, R.L.; Lawler, N.G.; Nitschke, P.; Bong, S.-H.; Morrison, D.L.; Begum, S.; et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19, 4442–4454. [Google Scholar] [CrossRef]
- San Juan, I.; Bruzzone, C.; Bizkarguenaga, M.; Bernardo-Seisdedos, G.; Laín, A.; Gil-Redondo, R.; Diercks, T.; Gil-Martínez, J.; Urquiza, P.; Arana, E.; et al. Abnormal Concentration of Porphyrins in Serum from COVID-19 Patients. Br. J. Haematol. 2020, 190, e265. [Google Scholar] [CrossRef]
- Caterino, M.; Costanzo, M.; Fedele, R.; Cevenini, A.; Gelzo, M.; Di Minno, A.; Andolfo, I.; Capasso, M.; Russo, R.; Annunziata, A.; et al. The Serum Metabolome of Moderate and Severe COVID-19 Patients Reflects Possible Liver Alterations Involving Carbon and Nitrogen Metabolism. Int. J. Mol. Sci. 2021, 22, 9548. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Rocha, J.; Gonzalez-Colin, C.; Bhattacharyya, S.; Madrigal, A.; Ottensmeier, C.H.; Ay, F.; Chandra, V.; Vijayanand, P. COVID-19 Genetic Risk Variants Are Associated with Expression of Multiple Genes in Diverse Immune Cell Types. Nat. Commun. 2021, 12, 6760. [Google Scholar] [CrossRef]
- Hasan, M.; Murtaz, S.B.; Islam, M.U.; Sadeq, M.J.; Uddin, J. Robust and Efficient COVID-19 Detection Techniques: A Machine Learning Approach. PLoS ONE 2022, 17, e0274538. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef]
- Holmes, E.; Wist, J.; Masuda, R.; Lodge, S.; Nitschke, P.; Kimhofer, T.; Loo, R.L.; Begum, S.; Boughton, B.; Yang, R.; et al. Incomplete Systemic Recovery and Metabolic Phenoreversion in Post-Acute-Phase Nonhospitalized COVID-19 Patients: Implications for Assessment of Post-Acute COVID-19 Syndrome. J. Proteome Res. 2021, 20, 3315–3329. [Google Scholar] [CrossRef] [PubMed]
- Bizkarguenaga, M.; Bruzzone, C.; Gil-Redondo, R.; SanJuan, I.; Martin-Ruiz, I.; Barriales, D.; Palacios, A.; Pasco, S.T.; González-Valle, B.; Laín, A.; et al. Uneven Metabolic and Lipidomic Profiles in Recovered COVID-19 Patients as Investigated by Plasma NMR Metabolomics. NMR Biomed. 2022, 35, e4637. [Google Scholar] [CrossRef] [PubMed]
- Ghini, V.; Meoni, G.; Pelagatti, L.; Celli, T.; Veneziani, F.; Petrucci, F.; Vannucchi, V.; Bertini, L.; Luchinat, C.; Landini, G.; et al. Profiling Metabolites and Lipoproteins in COMETA, an Italian Cohort of COVID-19 Patients. PLoS Pathog. 2022, 18, e1010443. [Google Scholar] [CrossRef] [PubMed]
- Sorg, A.-L.; Bergfeld, L.; Jank, M.; Corman, V.; Semmler, I.; Goertz, A.; Beyerlein, A.; Verjans, E.; Wagner, N.; Von Bernuth, H.; et al. Cross-Sectional Seroprevalence Surveys of SARS-CoV-2 Antibodies in Children in Germany, June 2020 to May 2021. Nat. Commun. 2022, 13, 3128. [Google Scholar] [CrossRef]
- Hasan, M.R.; Suleiman, M.; Pérez-López, A. Metabolomics in the Diagnosis and Prognosis of COVID-19. Front. Genet. 2021, 12, 1358. [Google Scholar] [CrossRef]
- Amberg, A.; Riefke, B.; Schlotterbeck, G.; Ross, A.; Senn, H.; Dieterle, F.; Keck, M. NMR and MS Methods for Metabolomics. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1641. [Google Scholar]
- Costa Dos Santos Junior, G.; Pereira, C.M.; Kelly Da Silva Fidalgo, T.; Valente, A.P. Saliva NMR-Based Metabolomics in the War against COVID-19. Anal. Chem. 2020, 92, 15688–15692. [Google Scholar] [CrossRef]
- Bruzzone, C.; Gil-Redondo, R.; Seco, M.; Barragán, R.; de la Cruz, L.; Cannet, C.; Schäfer, H.; Fang, F.; Diercks, T.; Bizkarguenaga, M.; et al. A Molecular Signature for the Metabolic Syndrome by Urine Metabolomics. Cardiovasc. Diabetol. 2021, 20, 155. [Google Scholar] [CrossRef]
- Loo, R.L.; Lodge, S.; Kimhofer, T.; Bong, S.H.; Begum, S.; Whiley, L.; Gray, N.; Lindon, J.C.; Nitschke, P.; Lawler, N.G.; et al. Quantitative In-Vitro Diagnostic NMR Spectroscopy for Lipoprotein and Metabolite Measurements in Plasma and Serum: Recommendations for Analytical Artifact Minimization with Special Reference to COVID-19/SARS-CoV-2 Samples. J. Proteome Res. 2020, 19, 4428–4441. [Google Scholar] [CrossRef]
- Nitschke, P.; Lodge, S.; Kimhofer, T.; Masuda, R.; Bong, S.-H.; Hall, D.; Schäfer, H.; Spraul, M.; Pompe, N.; Diercks, T.; et al. J-Edited DIffusional Proton Nuclear Magnetic Resonance Spectroscopic Measurement of Glycoprotein and Supramolecular Phospholipid Biomarkers of Inflammation in Human Serum. Anal. Chem. 2022, 94, 1333–1341. [Google Scholar] [CrossRef]
- Masuda, R.; Lodge, S.; Nitschke, P.; Spraul, M.; Schaefer, H.; Bong, S.H.; Kimhofer, T.; Hall, D.; Loo, R.L.; Bizkarguenaga, M.; et al. Integrative Modeling of Plasma Metabolic and Lipoprotein Biomarkers of SARS-CoV-2 Infection in Spanish and Australian COVID-19 Patient Cohorts. J. Proteome Res. 2021, 20, 4139–4152. [Google Scholar] [CrossRef]
- He, B.; Wang, J.; Wang, Y.; Zhao, J.; Huang, J.; Tian, Y.; Yang, C.; Zhang, H.; Zhang, M.; Gu, L.; et al. The Metabolic Changes and Immune Profiles in Patients with COVID-19. Front. Immunol. 2020, 11, 2075. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 Are Disease Severity Predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal Characteristics of Lymphocyte Responses and Cytokine Profiles in the Peripheral Blood of SARS-CoV-2 Infected Patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef] [PubMed]
- Mudd, P.A.; Crawford, J.C.; Turner, J.S.; Souquette, A.; Reynolds, D.; Bender, D.; Bosanquet, J.P.; Anand, N.J.; Striker, D.A.; Martin, R.S.; et al. Distinct Inflammatory Profiles Distinguish COVID-19 from Influenza with Limited Contributions from Cytokine Storm. Sci. Adv. 2020, 6, eabe3024. [Google Scholar] [CrossRef]
- Gray, N.; Lawler, N.G.; Zeng, A.X.; Ryan, M.; Bong, S.H.; Boughton, B.A.; Bizkarguenaga, M.; Bruzzone, C.; Embade, N.; Wist, J.; et al. Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute Sars-Cov-2 Infection. Metabolites 2021, 11, 467. [Google Scholar] [CrossRef]
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/Lipidomic Profiling of COVID-19 and Individual Response to Tocilizumab. PLoS Pathog. 2021, 17, e1009243. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-Term Cardiovascular Outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef]
- Gorog, D.A.; Storey, R.F.; Gurbel, P.A.; Tantry, U.S.; Berger, J.S.; Chan, M.Y.; Duerschmied, D.; Smyth, S.S.; Parker, W.A.E.; Ajjan, R.A.; et al. Current and Novel Biomarkers of Thrombotic Risk in COVID-19: A Consensus Statement from the International COVID-19 Thrombosis Biomarkers Colloquium. Nat. Rev. Cardiol. 2022, 19, 475–495. [Google Scholar] [CrossRef]
- Zhao, Y.; Qin, L.; Zhang, P.; Li, K.; Liang, L.; Sun, J.; Xu, B.; Dai, Y.; Li, X.; Zhang, C.; et al. Longitudinal COVID-19 Profiling Associates IL-1RA and IL-10 with Disease Severity and RANTES with Mild Disease. JCI Insight 2020, 5, e139834. [Google Scholar] [CrossRef]
- Georgakis, M.K.; Van Der Laan, S.W.; Asare, Y.; Mekke, J.M.; Haitjema, S.; Schoneveld, A.H.; De Jager, S.C.A.; Nurmohamed, N.S.; Kroon, J.; Stroes, E.S.G.; et al. Monocyte-Chemoattractant Protein-1 Levels in Human Atherosclerotic Lesions Associate With Plaque Vulnerability. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2038–2048. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Alshammary, H.; Amoako, A.A.; Awawda, M.H.; Beach, K.F.; Bermúdez-González, M.C.; Chernet, R.L.; et al. SARS-CoV-2 Spike E484K Mutation Reduces Antibody Neutralisation. Lancet Microbe 2021, 2, e283–e284. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant. Cell 2021, 184, 3426–3437.e8. [Google Scholar] [CrossRef] [PubMed]
- Begue, F.; Tanaka, S.; Mouktadi, Z.; Rondeau, P.; Veeren, B.; Diotel, N.; Tran-Dinh, A.; Robert, T.; Vélia, E.; Mavingui, P.; et al. Altered High-Density Lipoprotein Composition and Functions during Severe COVID-19. Sci. Rep. 2021, 11, 2291. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K. Molecular Phenomic Approaches to Deconvolving the Systemic Effects of SARS-CoV-2 Infection and Post-Acute COVID-19 Syndrome. Phenomics 2021, 1, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Zhu, H.; Witteles, R.M.; Wu, J.C.; Quertermous, T.; Wu, S.M.; Rhee, J.W. Cardiovascular Risks in Patients with COVID-19: Potential Mechanisms and Areas of Uncertainty. Curr. Cardiol. Rep. 2020, 22, 34. [Google Scholar] [CrossRef]
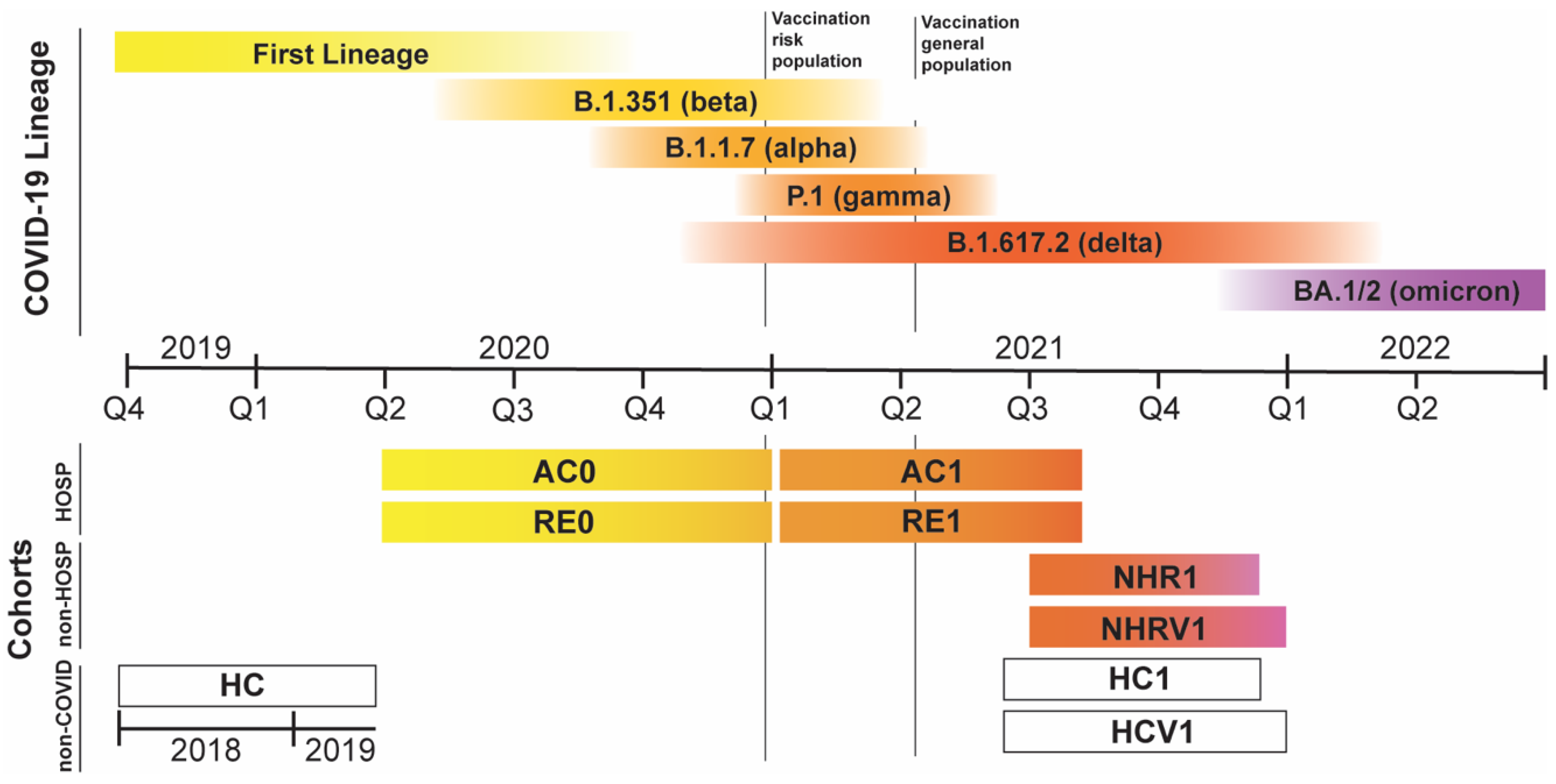

| Donor Type | Cohort | Days from Disease Onset | N | Recollection Time | Expected COVID-19 Variants | Vaccinated |
|---|---|---|---|---|---|---|
| Hospitalized COVID-19 patients | AC0 | 0 | 697 | Apr–Dec 2020 | FL, α, β | No |
| RE0 | (0,7], (7–14], (14–30], (30,60], >60, TOT | 137, 96, 104, 97, 10, 444 | FL, α, β | No | ||
| AC1 | 0 | 189 | Jan–Oct 2021 | γ, δ | Mixed | |
| RE1 | (0,7], (7–14], (14–30], (30,60], >60, TOT | 100,1,15, 158, 79, 353 | γ, δ | Mixed | ||
| General population | HC | no COVID | 8664 | Before 2020 | none | No |
| NHR1 | (7–14], (30,60], >60, TOT | 1, 16, 78, 95 | Jun–Nov 2021 | δ, ο | No | |
| NHRV1 | (0,7], (7–14], (14–30], (30,60], >60, TOT | 1, 2, 8, 27, 380, 418 | Jun–Dec 2021 | δ, ο | Yes | |
| HC1 | no COVID | 238 | May–Nov 2021 | none | No | |
| HCV1 | no COVID | 2322 | May–Dec 2021 | none | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil-Redondo, R.; Conde, R.; Bizkarguenaga, M.; Bruzzone, C.; Laín, A.; González-Valle, B.; Iriberri, M.; Ramos-Acosta, C.; Anguita, E.; Arriaga Lariz, J.I.; et al. An NMR-Based Model to Investigate the Metabolic Phenoreversion of COVID-19 Patients throughout a Longitudinal Study. Metabolites 2022, 12, 1206. https://doi.org/10.3390/metabo12121206
Gil-Redondo R, Conde R, Bizkarguenaga M, Bruzzone C, Laín A, González-Valle B, Iriberri M, Ramos-Acosta C, Anguita E, Arriaga Lariz JI, et al. An NMR-Based Model to Investigate the Metabolic Phenoreversion of COVID-19 Patients throughout a Longitudinal Study. Metabolites. 2022; 12(12):1206. https://doi.org/10.3390/metabo12121206
Chicago/Turabian StyleGil-Redondo, Rubén, Ricardo Conde, Maider Bizkarguenaga, Chiara Bruzzone, Ana Laín, Beatriz González-Valle, Milagros Iriberri, Carlos Ramos-Acosta, Eduardo Anguita, Juan Ignacio Arriaga Lariz, and et al. 2022. "An NMR-Based Model to Investigate the Metabolic Phenoreversion of COVID-19 Patients throughout a Longitudinal Study" Metabolites 12, no. 12: 1206. https://doi.org/10.3390/metabo12121206






