Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and RNA Preparation
2.2. Electrophoretic Mobility Shift Assay (EMSA)
2.3. Ribozyme Activity Assay
3. Results and Discussion
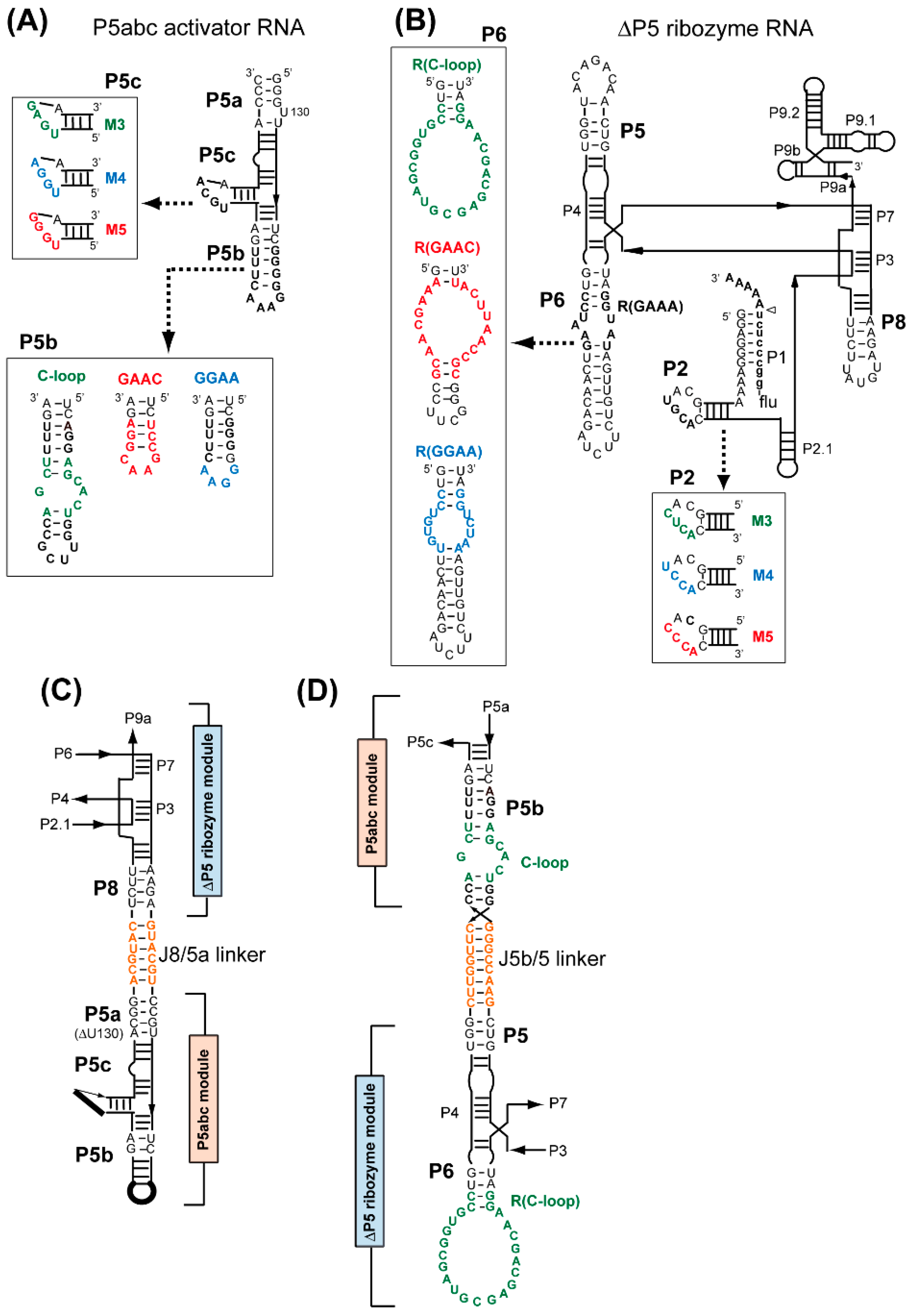
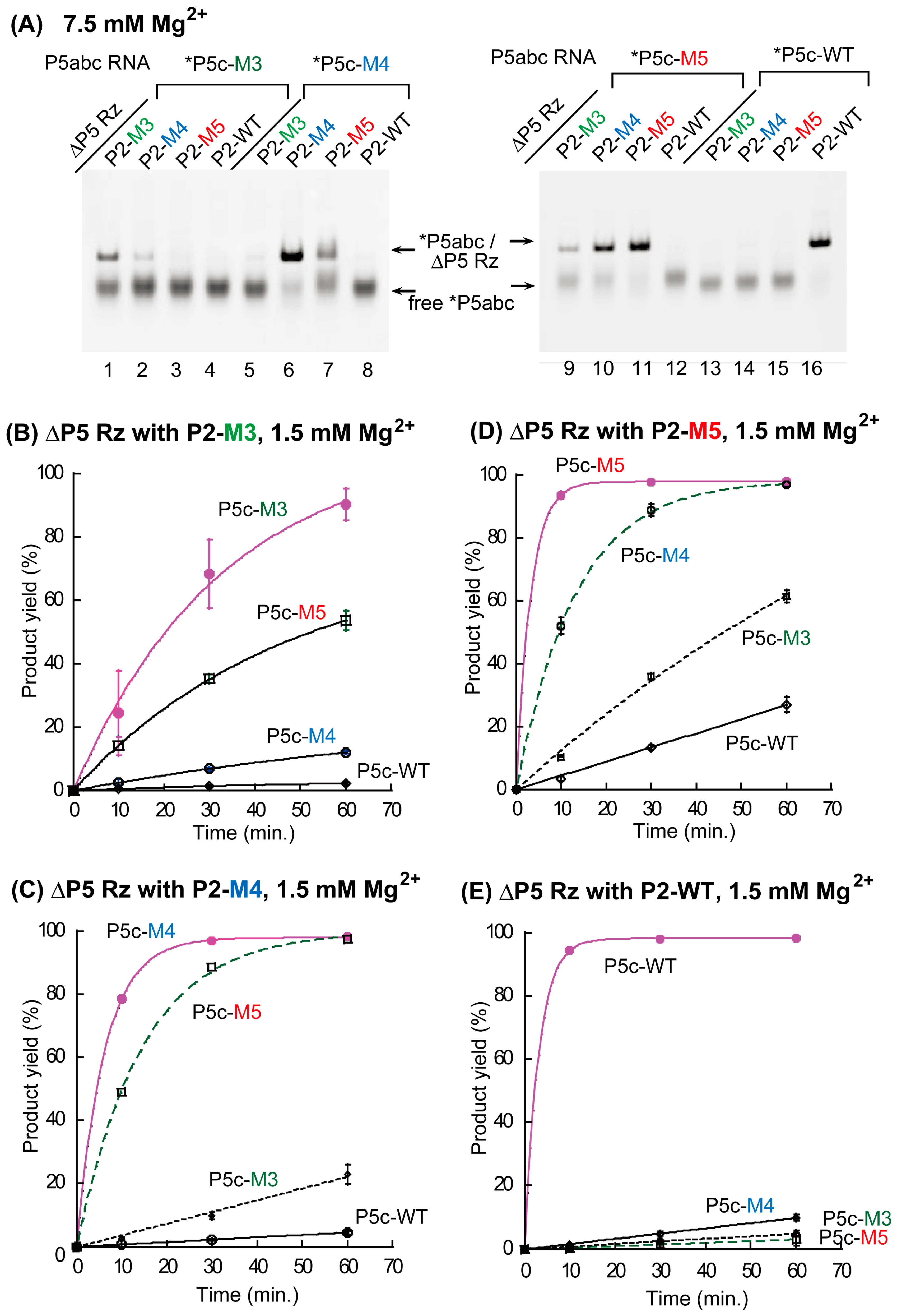
3.1. Assembly of Bimolecular Group I Ribozymes through Artificial RNA–RNA Interacting Motifs
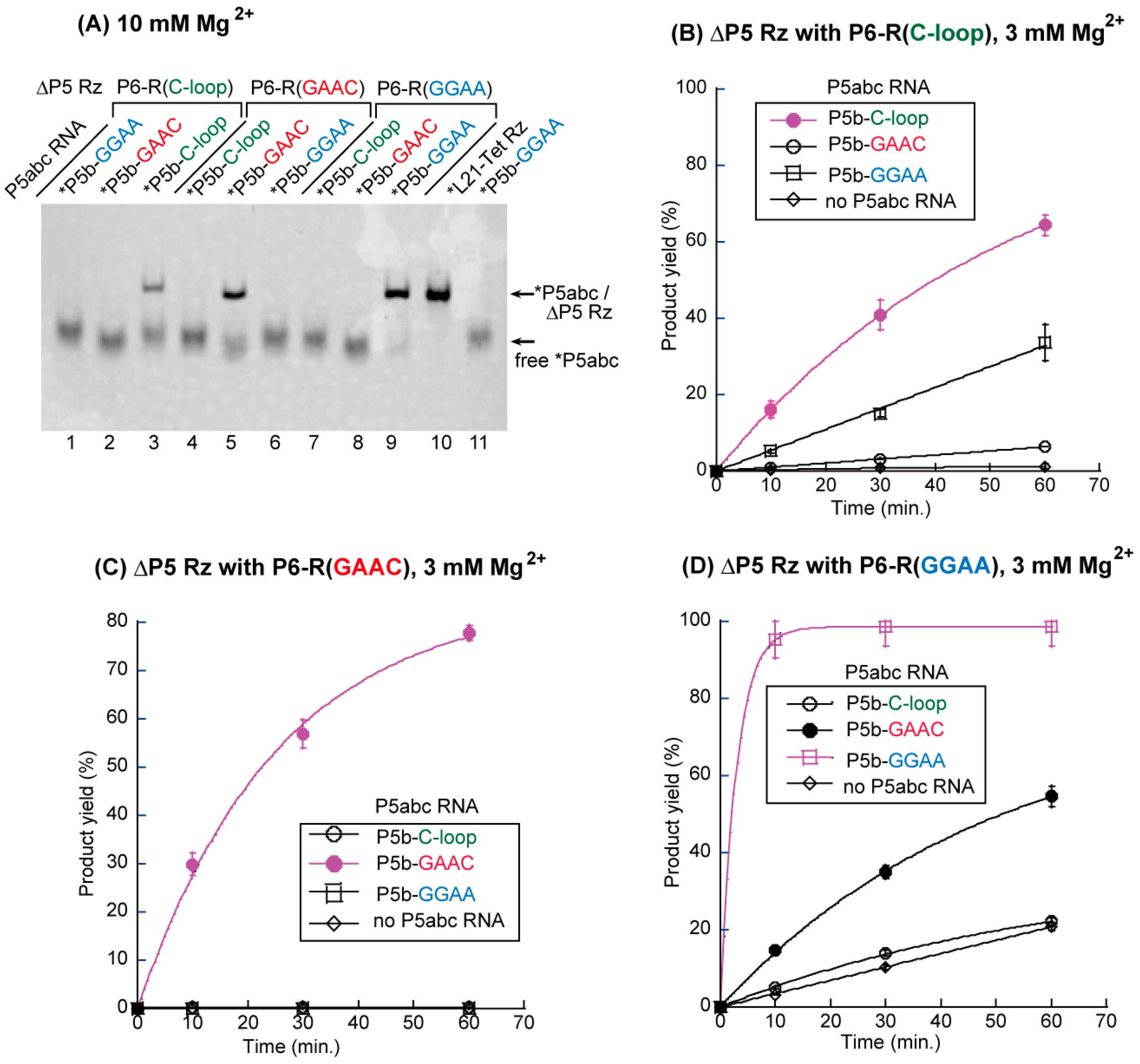
3.2. Artificial Kissing Loop Interactions to Assemble Bimolecular Group I Ribozymes
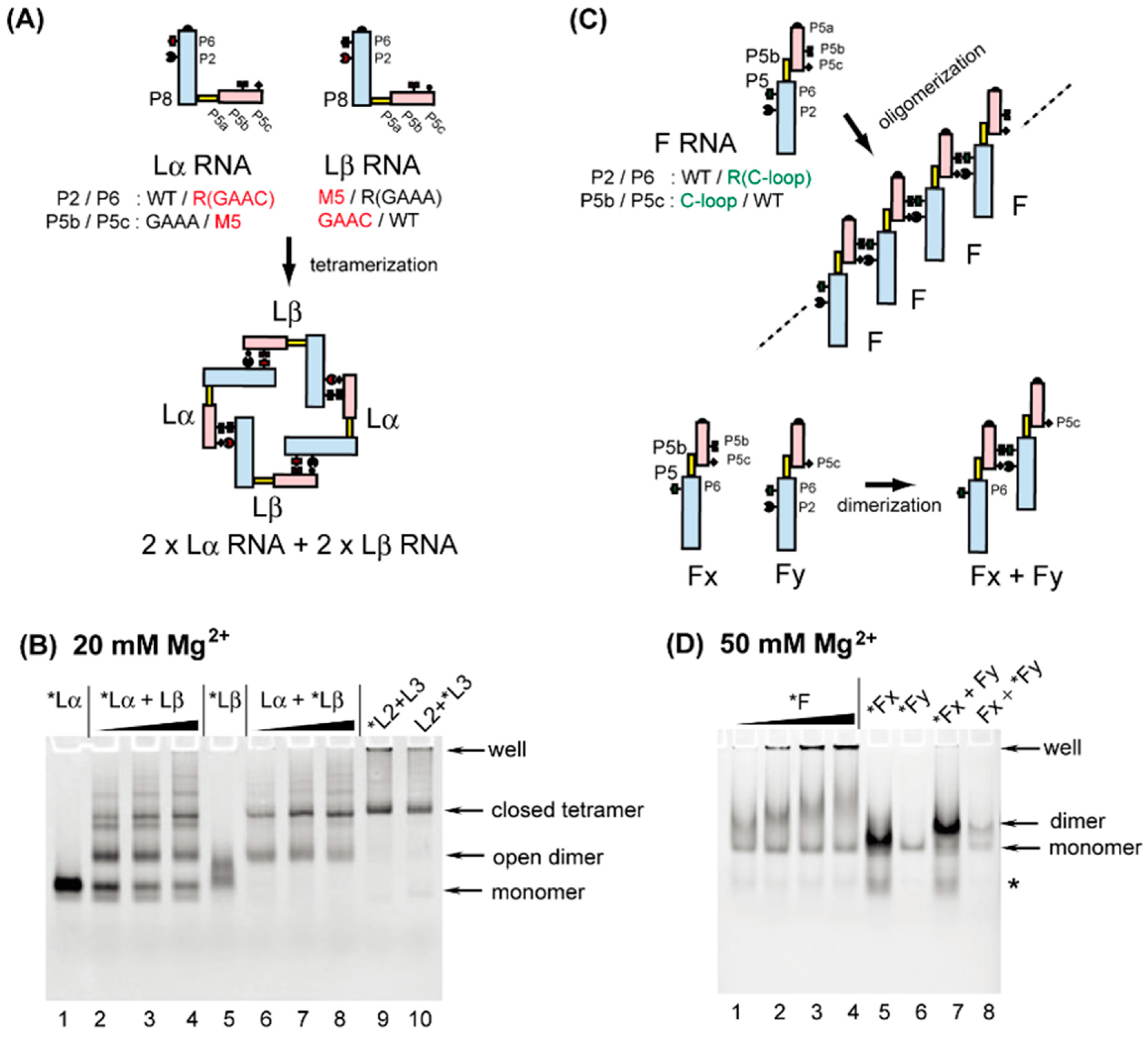
3.3. Formation of RNA Squares through Selective Assembly of Engineered Group I Ribozymes
3.4. Formation of Novel RNA 1D Array through Selective Oligomerization
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Simmel, S.S.; Nickels, P.C.; Liedl, T. Wireframe and tensegrity DNA nanostructures. Acc. Chem. Res. 2014, 47, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, L.; Li, J.; Fan, C. Precisely tailored DNA nanostructures and their theranostic applications. Chem. Rec. 2017. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Jaeger, L. RNA self-assembly and RNA nanotechnology. Acc. Chem. Res. 2014, 47, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for creating higher order structures. Chem. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Takeuchi, Y.; Emura, T.; Hidaka, K.; Sugiyama, H. Preparation of chemically modified RNA origami nanostructures. Chem. Eur. J. 2014, 20, 15330–15333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geary, C.; Rothemund, P.W.; Andersen, E.S. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 2014, 345, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Sparvath, S.L.; Geary, C.W.; Andersen, E.S. Computer-aided design of RNA origami structures. Methods Mol. Biol. 2017, 1500, 51–80. [Google Scholar] [PubMed]
- Kozyra, J.; Ceccarelli, A.; Torelli, E.; Lopiccolo, A.; Gu, J.Y.; Fellermann, H.; Stimming, U.; Krasnogor, N. Designing uniquely addressable bio-orthogonal synthetic scaffolds for DNA and RNA origami. ACS Synth. Biol. 2017, 6, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Chworos, A.; Severcan, I.; Koyfman, A.Y.; Weinkam, P.; Oroudjev, E.; Hansma, H.G.; Jaeger, L. Building programmable jigsaw puzzles with RNA. Science 2004, 306, 2068–2072. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Zakrevsky, P.; Afonin, K.A.; Chworos, A.; Shapiro, B.A.; Jaeger, L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011, 11, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Bui, M.N.; Brittany Johnson, M.; Viard, M.; Satterwhite, E.; Martins, A.N.; Li, Z.; Marriott, I.; Afonin, K.A.; Khisamutdinov, E.F. Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine 2017, 13, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.B.; Halman, J.R.; Satterwhite, E.; Zakharov, A.V.; Bui, M.N.; Benkato, K.; Goldsworthy, V.; Kim, T.; Hong, E.; Dobrovolskaia, M.A.; et al. Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling. Small 2017. [Google Scholar] [CrossRef] [PubMed]
- Delebecque, C.J.; Lindner, A.B.; Silver, P.A.; Aldaye, F.A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011, 333, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Oi, H.; Fujita, D.; Suzuki, Y.; Sugiyama, H.; Endo, M.; Matsumura, S.; Ikawa, Y. Programmable formation of catalytic RNA triangles and squares by assembling modular RNA enzymes. J. Biochem. 2017, 161, 451–462. [Google Scholar] [PubMed]
- Van der Horst, G.; Christian, A.; Inoue, T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. USA. 1991, 88, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.A.; Herschlag, D.; Doudna, J.A. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry 1999, 38, 2982–2990. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Furuta, H.; Ikawa, Y. Installation of orthogonality to the interface that assembles two modular domains in the Tetrahymena group I ribozyme. J. Biosci. Bioeng. 2014, 117, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.; Baudrey, S.; Jaeger, L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008, 36, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Fujita, Y.; Maeda, Y.; Furuta, H.; Ikawa, Y. GNRA/receptor interacting modules: Versatile modular units for natural and artificial RNA architectures. Methods 2011, 54, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Shiraishi, H.; Inoue, T. P5abc of the Tetrahymena ribozyme consists of three functionally independent elements. RNA 1998, 4, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, Y.; Moriyama, S.; Furuta, H. Facile syntheses of BODIPY derivatives for fluorescent labeling of the 3′ and 5′ ends of RNAs. Anal. Biochem. 2008, 378, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, M.A.; Doherty, E.A.; Knitt, D.S.; Doudna, J.A.; Herschlag, D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry 2000, 39, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S.P.; Ikawa, Y.; Nakamura, Y. Selection of a novel class of RNA-RNA interaction motifs based on the ligase ribozyme with defined modular architecture. Nucleic Acids Res. 2008, 36, 3600–3607. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Matsumura, S.; Jaeger, L.; Inoue, T.; Furuta, H.; Ikawa, Y. Rational optimization of the DSL ligase ribozyme with GNRA/receptor interacting modules. Arch. Biochem. Biophys. 2009, 490, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Furuta, H.; Ikawa, Y. An in vitro-selected RNA receptor for the GAAC loop: Modular receptor for non-GNRA-type tetraloop. Nucleic Acids Res. 2013, 41, 3748–3759. [Google Scholar] [CrossRef] [PubMed]
- Serra, M.J.; Baird, J.D.; Dale, T.; Fey, B.L.; Retatagos, K.; Westhof, E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA 2002, 8, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.J.; Chen, S.J. Importance of diffuse metal ion binding to RNA. Met. Ions Life Sci. 2011, 9, 101–124. [Google Scholar] [PubMed]
- Lehnert, V.; Jaeger, L.; Michel, F.; Westhof, E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: A complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996, 3, 993–1009. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hassan, B.H.; Mirzoyan, M.G.; Leontis, N.B. Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res. 2011, 39, 2903–2917. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.; Westhof, E.; Leontis, N.B. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001, 29, 455–463. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.M.; Matsumura, S.; Ikawa, Y. Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design. Biology 2017, 6, 37. https://doi.org/10.3390/biology6040037
Rahman MM, Matsumura S, Ikawa Y. Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design. Biology. 2017; 6(4):37. https://doi.org/10.3390/biology6040037
Chicago/Turabian StyleRahman, Md. Motiar, Shigeyoshi Matsumura, and Yoshiya Ikawa. 2017. "Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design" Biology 6, no. 4: 37. https://doi.org/10.3390/biology6040037





