A Molecular View of Kinetochore Assembly and Function
Abstract
:1. An Overview of Kinetochore Structure and Functions
2. The Centromere
3. The Inner Kinetochore
3.1. The CCAN
3.2. Structural Organization of the CENP-A Nucleosome
3.3. Recognition of CENP-A by CCAN Subunits
3.4. The CENP-TWSX Complex
3.5. The CENP-OPQRU Complex
3.6. CENP-B
3.7. Summary
4. The Outer Kinetochore
4.1. The Ndc80 Complex
4.2. The Mis12 and Knl1 Complexes
4.3. Complexity of the Kinetochore Microtubule Interfaces
5. Linkages between the Inner and the Outer Kinetochore
5.1. Two Mechanisms Link Inner and Outer Kinetochores
5.2. Stoichiometry of Kinetochore Subunits
5.3. Temporal Framework of Kinetochore Assembly and Disassembly
6. Organization of the Chromatin Foundation of the Kinetochore in Regional Centromeres
7. The Propagation of Centromeric Chromatin
8. Unconventional Kinetochores
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cheeseman, I.M. The kinetochore. Cold Spring Harb. Perspect. Biol. 2014, 6, a015826. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, H.; Fukagawa, T. Kinetochore assembly and function through the cell cycle. Chromosoma 2016, 125, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Pesenti, M.E.; Weir, J.R.; Musacchio, A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 2016, 37, 152–163. [Google Scholar] [CrossRef] [PubMed]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Almouzni, G. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochim. Biophys. Acta 2014, 1839, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Earnshaw, W.C. The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 2014, 30, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982, 79, 1–58. [Google Scholar] [PubMed]
- Magidson, V.; Paul, R.; Yang, N.; Ault, J.G.; O’Connell, C.B.; Tikhonenko, I.; McEwen, B.F.; Mogilner, A.; Khodjakov, A. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 2015, 17, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B.; Koch, C.A. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 1969, 43, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.S.; Takagaki, K.; Kumada, K.; Hirayama, Y.; Noda, T.; Hirota, T. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009, 184, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Maresca, T.J.; Salmon, E.D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009, 184, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Maresca, T.J.; Salmon, E.D. Welcome to a new kind of tension: Translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 2010, 123, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Lampson, M.A.; Grishchuk, E.L. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology (Basel) 2017, 6. [Google Scholar] [CrossRef]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Biggins, S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 2014, 15, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology (Basel) 2016, 5, E44. [Google Scholar] [CrossRef] [PubMed]
- Brinkley, B.R.; Stubblefield, E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma 1966, 19, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Alexander, S.P. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 1990, 110, 81–95. [Google Scholar] [CrossRef] [PubMed]
- McEwen, B.F.; Arena, J.T.; Frank, J.; Rieder, C.L. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J. Cell Biol. 1993, 120, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Jokelainen, P.T. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 1967, 19, 19–44. [Google Scholar] [CrossRef]
- Yao, X.; Anderson, K.L.; Cleveland, D.W. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 1997, 139, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.A.; Schaar, B.; Yen, T.J.; Earnshaw, W.C. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 1997, 106, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.B.; Pearson, C.G.; Yen, T.J.; Howell, B.J.; Salmon, E.D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 2001, 12, 1995–2009. [Google Scholar] [CrossRef] [PubMed]
- Wynne, D.J.; Funabiki, H. Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J. Cell Biol. 2015, 210, 899–916. [Google Scholar] [CrossRef] [PubMed]
- Thrower, D.A.; Jordan, M.A.; Wilson, L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskelet. 1996, 35, 121–133. [Google Scholar] [CrossRef]
- Magidson, V.; He, J.; Ault, J.G.; O’Connell, C.B.; Yang, N.; Tikhonenko, I.; McEwen, B.F.; Sui, H.; Khodjakov, A. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J. Cell Biol. 2016, 212, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Vanden Beldt, K.J.; Meng, X.; Khodjakov, A.; McEwen, B.F. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 2007, 9, 516–522. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Grishchuk, E.L.; Morphew, M.K.; Efremov, A.K.; Zhudenkov, K.; Volkov, V.A.; Cheeseman, I.M.; Desai, A.; Mastronarde, D.N.; Ataullakhanov, F.I. Fibrils connect microtubule tips with kinetochores: A mechanism to couple tubulin dynamics to chromosome motion. Cell 2008, 135, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Asbury, C.L. Anaphase A: Melting microtubules give mitosis its meaning. Biology (Basel) Submitted. 2017. [Google Scholar]
- Clarke, L.; Carbon, J. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 1983, 305, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.; Carbon, J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 1980, 287, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald-Hayes, M.; Clarke, L.; Carbon, J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 1982, 29, 235–244. [Google Scholar] [CrossRef]
- Lechner, J.; Carbon, J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 1991, 64, 717–725. [Google Scholar] [CrossRef]
- Hieter, P.; Pridmore, D.; Hegemann, J.H.; Thomas, M.; Davis, R.W.; Philippsen, P. Functional selection and analysis of yeast centromeric DNA. Cell 1985, 42, 913–921. [Google Scholar] [CrossRef]
- Pluta, A.F.; Mackay, A.M.; Ainsztein, A.M.; Goldberg, I.G.; Earnshaw, W.C. The centromere: Hub of chromosomal activities. Science 1995, 270, 1591–1594. [Google Scholar] [CrossRef] [PubMed]
- Catania, S.; Allshire, R.C. Anarchic centromeres: Deciphering order from apparent chaos. Curr. Opin. Cell Biol. 2014, 26, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Aldrup-Macdonald, M.E.; Sullivan, B.A. The past, present, and future of human centromere genomics. Genes (Basel) 2014, 5, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Steiner, N.C.; Clarke, L. A novel epigenetic effect can alter centromere function in fission yeast. Cell 1994, 79, 865–874. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Ratrie, H., 3rd; Stetten, G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 1989, 98, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.A.; Schwartz, S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 1995, 4, 2189–2197. [Google Scholar] [CrossRef] [PubMed]
- Voullaire, L.E.; Slater, H.R.; Petrovic, V.; Choo, K.H. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am. J. Hum. Genet. 1993, 52, 1153–1163. [Google Scholar] [PubMed]
- Du Sart, D.; Cancilla, M.R.; Earle, E.; Mao, J.I.; Saffery, R.; Tainton, K.M.; Kalitsis, P.; Martyn, J.; Barry, A.E.; Choo, K.H. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 1997, 16, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Amor, D.J.; Bentley, K.; Ryan, J.; Perry, J.; Wong, L.; Slater, H.; Choo, K.H. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 2004, 101, 6542–6547. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.D.; Karpen, G.H. Localization of centromere function in a Drosophila minichromosome. Cell 1995, 82, 599–609. [Google Scholar] [CrossRef]
- Shang, W.H.; Hori, T.; Toyoda, A.; Kato, J.; Popendorf, K.; Sakakibara, Y.; Fujiyama, A.; Fukagawa, T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010, 20, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Locke, D.P.; Hillier, L.W.; Warren, W.C.; Worley, K.C.; Nazareth, L.V.; Muzny, D.M.; Yang, S.P.; Wang, Z.; Chinwalla, A.T.; Minx, P.; et al. Comparative and demographic analysis of orang-utan genomes. Nature 2011, 469, 529–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piras, F.M.; Nergadze, S.G.; Magnani, E.; Bertoni, L.; Attolini, C.; Khoriauli, L.; Raimondi, E.; Giulotto, E. Uncoupling of satellite DNA and centromeric function in the genus equus. PLoS Genet. 2010, 6, e1000845. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, K.; Baum, M.; Carbon, J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA 2004, 101, 11374–11379. [Google Scholar] [CrossRef] [PubMed]
- Montefalcone, G.; Tempesta, S.; Rocchi, M.; Archidiacono, N. Centromere repositioning. Genome Res. 1999, 9, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity (Edinb) 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Antonacci, F.; Cardone, M.F.; Stanyon, R.; D’Addabbo, P.; Cellamare, A.; Sprague, L.J.; Eichler, E.E.; Archidiacono, N.; Rocchi, M. Evolutionary formation of new centromeres in macaque. Science 2007, 316, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Wiens, G.R.; Sorger, P.K. Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell 1998, 93, 313–316. [Google Scholar] [CrossRef]
- Karpen, G.H.; Allshire, R.C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997, 13, 489–496. [Google Scholar] [CrossRef]
- Murphy, T.D.; Karpen, G.H. Centromeres take flight: Alpha satellite and the quest for the human centromere. Cell 1998, 93, 317–320. [Google Scholar] [CrossRef]
- Earnshaw, W.C.; Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 1985, 91, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C. Discovering centromere proteins: From cold white hands to the A, B, C of CENPs. Nat. Rev. Mol. Cell Biol. 2015, 16, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; O’Day, K.; Wener, M.H.; Andrews, B.S.; Margolis, R.L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987, 104, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; O’Day, K.; Trong, H.L.; Charbonneau, H.; Margolis, R.L. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 1991, 88, 3734–3738. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.F.; Hechenberger, M.; Masri, K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994, 127, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; deYoung, D.; Henikoff, S.; Malik, H.S. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Warburton, P.E.; Cooke, C.A.; Bourassa, S.; Vafa, O.; Sullivan, B.A.; Stetten, G.; Gimelli, G.; Warburton, D.; Tyler-Smith, C.; Sullivan, K.F.; et al. Immunolocalization of cenp-a suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997, 7, 901–904. [Google Scholar] [CrossRef]
- Stoler, S.; Keith, K.C.; Curnick, K.E.; Fitzgerald-Hayes, M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995, 9, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Oegema, K.; Desai, A.; Rybina, S.; Kirkham, M.; Hyman, A.A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001, 153, 1209–1226. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.L.; Roth, M.B. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 2001, 153, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Howman, E.V.; Fowler, K.J.; Newson, A.J.; Redward, S.; MacDonald, A.C.; Kalitsis, P.; Choo, K.H. Early disruption of centromeric chromatin organization in centromere protein a (cenpa) null mice. Proc. Natl. Acad. Sci. USA 2000, 97, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.T.; Rattner, J.B.; Jablonski, S.A.; Yen, T.J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006, 175, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Folco, H.D.; Nechemia-Arbely, Y.; Valente, L.P.; Nguyen, K.; Wong, A.J.; Zhu, Q.; Holland, A.J.; Desai, A.; Jansen, L.E.; et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013, 15, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Barrey, E.J.; Bassett, E.A.; DeNizio, J.E.; Guo, L.Y.; Panchenko, T.; Dawicki-McKenna, J.M.; Heun, P.; Black, B.E. Both tails and the centromere targeting domain of cenp-a are required for centromere establishment. J. Cell Biol. 2015, 208, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Heun, P.; Erhardt, S.; Blower, M.D.; Weiss, S.; Skora, A.D.; Karpen, G.H. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 2006, 10, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Mendiburo, M.J.; Padeken, J.; Fulop, S.; Schepers, A.; Heun, P. Drosophila CENH3 is sufficient for centromere formation. Science 2011, 334, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.C.; Kuich, P.H.; Stellfox, M.E.; Ward, J.A.; Bassett, E.A.; Black, B.E.; Foltz, D.R. Hjurp is a cenp-a chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011, 194, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Van Hooser, A.A.; Ouspenski, I.I.; Gregson, H.C.; Starr, D.A.; Yen, T.J.; Goldberg, M.L.; Yokomori, K.; Earnshaw, W.C.; Sullivan, K.F.; Brinkley, B.R. Specification of kinetochore-forming chromatin by the histone h3 variant cenp-a. J. Cell Sci. 2001, 114, 3529–3542. [Google Scholar] [PubMed]
- Hori, T.; Shang, W.H.; Takeuchi, K.; Fukagawa, T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 2013, 200, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Guse, A.; Carroll, C.W.; Moree, B.; Fuller, C.J.; Straight, A.F. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 2011, 477, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, J.G.; Thakur, J.; Kasinathan, S.; Henikoff, S. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci. Adv. 2015, 1, e1400234. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.; Henikoff, S. CENPT bridges adjacent CENPA nucleosomes on young human alpha-satellite dimers. Genome Res. 2016, 26, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Jiang, J.; Zhou, B.R.; Rozendaal, M.; Feng, H.; Ghirlando, R.; Xiao, T.S.; Straight, A.F.; Bai, Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein cenp-c. Science 2013, 340, 1110–1113. [Google Scholar] [CrossRef] [PubMed]
- Klare, K.; Weir, J.R.; Basilico, F.; Zimniak, T.; Massimiliano, L.; Ludwigs, N.; Herzog, F.; Musacchio, A. Cenp-c is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 2015, 210, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 2002, 12, 711–718. [Google Scholar] [CrossRef]
- Talbert, P.B.; Bryson, T.D.; Henikoff, S. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 2004, 3, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schueler, M.G.; Swanson, W.; Thomas, P.J.; Program, N.C.S.; Green, E.D. Adaptive evolution of foundation kinetochore proteins in primates. Mol. Biol. Evol. 2010, 27, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Chmatal, L.; Gabriel, S.I.; Mitsainas, G.P.; Martinez-Vargas, J.; Ventura, J.; Searle, J.B.; Schultz, R.M.; Lampson, M.A. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 2014, 24, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Bassett, E.A.; Wood, S.; Salimian, K.J.; Ajith, S.; Foltz, D.R.; Black, B.E. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol. 2010, 190, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, N.; Kitajima, T.; Yokobayashi, S.; Xiao, G.; Yamamoto, M.; Grewal, S.I.; Watanabe, Y. Recruitment of cohesin to heterochromatic regions by swi6/hp1 in fission yeast. Nat. Cell Biol. 2002, 4, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.; Maure, J.F.; Partridge, J.F.; Genier, S.; Javerzat, J.P.; Allshire, R.C. Requirement of heterochromatin for cohesion at centromeres. Science 2001, 294, 2539–2542. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Schleiffer, A. Family matters: Structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 2013, 23, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Meraldi, P.; McAinsh, A.D.; Rheinbay, E.; Sorger, P.K. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006, 7, R23. [Google Scholar] [CrossRef] [PubMed]
- Moroi, Y.; Peebles, C.; Fritzler, M.J.; Steigerwald, J.; Tan, E.M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA 1980, 77, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Sullivan, K.F.; Machlin, P.S.; Cooke, C.A.; Kaiser, D.A.; Pollard, T.D.; Rothfield, N.F.; Cleveland, D.W. Molecular cloning of cdna for cenp-b, the major human centromere autoantigen. J. Cell Biol. 1987, 104, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Tomkiel, J.; Cooke, C.A.; Ratrie, H., 3rd; Maurer, M.; Rothfield, N.F.; Earnshaw, W.C. Cenp-c, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 1992, 70, 115–125. [Google Scholar] [CrossRef]
- Nishihashi, A.; Haraguchi, T.; Hiraoka, Y.; Ikemura, T.; Regnier, V.; Dodson, H.; Earnshaw, W.C.; Fukagawa, T. Cenp-i is essential for centromere function in vertebrate cells. Dev. Cell 2002, 2, 463–476. [Google Scholar] [CrossRef]
- Sugata, N.; Munekata, E.; Todokoro, K. Characterization of a novel kinetochore protein, cenp-h. J. Biol. Chem. 1999, 274, 27343–27346. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, S.; Takahashi, K.; Yanagida, M. Mis6, a fission yeast inner centromere protein, acts during g1/s and forms specialized chromatin required for equal segregation. Cell 1997, 90, 131–143. [Google Scholar] [CrossRef]
- Okada, M.; Cheeseman, I.M.; Hori, T.; Okawa, K.; McLeod, I.X.; Yates, J.R., 3rd; Desai, A.; Fukagawa, T. The cenp-h-i complex is required for the efficient incorporation of newly synthesized cenp-a into centromeres. Nat. Cell Biol. 2006, 8, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Foltz, D.R.; Jansen, L.E.; Black, B.E.; Bailey, A.O.; Yates, J.R., 3rd; Cleveland, D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006, 8, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Obuse, C.; Yang, H.; Nozaki, N.; Goto, S.; Okazaki, T.; Yoda, K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 2004, 9, 105–120. [Google Scholar] [CrossRef]
- Izuta, H.; Ikeno, M.; Suzuki, N.; Tomonaga, T.; Nozaki, N.; Obuse, C.; Kisu, Y.; Goshima, N.; Nomura, F.; Nomura, N.; et al. Comprehensive analysis of the icen (interphase centromere complex) components enriched in the cenp-a chromatin of human cells. Genes Cells 2006, 11, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Amano, M.; Suzuki, A.; Backer, C.B.; Welburn, J.P.; Dong, Y.; McEwen, B.F.; Shang, W.H.; Suzuki, E.; Okawa, K.; et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 2008, 135, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Suzuki, A.; Hori, T.; Backer, C.; Okawa, K.; Cheeseman, I.M.; Fukagawa, T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 2009, 186, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Bloom, K.; Salmon, E.D. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 2009, 19, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; O’Quinn, R.P.; Pierce, H.L.; Joglekar, A.P.; Gall, W.E.; DeLuca, J.G.; Carroll, C.W.; Liu, S.T.; Yen, T.J.; McEwen, B.F.; et al. Protein architecture of the human kinetochore microtubule attachment site. Cell 2009, 137, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Badger, B.L.; Wan, X.; DeLuca, J.G.; Salmon, E.D. The architecture of ccan proteins creates a structural integrity to resist spindle forces and achieve proper intrakinetochore stretch. Dev. Cell 2014, 30, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Bouck, D.C.; Molk, J.N.; Bloom, K.S.; Salmon, E.D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006, 8, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Pot, I.; Measday, V.; Snydsman, B.; Cagney, G.; Fields, S.; Davis, T.N.; Muller, E.G.; Hieter, P. Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 2003, 14, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, S.M.; Harrison, S.C. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep. 2013, 5, 29–36. [Google Scholar] [CrossRef] [PubMed]
- McKinley, K.L.; Sekulic, N.; Guo, L.Y.; Tsinman, T.; Black, B.E.; Cheeseman, I.M. The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol. Cell 2015, 60, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Weir, J.R.; Faesen, A.C.; Klare, K.; Petrovic, A.; Basilico, F.; Fischbock, J.; Pentakota, S.; Keller, J.; Pesenti, M.E.; Pan, D.; et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 2016, 537, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Basilico, F.; Maffini, S.; Weir, J.R.; Prumbaum, D.; Rojas, A.M.; Zimniak, T.; De Antoni, A.; Jeganathan, S.; Voss, B.; van Gerwen, S.; et al. The pseudo gtpase cenp-m drives human kinetochore assembly. eLife 2014, 3, e02978. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Okawa, K.; Isobe, T.; Fukagawa, T. Cenp-h-containing complex facilitates centromere deposition of cenp-a in cooperation with fact and chd1. Mol. Biol. Cell 2009, 20, 3986–3995. [Google Scholar] [CrossRef] [PubMed]
- Pekgoz Altunkaya, G.; Malvezzi, F.; Demianova, Z.; Zimniak, T.; Litos, G.; Weissmann, F.; Mechtler, K.; Herzog, F.; Westermann, S. Ccan assembly configures composite binding interfaces to promote cross-linking of ndc80 complexes at the kinetochore. Curr. Biol. 2016, 26, 2370–2378. [Google Scholar] [CrossRef] [PubMed]
- Measday, V.; Hailey, D.W.; Pot, I.; Givan, S.A.; Hyland, K.M.; Cagney, G.; Fields, S.; Davis, T.N.; Hieter, P. Ctf3p, the mis6 budding yeast homolog, interacts with mcm22p and mcm16p at the yeast outer kinetochore. Genes Dev. 2002, 16, 101–113. [Google Scholar] [CrossRef] [PubMed]
- De Wulf, P.; McAinsh, A.D.; Sorger, P.K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003, 17, 2902–2921. [Google Scholar] [CrossRef] [PubMed]
- Schmitzberger, F.; Harrison, S.C. RWD domain: A recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012, 13, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, J.; Stemmann, O.; Rank, S.; Lechner, J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999, 13, 1140–1155. [Google Scholar] [CrossRef] [PubMed]
- Meluh, P.B.; Koshland, D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 1995, 6, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, B.; Nelson, C.R.; Ranish, J.A.; Biggins, S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009, 23, 2887–2899. [Google Scholar] [CrossRef] [PubMed]
- Cole, H.A.; Howard, B.H.; Clark, D.J. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl. Acad. Sci. USA 2011, 108, 12687–12692. [Google Scholar] [CrossRef] [PubMed]
- Meluh, P.B.; Yang, P.; Glowczewski, L.; Koshland, D.; Smith, M.M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 1998, 94, 607–613. [Google Scholar] [CrossRef]
- Furuyama, S.; Biggins, S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 2007, 104, 14706–14711. [Google Scholar] [CrossRef] [PubMed]
- Bram, R.J.; Kornberg, R.D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol. Cell. Biol. 1987, 7, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Espelin, C.W.; Kaplan, K.B.; Sorger, P.K. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 1997, 139, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Drubin, D.G.; Barnes, G. Simple centromere, complex kinetochore: Linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 2002, 157, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Hemmerich, P.; Stoyan, T.; Wieland, G.; Koch, M.; Lechner, J.; Diekmann, S. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc. Natl. Acad. Sci. USA 2000, 97, 12583–12588. [Google Scholar] [CrossRef] [PubMed]
- Krassovsky, K.; Henikoff, J.G.; Henikoff, S. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 2012, 109, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Cho, U.S.; Harrison, S.C. Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat. Struct. Mol. Biol. 2012, 19, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Mizuguchi, G.; Wisniewski, J.; Huang, Y.; Wei, D.; Wu, C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell 2011, 43, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Furuyama, T.; Henikoff, S. Centromeric nucleosomes induce positive DNA supercoils. Cell 2009, 138, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Chang, K.M.; Cui, H.; Jayaram, M. Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc. Natl. Acad. Sci. USA 2011, 108, 13671–13676. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Ingelmo, O.; Martinez-Garcia, B.; Segura, J.; Valdes, A.; Roca, J. DNA Topology and Global Architecture of Point Centromeres. Cell Rep. 2015, 13, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Henikoff, S.; Malik, H.S. Evolutionary Turnover of Kinetochore Proteins: A Ship of Theseus? Trends Cell Biol. 2016, 26, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.K.; Margolis, R.L. Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol. Cell. Biol. 1985, 5, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Shelby, R.D.; Vafa, O.; Sullivan, K.F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997, 136, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Yoda, K.; Ando, S.; Morishita, S.; Houmura, K.; Hashimoto, K.; Takeyasu, K.; Okazaki, T. Human centromere protein a (cenp-a) can replace histone h3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 2000, 97, 7266–7271. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Kagawa, W.; Shiga, T.; Osakabe, A.; Miya, Y.; Saito, K.; Hayashi-Takanaka, Y.; Oda, T.; Sato, M.; Park, S.Y.; et al. Crystal structure of the human centromeric nucleosome containing cenp-a. Nature 2011, 476, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Kingston, I.J.; Yung, J.S.; Singleton, M.R. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J. Biol. Chem. 2011, 286, 4021–4026. [Google Scholar] [CrossRef] [PubMed]
- Black, B.E.; Foltz, D.R.; Chakravarthy, S.; Luger, K.; Woods, V.L., Jr.; Cleveland, D.W. Structural determinants for generating centromeric chromatin. Nature 2004, 430, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Falk, S.J.; Lee, J.; Sekulic, N.; Sennett, M.A.; Lee, T.H.; Black, B.E. CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat. Struct. Mol. Biol. 2016, 23, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Falk, S.J.; Guo, L.Y.; Sekulic, N.; Smoak, E.M.; Mani, T.; Logsdon, G.A.; Gupta, K.; Jansen, L.E.; Van Duyne, G.D.; Vinogradov, S.A.; et al. Chromosomes. Cenp-c reshapes and stabilizes cenp-a nucleosomes at the centromere. Science 2015, 348, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Sekulic, N.; Bassett, E.A.; Rogers, D.J.; Black, B.E. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 2010, 467, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Conde e Silva, N.; Black, B.E.; Sivolob, A.; Filipski, J.; Cleveland, D.W.; Prunell, A. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007, 370, 555–573. [Google Scholar] [CrossRef] [PubMed]
- Black, B.E.; Brock, M.A.; Bedard, S.; Woods, V.L., Jr.; Cleveland, D.W. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 5008–5013. [Google Scholar] [CrossRef] [PubMed]
- Panchenko, T.; Sorensen, T.C.; Woodcock, C.L.; Kan, Z.Y.; Wood, S.; Resch, M.G.; Luger, K.; Englander, S.W.; Hansen, J.C.; Black, B.E. Replacement of histone h3 with cenp-a directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl. Acad. Sci. USA 2011, 108, 16588–16593. [Google Scholar] [CrossRef] [PubMed]
- Hasson, D.; Panchenko, T.; Salimian, K.J.; Salman, M.U.; Sekulic, N.; Alonso, A.; Warburton, P.E.; Black, B.E. The octamer is the major form of cenp-a nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013, 20, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Miell, M.D.; Fuller, C.J.; Guse, A.; Barysz, H.M.; Downes, A.; Owen-Hughes, T.; Rappsilber, J.; Straight, A.F.; Allshire, R.C. Cenp-a confers a reduction in height on octameric nucleosomes. Nat. Struct. Mol. Biol. 2013, 20, 763–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, B.E.; Cleveland, D.W. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 2011, 144, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Padeganeh, A.; De Rop, V.; Maddox, P.S. Nucleosomal composition at the centromere: A numbers game. Chromosome Res. 2013, 21, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Silva, M.C.; Godek, K.M.; Jansen, L.E.; Straight, A.F. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009, 11, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Milks, K.J.; Straight, A.F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010, 189, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Smoak, E.M.; Stein, P.; Schultz, R.M.; Lampson, M.A.; Black, B.E. Long-Term Retention of CENP-A Nucleosomes in Mammalian Oocytes Underpins Transgenerational Inheritance of Centromere Identity. Curr. Biol. 2016, 26, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.E.; Black, B.E.; Foltz, D.R.; Cleveland, D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007, 176, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Westhorpe, F.G.; Fuller, C.J.; Straight, A.F. A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J. Cell Biol. 2015, 209, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Armache, K.J.; Garlick, J.D.; Canzio, D.; Narlikar, G.J.; Kingston, R.E. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 2011, 334, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Barbera, A.J.; Chodaparambil, J.V.; Kelley-Clarke, B.; Joukov, V.; Walter, J.C.; Luger, K.; Kaye, K.M. The nucleosomal surface as a docking station for kaposi’s sarcoma herpesvirus lana. Science 2006, 311, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Makde, R.D.; England, J.R.; Yennawar, H.P.; Tan, S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 2010, 467, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Muller, S.; Blumer, J.; Klare, K.; Musacchio, A.; Almouzni, G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015, 11, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Nakano, M.; Nozaki, N.; Egashira, S.; Okazaki, T.; Masumoto, H. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J. Biol. Chem. 2004, 279, 5934–5946. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Tomkiel, J.; Saitoh, H.; Johnson, D.H.; Earnshaw, W.C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 1996, 16, 3576–3586. [Google Scholar] [CrossRef] [PubMed]
- Politi, V.; Perini, G.; Trazzi, S.; Pliss, A.; Raska, I.; Earnshaw, W.C.; Della Valle, G. Cenp-c binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 2002, 115, 2317–2327. [Google Scholar] [PubMed]
- Trazzi, S.; Bernardoni, R.; Diolaiti, D.; Politi, V.; Earnshaw, W.C.; Perini, G.; Della Valle, G. In vivo functional dissection of human inner kinetochore protein cenp-c. J. Struct. Biol. 2002, 140, 39–48. [Google Scholar] [CrossRef]
- Song, K.; Gronemeyer, B.; Lu, W.; Eugster, E.; Tomkiel, J.E. Mutational analysis of the central centromere targeting domain of human centromere protein C, (CENP-C). Exp. Cell Res. 2002, 275, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Lanini, L.; McKeon, F. Domains required for CENP-C assembly at the kinetochore. Mol. Biol. Cell 1995, 6, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Milks, K.J.; Moree, B.; Straight, A.F. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell 2009, 20, 4246–4255. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Chang, H.L.; Kagami, A.; Watanabe, Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 2009, 17, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.L.; Espelin, C.W.; De Wulf, P.; Sorger, P.K.; Harrison, S.C.; Simons, K.T. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell 2008, 19, 4480–4491. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Kuriyama, K.; Shibata, A.; Himeno, M. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: Role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 1997, 5, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Bodor, D.L.; Mata, J.F.; Sergeev, M.; David, A.F.; Salimian, K.J.; Panchenko, T.; Cleveland, D.W.; Black, B.E.; Shah, J.V.; Jansen, L.E. The quantitative architecture of centromeric chromatin. eLife 2014, 3, e02137. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Shang, W.H.; Toyoda, A.; Misu, S.; Monma, N.; Ikeo, K.; Molina, O.; Vargiu, G.; Fujiyama, A.; Kimura, H.; et al. Histone h4 lys 20 monomethylation of the cenp-a nucleosome is essential for kinetochore assembly. Dev. Cell 2014, 29, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.O.; Panchenko, T.; Sathyan, K.M.; Petkowski, J.J.; Pai, P.J.; Bai, D.L.; Russell, D.H.; Macara, I.G.; Shabanowitz, J.; Hunt, D.F.; et al. Posttranslational modification of cenp-a influences the conformation of centromeric chromatin. Proc. Natl. Acad. Sci. USA 2013, 110, 11827–11832. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Delannoy, M.; Ling, C.; Daee, D.; Osman, F.; Muniandy, P.A.; Shen, X.; Oostra, A.B.; Du, H.; Steltenpool, J.; et al. A histone-fold complex and fancm form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell 2010, 37, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.R.; Saro, D.; Ali, A.M.; Zheng, X.F.; Du, C.H.; Killen, M.W.; Sachpatzidis, A.; Wahengbam, K.; Pierce, A.J.; Xiong, Y.; et al. Mhf1-mhf2, a histone-fold-containing protein complex, participates in the fanconi anemia pathway via fancm. Mol. Cell 2010, 37, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Kim, J.M.; Shiotani, B.; Yang, K.; Zou, L.; D’Andrea, A.D. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol. Cell 2010, 39, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Takeuchi, K.; Gascoigne, K.E.; Suzuki, A.; Hori, T.; Oyama, T.; Morikawa, K.; Cheeseman, I.M.; Fukagawa, T. Cenp-t-w-s-x forms a unique centromeric chromatin structure with a histone-like fold. Cell 2012, 148, 487–501. [Google Scholar] [CrossRef]
- Takeuchi, K.; Nishino, T.; Mayanagi, K.; Horikoshi, N.; Osakabe, A.; Tachiwana, H.; Hori, T.; Kurumizaka, H.; Fukagawa, T. The centromeric nucleosome-like cenp-t-w-s-x complex induces positive supercoils into DNA. Nucleic Acids Res. 2014, 42, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, L.; van Vuuren, C.; Kaczmarczyk, A.; Doering, V.; Hellwig, D.; Quinn, N.; Hoischen, C.; Diekmann, S.; Sullivan, K.F. Premitotic assembly of human cenps -t and -w switches centromeric chromatin to a mitotic state. PLoS Biol. 2011, 9, e1001082. [Google Scholar] [CrossRef] [PubMed]
- Samejima, I.; Spanos, C.; Alves Fde, L.; Hori, T.; Perpelescu, M.; Zou, J.; Rappsilber, J.; Fukagawa, T.; Earnshaw, W.C. Whole-proteome genetic analysis of dependencies in assembly of a vertebrate kinetochore. J. Cell Biol. 2015, 211, 1141–1156. [Google Scholar] [CrossRef] [PubMed]
- Folco, H.D.; Campbell, C.S.; May, K.M.; Espinoza, C.A.; Oegema, K.; Hardwick, K.G.; Grewal, S.I.; Desai, A. The cenp-a n-tail confers epigenetic stability to centromeres via the cenp-t branch of the ccan in fission yeast. Curr. Biol. 2015, 25, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Okada, M.; Maenaka, K.; Fukagawa, T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell 2008, 19, 843–854. [Google Scholar] [CrossRef]
- Kagawa, N.; Hori, T.; Hoki, Y.; Hosoya, O.; Tsutsui, K.; Saga, Y.; Sado, T.; Fukagawa, T. The cenp-o complex requirement varies among different cell types. Chromosome Res. 2014, 22, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, J.; Auckland, P.; Samora, C.P.; McAinsh, A.D. Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J. Cell Sci. 2015, 128, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Wang, Z.; Jiang, K.; Huang, Y.; Ward, T.; Zhao, L.; Dou, Z.; Yao, X. Cenp-u cooperates with hec1 to orchestrate kinetochore-microtubule attachment. J. Biol. Chem. 2011, 286, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.C.; Samora, C.P.; Holtackers, R.; Wang, E.; Kingston, I.J.; Alonso, M.; Lampson, M.; McAinsh, A.D.; Meraldi, P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 2010, 12, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Park, C.H.; Kim, T.S.; Soung, N.K.; Bang, J.K.; Kim, B.Y.; Park, J.E.; Lee, K.S. Mammalian polo-like kinase 1-dependent regulation of the pbip1-cenp-q complex at kinetochores. J. Biol. Chem. 2011, 286, 19744–19757. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Park, J.E.; Yu, L.R.; Soung, N.K.; Yun, S.M.; Bang, J.K.; Seong, Y.S.; Yu, H.; Garfield, S.; Veenstra, T.D.; et al. Self-regulated plk1 recruitment to kinetochores by the plk1-pbip1 interaction is critical for proper chromosome segregation. Mol. Cell 2006, 24, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Hyland, K.M.; Kingsbury, J.; Koshland, D.; Hieter, P. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 1999, 145, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Fernius, J.; Marston, A.L. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009, 5, e1000629. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.M.; Waples, W.G.; Lavoie, B.D.; Biggins, S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell 2009, 20, 3818–3827. [Google Scholar] [CrossRef] [PubMed]
- Natsume, T.; Muller, C.A.; Katou, Y.; Retkute, R.; Gierlinski, M.; Araki, H.; Blow, J.J.; Shirahige, K.; Nieduszynski, C.A.; Tanaka, T.U. Kinetochores coordinate pericentromeric cohesion and early DNA replication by cdc7-dbf4 kinase recruitment. Mol. Cell 2013, 50, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Hornung, P.; Troc, P.; Malvezzi, F.; Maier, M.; Demianova, Z.; Zimniak, T.; Litos, G.; Lampert, F.; Schleiffer, A.; Brunner, M.; et al. A cooperative mechanism drives budding yeast kinetochore assembly downstream of cenp-a. J. Cell Biol. 2014, 206, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, H.; Masukata, H.; Muro, Y.; Nozaki, N.; Okazaki, T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989, 109, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.; Riggs, A.D. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA 1996, 93, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.F.; Fowler, K.J.; Earle, E.; Saffery, R.; Kalitsis, P.; Trowell, H.; Hill, J.; Wreford, N.G.; de Kretser, D.M.; Cancilla, M.R.; et al. Centromere protein b null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 1998, 141, 309–319. [Google Scholar] [CrossRef]
- Kapoor, M.; Montes de Oca Luna, R.; Liu, G.; Lozano, G.; Cummings, C.; Mancini, M.; Ouspenski, I.; Brinkley, B.R.; May, G.S. The cenpb gene is not essential in mice. Chromosoma 1998, 107, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Perez-Castro, A.V.; Shamanski, F.L.; Meneses, J.J.; Lovato, T.L.; Vogel, K.G.; Moyzis, R.K.; Pedersen, R. Centromeric protein b null mice are viable with no apparent abnormalities. Dev. Biol. 1998, 201, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ohzeki, J.; Nakano, M.; Yoda, K.; Brinkley, W.R.; Larionov, V.; Masumoto, H. Cenp-b controls centromere formation depending on the chromatin context. Cell 2007, 131, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
- Ohzeki, J.; Nakano, M.; Okada, T.; Masumoto, H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002, 159, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Han, J.S.; McMahon, M.A.; Ly, P.; Abdullah, A.; Wong, A.J.; Cleveland, D.W. DNA sequence-specific binding of cenp-b enhances the fidelity of human centromere function. Dev. Cell 2015, 33, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.; Dumont, M.; Barra, V.; Ly, P.; Nechemia-Arbely, Y.; McMahon, M.A.; Herve, S.; Cleveland, D.W.; Fachinetti, D. Cenp-a is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep. 2016, 17, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Fujita, R.; Otake, K.; Arimura, Y.; Horikoshi, N.; Miya, Y.; Shiga, T.; Osakabe, A.; Tachiwana, H.; Ohzeki, J.; Larionov, V.; et al. Stable complex formation of cenp-b with the cenp-a nucleosome. Nucleic Acids Res. 2015, 43, 4909–4922. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, R.; Qi, W.; Yu, H. Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 2004, 279, 13076–13085. [Google Scholar] [CrossRef] [PubMed]
- McCleland, M.L.; Gardner, R.D.; Kallio, M.J.; Daum, J.R.; Gorbsky, G.J.; Burke, D.J.; Stukenberg, P.T. The highly conserved ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003, 17, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Rybina, S.; Muller-Reichert, T.; Shevchenko, A.; Shevchenko, A.; Hyman, A.; Oegema, K. Knl-1 directs assembly of the microtubule-binding interface of the kinetochore in c. Elegans. Genes Dev. 2003, 17, 2421–2435. [Google Scholar] [CrossRef] [PubMed]
- Obuse, C.; Iwasaki, O.; Kiyomitsu, T.; Goshima, G.; Toyoda, Y.; Yanagida, M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004, 6, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A.; Kilmartin, J.V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001, 152, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Cheeseman, I.M.; Anderson, S.; Yates, J.R., 3rd; Drubin, D.G.; Barnes, G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003, 163, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Niessen, S.; Anderson, S.; Hyndman, F.; Yates, J.R., 3rd; Oegema, K.; Desai, A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004, 18, 2255–2268. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Chappie, J.S.; Wilson-Kubalek, E.M.; Desai, A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006, 127, 983–997. [Google Scholar] [CrossRef]
- Nekrasov, V.S.; Smith, M.A.; Peak-Chew, S.; Kilmartin, J.V. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 4931–4946. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, B.A.; Tatsutani, S.Y.; Collins, K.A.; Biggins, S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 2003, 5, 735–745. [Google Scholar] [CrossRef]
- Kline, S.L.; Cheeseman, I.M.; Hori, T.; Fukagawa, T.; Desai, A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006, 173, 9–17. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006, 127, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Schnell, J.R.; Larsen, N.A.; Sorger, P.K.; Chou, J.J.; Harrison, S.C. Structure of a central component of the yeast kinetochore: The Spc24p/Spc25p globular domain. Structure 2006, 14, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Sorger, P.K.; Harrison, S.C. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA 2005, 102, 5363–5367. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Al-Bassam, J.; Harrison, S.C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007, 14, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, C.; De Luca, J.; Monzani, S.; Ferrari, K.J.; Ristic, D.; Wyman, C.; Stark, H.; Kilmartin, J.; Salmon, E.D.; Musacchio, A. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 2005, 280, 29088–29095. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, C.; Pasqualato, S.; Screpanti, E.; Varetti, G.; Santaguida, S.; Dos Reis, G.; Maiolica, A.; Polka, J.; De Luca, J.G.; De Wulf, P.; et al. Implications for kinetochore-microtubule attachment from the structure of an engineered ndc80 complex. Cell 2008, 133, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.; Ingram, J.; Harrison, S.C. Conserved Tetramer Junction in the Kinetochore Ndc80 Complex. Cell Rep. 2016, 17, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Schou, K.B.; Andersen, J.S.; Pedersen, L.B. A divergent calponin homology (NN-CH) domain defines a novel family: Implications for evolution of ciliary IFT complex B proteins. Bioinformatics 2014, 30, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Alushin, G.M.; Ramey, V.H.; Pasqualato, S.; Ball, D.A.; Grigorieff, N.; Musacchio, A.; Nogales, E. The ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 2010, 467, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Alushin, G.M.; Musinipally, V.; Matson, D.; Tooley, J.; Stukenberg, P.T.; Nogales, E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat. Struct. Mol. Biol. 2012, 19, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, J.G.; Musacchio, A. Structural organization of the kinetochore-microtubule interface. Curr. Opin. Cell Biol. 2012, 24, 48–56. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, K.F.; Lens, S.M.; DeLuca, J.G. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 2011, 124, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Johnson, M.L.; Stukenberg, P.T. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr. Biol. 2008, 18, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, G.J.; Dong, Y.; McEwen, B.F.; Deluca, J.G. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 2008, 18, 1778–1784. [Google Scholar] [CrossRef] [PubMed]
- Umbreit, N.T.; Gestaut, D.R.; Tien, J.F.; Vollmar, B.S.; Gonen, T.; Asbury, C.L.; Davis, T.N. The ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA 2012, 109, 16113–16118. [Google Scholar] [CrossRef] [PubMed]
- Aravamudhan, P.; Felzer-Kim, I.; Gurunathan, K.; Joglekar, A.P. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr. Biol. 2014, 24, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Zaytsev, A.V.; Mick, J.E.; Maslennikov, E.; Nikashin, B.; DeLuca, J.G.; Grishchuk, E.L. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell 2015, 26, 1829–1844. [Google Scholar] [CrossRef] [PubMed]
- Kemmler, S.; Stach, M.; Knapp, M.; Ortiz, J.; Pfannstiel, J.; Ruppert, T.; Lechner, J. Mimicking ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009, 28, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Lampert, F.; Mieck, C.; Alushin, G.M.; Nogales, E.; Westermann, S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J. Cell Biol. 2013, 200, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Cheerambathur, D.K.; Gassmann, R.; Cook, B.; Oegema, K.; Desai, A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 2013, 342, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Zaytsev, A.V.; Sundin, L.J.; DeLuca, K.F.; Grishchuk, E.L.; DeLuca, J.G. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J. Cell Biol. 2014, 206, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.; Pasqualato, S.; Dube, P.; Krenn, V.; Santaguida, S.; Cittaro, D.; Monzani, S.; Massimiliano, L.; Keller, J.; Tarricone, A.; et al. The mis12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 2010, 190, 835–852. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.; Mosalaganti, S.; Keller, J.; Mattiuzzo, M.; Overlack, K.; Krenn, V.; De Antoni, A.; Wohlgemuth, S.; Cecatiello, V.; Pasqualato, S.; et al. Modular assembly of rwd domains on the mis12 complex underlies outer kinetochore organization. Mol. Cell 2014, 53, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Maskell, D.P.; Hu, X.W.; Singleton, M.R. Molecular architecture and assembly of the yeast kinetochore mind complex. J. Cell Biol. 2010, 190, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Hornung, P.; Maier, M.; Alushin, G.M.; Lander, G.C.; Nogales, E.; Westermann, S. Molecular architecture and connectivity of the budding yeast mtw1 kinetochore complex. J. Mol. Biol. 2011, 405, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Screpanti, E.; De Antoni, A.; Alushin, G.M.; Petrovic, A.; Melis, T.; Nogales, E.; Musacchio, A. Direct binding of cenp-c to the mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011, 21, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Huis in ’t Veld, P.J.; Jeganathan, S.; Petrovic, A.; John, J.; Singh, P.; Weissmann, F.; Bange, T.; Musacchio, A. Molecular basis of outer kinetochore assembly on cenp-t. eLife 2016. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, Y.N.; Jenni, S.; Valverde, R.; Khin, Y.; Harrison, S.C. Structure of the mind complex defines a regulatory focus for yeast kinetochore assembly. Cell 2016, 167, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.; Keller, J.; Liu, Y.; Overlack, K.; John, J.; Dimitrova, Y.N.; Jenni, S.; van Gerwen, S.; Stege, P.; Wohlgemuth, S.; et al. Structure of the mis12 complex and molecular basis of its interaction with cenp-c at human kinetochores. Cell 2016, 167, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Malvezzi, F.; Litos, G.; Schleiffer, A.; Heuck, A.; Mechtler, K.; Clausen, T.; Westermann, S. A structural basis for kinetochore recruitment of the ndc80 complex via two distinct centromere receptors. EMBO J. 2013, 32, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Przewloka, M.R.; Zhang, W.; Costa, P.; Archambault, V.; D’Avino, P.P.; Lilley, K.S.; Laue, E.D.; McAinsh, A.D.; Glover, D.M. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2007, 2, e478. [Google Scholar] [CrossRef] [PubMed]
- Schittenhelm, R.B.; Heeger, S.; Althoff, F.; Walter, A.; Heidmann, S.; Mechtler, K.; Lehner, C.F. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in drosophila chromosomes. Chromosoma 2007, 116, 385–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Petrovic, A.; Rombaut, P.; Mosalaganti, S.; Keller, J.; Raunser, S.; Herzog, F.; Musacchio, A. Insights from the reconstitution of the divergent outer kinetochore of Drosophila melanogaster. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.M.; Poznanski, J.; Zdziarska, A.; Czarnocki-Cieciura, M.; Lipinszki, Z.; Dadlez, M.; Glover, D.M.; Przewloka, M.R. Network of protein interactions within the drosophila inner kinetochore. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Morelli, E.; Mastrodonato, V.; Beznoussenko, G.V.; Mironov, A.A.; Tognon, E.; Vaccari, T. An essential step of kinetochore formation controlled by the snare protein snap29. EMBO J. 2016, 35, 2223–2237. [Google Scholar] [CrossRef] [PubMed]
- Tromer, E.; Snel, B.; Kops, G.J. Widespread recurrent patterns of rapid repeat evolution in the kinetochore scaffold knl1. Genome Biol. Evol. 2015, 7, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E.; Mitchison, T.J.; Kirschner, M.W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 1988, 331, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Coue, M.; Lombillo, V.A.; McIntosh, J.R. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991, 112, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, E.L.; Molodtsov, M.I.; Ataullakhanov, F.I.; McIntosh, J.R. Force production by disassembling microtubules. Nature 2005, 438, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.F.; Franck, A.D.; Gestaut, D.R.; Cooper, J.; Gracyzk, B.; Wei, R.R.; Wordeman, L.; Davis, T.N.; Asbury, C.L. The ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 2009, 136, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.C.; Arthanari, H.; Boeszoermenyi, A.; Dashkevich, N.M.; Wilson-Kubalek, E.M.; Monnier, N.; Markus, M.; Oberer, M.; Milligan, R.A.; Bathe, M.; et al. The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell 2012, 23, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Maiato, H.; DeLuca, J.; Salmon, E.D.; Earnshaw, W.C. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004, 117, 5461–5477. [Google Scholar] [CrossRef] [PubMed]
- Maiato, H.; Gomes, A.M.; Sousa, F.; Barisic, M. Mechanisms of chromosome congression during mitosis. Biology (Basel) 2017. [Google Scholar]
- Gaitanos, T.N.; Santamaria, A.; Jeyaprakash, A.A.; Wang, B.; Conti, E.; Nigg, E.A. Stable kinetochore-microtubule interactions depend on the ska complex and its new component ska3/c13orf3. EMBO J. 2009, 28, 1442–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welburn, J.P.; Grishchuk, E.L.; Backer, C.B.; Wilson-Kubalek, E.M.; Yates, J.R., 3rd; Cheeseman, I.M. The human kinetochore ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell 2009, 16, 374–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theis, M.; Slabicki, M.; Junqueira, M.; Paszkowski-Rogacz, M.; Sontheimer, J.; Kittler, R.; Heninger, A.K.; Glatter, T.; Kruusmaa, K.; Poser, I.; et al. Comparative profiling identifies c13orf3 as a component of the ska complex required for mammalian cell division. EMBO J. 2009, 28, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, C.; Cheeseman, I.M.; Goode, B.L.; McDonald, K.L.; Barnes, G.; Drubin, D.G. Saccharomyces cerevisiae duo1p and dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 1998, 143, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Asbury, C.L.; Gestaut, D.R.; Powers, A.F.; Franck, A.D.; Davis, T.N. The dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA 2006, 103, 9873–9878. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, E.L.; Efremov, A.K.; Volkov, V.A.; Spiridonov, I.S.; Gudimchuk, N.; Westermann, S.; Drubin, D.; Barnes, G.; McIntosh, J.R.; Ataullakhanov, F.I. The dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA 2008, 105, 15423–15428. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, E.L.; Spiridonov, I.S.; Volkov, V.A.; Efremov, A.; Westermann, S.; Drubin, D.; Barnes, G.; Ataullakhanov, F.I.; McIntosh, J.R. Different assemblies of the dam1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA 2008, 105, 6918–6923. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Wang, H.W.; Avila-Sakar, A.; Drubin, D.G.; Nogales, E.; Barnes, G. The dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 2006, 440, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Tien, J.F.; Umbreit, N.T.; Gestaut, D.R.; Franck, A.D.; Cooper, J.; Wordeman, L.; Gonen, T.; Asbury, C.L.; Davis, T.N. Cooperation of the dam1 and ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora b. J. Cell Biol. 2010, 189, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Lampert, F.; Hornung, P.; Westermann, S. The dam1 complex confers microtubule plus end-tracking activity to the ndc80 kinetochore complex. J. Cell Biol. 2010, 189, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Anderson, S.; Jwa, M.; Green, E.M.; Kang, J.; Yates, J.R., 3rd; Chan, C.S.; Drubin, D.G.; Barnes, G. Phospho-regulation of kinetochore-microtubule attachments by the aurora kinase ipl1p. Cell 2002, 111, 163–172. [Google Scholar] [CrossRef]
- Chan, Y.W.; Jeyaprakash, A.A.; Nigg, E.A.; Santamaria, A. Aurora b controls kinetochore-microtubule attachments by inhibiting ska complex-kmn network interaction. J. Cell Biol. 2012, 196, 563–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redli, P.M.; Gasic, I.; Meraldi, P.; Nigg, E.A.; Santamaria, A. The ska complex promotes aurora b activity to ensure chromosome biorientation. J. Cell Biol. 2016, 215, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Jeyaprakash, A.A.; Santamaria, A.; Jayachandran, U.; Chan, Y.W.; Benda, C.; Nigg, E.A.; Conti, E. Structural and functional organization of the ska complex, a key component of the kinetochore-microtubule interface. Mol. Cell 2012, 46, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.A.; Medina, B.; Santamaria, A.; Zou, J.; Plasberg-Hill, C.; Madhumalar, A.; Jayachandran, U.; Redli, P.M.; Rappsilber, J.; Nigg, E.A.; et al. Structural basis for microtubule recognition by the human kinetochore ska complex. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.A.; Zou, J.; Medina-Pritchard, B.; Nigg, E.A.; Rappsilber, J.; Santamaria, A.; Jeyaprakash, A.A. Ska3 ensures timely mitotic progression by interacting directly with microtubules and ska1 microtubule binding domain. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.J.; De Wulf, P.; Sorger, P.K.; Harrison, S.C. The yeast dash complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005, 12, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Avila-Sakar, A.; Wang, H.W.; Niederstrasser, H.; Wong, J.; Drubin, D.G.; Nogales, E.; Barnes, G. Formation of a dynamic kinetochore- microtubule interface through assembly of the dam1 ring complex. Mol. Cell 2005, 17, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Ramey, V.H.; Wang, H.W.; Nakajima, Y.; Wong, A.; Liu, J.; Drubin, D.; Barnes, G.; Nogales, E. The dam1 ring binds to the e-hook of tubulin and diffuses along the microtubule. Mol. Biol. Cell 2011, 22, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Legal, T.; Zou, J.; Sochaj, A.; Rappsilber, J.; Welburn, J.P. Molecular architecture of the dam1 complex-microtubule interaction. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Ramey, V.H.; Westermann, S.; Leschziner, A.E.; Welburn, J.P.; Nakajima, Y.; Drubin, D.G.; Barnes, G.; Nogales, E. Architecture of the dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat. Struct. Mol. Biol. 2007, 14, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Ramey, V.H.; Wong, A.; Fang, J.; Howes, S.; Barnes, G.; Nogales, E. Subunit organization in the dam1 kinetochore complex and its ring around microtubules. Mol. Biol. Cell 2011, 22, 4335–4342. [Google Scholar] [CrossRef] [PubMed]
- Przewloka, M.R.; Venkei, Z.; Bolanos-Garcia, V.M.; Debski, J.; Dadlez, M.; Glover, D.M. Cenp-c is a structural platform for kinetochore assembly. Curr. Biol. 2011, 21, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Gascoigne, K.E.; Takeuchi, K.; Suzuki, A.; Hori, T.; Fukagawa, T.; Cheeseman, I.M. Induced ectopic kinetochore assembly bypasses the requirement for cenp-a nucleosomes. Cell 2011, 145, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Rago, F.; Gascoigne, K.E.; Cheeseman, I.M. Distinct organization and regulation of the outer kinetochore kmn network downstream of cenp-c and cenp-t. Curr. Biol. 2015, 25, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Rago, F.; Hori, T.; Tomii, K.; Cheeseman, I.M.; Fukagawa, T. Cenp-t provides a structural platform for outer kinetochore assembly. EMBO J. 2013, 32, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Schleiffer, A.; Maier, M.; Litos, G.; Lampert, F.; Hornung, P.; Mechtler, K.; Westermann, S. Cenp-t proteins are conserved centromere receptors of the ndc80 complex. Nat. Cell Biol. 2012, 14, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Welburn, J.P.; Vleugel, M.; Liu, D.; Yates, J.R., 3rd; Lampson, M.A.; Fukagawa, T.; Cheeseman, I.M. Aurora b phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 2010, 38, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyoshi, B.; Nelson, C.R.; Biggins, S. The aurora b kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics 2013, 194, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, F.; Ward, T.; Yan, F.; Wu, Q.; Wang, Z.; McGlothen, T.; Peng, W.; You, T.; Sun, M.; et al. Phosphorylation of hsmis13 by aurora b kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 2008, 283, 26726–26736. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, M.J.; Lan, W.; Jwa, M.; Miller, S.A.; Chan, C.S.; Stukenberg, P.T. Aurora b kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 2008, 181, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yu, H. Multiple assembly mechanisms anchor the kmn spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 2015, 208, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Badger, B.L.; Salmon, E.D. A quantitative description of ndc80 complex linkage to human kinetochores. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.J.; Pagliuca, C.; Kobayashi, N.; Grove, R.A.; Oku, Y.; Shrestha, K.; Alfieri, C.; Golfieri, C.; Oldani, A.; Dal Maschio, M.; et al. Cnn1 inhibits the interactions between the kmn complexes of the yeast kinetochore. Nat. Cell Biol. 2012, 14, 614–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, J.A.; Asbury, C.L.; Biggins, S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 2010, 468, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Gonen, S.; Akiyoshi, B.; Iadanza, M.G.; Shi, D.; Duggan, N.; Biggins, S.; Gonen, T. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 2012, 19, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Gascoigne, K.E.; Cheeseman, I.M. Cdk-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol. 2013, 201, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Mikami, Y.; Nishihashi, A.; Regnier, V.; Haraguchi, T.; Hiraoka, Y.; Sugata, N.; Todokoro, K.; Brown, W.; Ikemura, T. Cenp-h, a constitutive centromere component, is required for centromere targeting of cenp-c in vertebrate cells. EMBO J. 2001, 20, 4603–4617. [Google Scholar] [CrossRef] [PubMed]
- McClelland, S.E.; Borusu, S.; Amaro, A.C.; Winter, J.R.; Belwal, M.; McAinsh, A.D.; Meraldi, P. The cenp-a nac/cad kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007, 26, 5033–5047. [Google Scholar] [CrossRef] [PubMed]
- Hemmerich, P.; Weidtkamp-Peters, S.; Hoischen, C.; Schmiedeberg, L.; Erliandri, I.; Diekmann, S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008, 180, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.S.; Hori, T.; Okada, M.; Fukagawa, T. Cenp-c is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell 2007, 18, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- Eskat, A.; Deng, W.; Hofmeister, A.; Rudolphi, S.; Emmerth, S.; Hellwig, D.; Ulbricht, T.; Doring, V.; Bancroft, J.M.; McAinsh, A.D.; et al. Step-wise assembly, maturation and dynamic behavior of the human cenp-p/o/r/q/u kinetochore sub-complex. PLoS ONE 2012, 7, e44717. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, D.; Emmerth, S.; Ulbricht, T.; Doring, V.; Hoischen, C.; Martin, R.; Samora, C.P.; McAinsh, A.D.; Carroll, C.W.; Straight, A.F.; et al. Dynamics of cenp-n kinetochore binding during the cell cycle. J. Cell Sci. 2011, 124, 3871–3883. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, D.; Munch, S.; Orthaus, S.; Hoischen, C.; Hemmerich, P.; Diekmann, S. Live-cell imaging reveals sustained centromere binding of cenp-t via cenp-a and cenp-b. J. Biophotonics 2008, 1, 245–254. [Google Scholar] [CrossRef] [PubMed]
- McAinsh, A.D.; Meraldi, P.; Draviam, V.M.; Toso, A.; Sorger, P.K. The human kinetochore proteins nnf1r and mcm21r are required for accurate chromosome segregation. EMBO J. 2006, 25, 4033–4049. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Kimura, M.; Takagi, S.; Toramoto, I.; Ishihama, Y. Identification of mitosis-specific phosphorylation in mitotic chromosome-associated proteins. J Proteome Res 2016, 15, 3331–3341. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Hori, T.; Fukagawa, T.; Desai, A. Knl1 and the cenp-h/i/k complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell 2008, 19, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Venkei, Z.; Przewloka, M.R.; Ladak, Y.; Albadri, S.; Sossick, A.; Juhasz, G.; Novak, B.; Glover, D.M. Spatiotemporal dynamics of spc105 regulates the assembly of the drosophila kinetochore. Open Biol. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Magidson, V.; O’Connell, C.B.; Loncarek, J.; Paul, R.; Mogilner, A.; Khodjakov, A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 2011, 146, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Karess, R. Rod-zw10-zwilch: A key player in the spindle checkpoint. Trends Cell Biol. 2005, 15, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Civril, F.; Wehenkel, A.; Giorgi, F.M.; Santaguida, S.; Di Fonzo, A.; Grigorean, G.; Ciccarelli, F.D.; Musacchio, A. Structural analysis of the rzz complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure 2010, 18, 616–626. [Google Scholar] [CrossRef] [PubMed]
- McKenney, R.J.; Huynh, W.; Tanenbaum, M.E.; Bhabha, G.; Vale, R.D. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 2014, 345, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; Sohm, B.; Mikolcevic, P.; Wandke, C.; Rauch, V.; Ringer, T.; Hess, M.; Bonn, G.; Geley, S. Spindly/ccdc99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell 2010, 21, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.W.; Fava, L.L.; Uldschmid, A.; Schmitz, M.H.; Gerlich, D.W.; Nigg, E.A.; Santamaria, A. Mitotic control of kinetochore-associated dynein and spindle orientation by human spindly. J. Cell Biol. 2009, 185, 859–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gassmann, R.; Essex, A.; Hu, J.S.; Maddox, P.S.; Motegi, F.; Sugimoto, A.; O’Rourke, S.M.; Bowerman, B.; McLeod, I.; Yates, J.R., 3rd; et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: Spdl-1 and the rod/zwilch/zw10 complex. Genes Dev. 2008, 22, 2385–2399. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A.; Williams, B.C.; Hays, T.S.; Goldberg, M.L. Zw10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998, 142, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Griffis, E.R.; Stuurman, N.; Vale, R.D. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 2007, 177, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.G.; Watanabe, S.; Essex, A.; Kitagawa, R. Spdl-1 functions as a kinetochore receptor for mdf-1 in caenorhabditis elegans. J. Cell Biol. 2008, 183, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Reis, R.M.; Niessen, S.; Pereira, C.; Andres, D.A.; Spielmann, H.P.; Cleveland, D.W.; Desai, A.; Gassmann, R. Preventing farnesylation of the dynein adaptor spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol. Biol. Cell 2015, 26, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Moudgil, D.K.; Westcott, N.; Famulski, J.K.; Patel, K.; Macdonald, D.; Hang, H.; Chan, G.K. A novel role of farnesylation in targeting a mitotic checkpoint protein, human spindly, to kinetochores. J. Cell Biol. 2015, 208, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Hussein, D.; Taylor, S.S. Farnesylation of cenp-f is required for g2/m progression and degradation after mitosis. J. Cell Sci. 2002, 115, 3403–3414. [Google Scholar] [PubMed]
- Ashar, H.R.; James, L.; Gray, K.; Carr, D.; Black, S.; Armstrong, L.; Bishop, W.R.; Kirschmeier, P. Farnesyl transferase inhibitors block the farnesylation of cenp-e and cenp-f and alter the association of cenp-e with the microtubules. J. Biol. Chem. 2000, 275, 30451–30457. [Google Scholar] [CrossRef] [PubMed]
- Basto, R.; Scaerou, F.; Mische, S.; Wojcik, E.; Lefebvre, C.; Gomes, R.; Hays, T.; Karess, R. In vivo dynamics of the rough deal checkpoint protein during drosophila mitosis. Curr. Biol. 2004, 14, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Howell, B.J.; McEwen, B.F.; Canman, J.C.; Hoffman, D.B.; Farrar, E.M.; Rieder, C.L.; Salmon, E.D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001, 155, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Mische, S.; He, Y.; Ma, L.; Li, M.; Serr, M.; Hays, T.S. Dynein light intermediate chain: An essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell 2008, 19, 4918–4929. [Google Scholar] [CrossRef] [PubMed]
- Sivaram, M.V.; Wadzinski, T.L.; Redick, S.D.; Manna, T.; Doxsey, S.J. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 2009, 28, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Varma, D.; Monzo, P.; Stehman, S.A.; Vallee, R.B. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J. Cell Biol. 2008, 182, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.C.; Gatti, M.; Goldberg, M.L. Bipolar spindle attachments affect redistributions of zw10, a drosophila centromere/kinetochore component required for accurate chromosome segregation. J. Cell Biol. 1996, 134, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, E.; Basto, R.; Serr, M.; Scaerou, F.; Karess, R.; Hays, T. Kinetochore dynein: Its dynamics and role in the transport of the rough deal checkpoint protein. Nat. Cell Biol. 2001, 3, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, R.; Holland, A.J.; Varma, D.; Wan, X.; Civril, F.; Cleveland, D.W.; Oegema, K.; Salmon, E.D.; Desai, A. Removal of spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010, 24, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Buffin, E.; Lefebvre, C.; Huang, J.; Gagou, M.E.; Karess, R.E. Recruitment of mad2 to the kinetochore requires the rod/zw10 complex. Curr. Biol. 2005, 15, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Caldas, G.V.; Lynch, T.R.; Anderson, R.; Afreen, S.; Varma, D.; DeLuca, J.G. The rzz complex requires the n-terminus of knl1 to mediate optimal mad1 kinetochore localization in human cells. Open Biol. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.; Kim, Y.; Weaver, B.A.; Mao, Y.; McLeod, I.; Yates, J.R., 3rd; Tagaya, M.; Cleveland, D.W. Zw10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005, 169, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Matson, D.R.; Stukenberg, P.T. Cenp-i and aurora b act as a molecular switch that ties rzz/mad1 recruitment to kinetochore attachment status. J. Cell Biol. 2014, 205, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Silio, V.; McAinsh, A.D.; Millar, J.B. Knl1-bubs and rzz provide two separable pathways for checkpoint activation at human kinetochores. Dev. Cell 2015, 35, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Lischetti, T.; Hayward, D.G.; Nilsson, J. Distinct domains in bub1 localize rzz and bubr1 to kinetochores to regulate the checkpoint. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Zinkowski, R.P.; Meyne, J.; Brinkley, B.R. The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 1991, 113, 1091–1110. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Bouck, D.; Finley, K.; Liu, X.; Wan, Y.; Berman, J.; He, X.; Salmon, E.D.; Bloom, K.S. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008, 181, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Bloom, K.S.; Salmon, E.D. Point centromeres contain more than a single centromere-specific cse4 (cenp-a) nucleosome. J. Cell Biol. 2011, 195, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lando, D.; Endesfelder, U.; Berger, H.; Subramanian, L.; Dunne, P.D.; McColl, J.; Klenerman, D.; Carr, A.M.; Sauer, M.; Allshire, R.C.; et al. Quantitative single-molecule microscopy reveals that cenp-a(cnp1) deposition occurs during g2 in fission yeast. Open Biol. 2012, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blower, M.D.; Sullivan, B.A.; Karpen, G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2002, 2, 319–330. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Karpen, G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004, 11, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.A.; Vagnarelli, P.; Dong, Y.; Hori, T.; McEwen, B.F.; Fukagawa, T.; Flors, C.; Earnshaw, W.C. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA 2010, 107, 10484–10489. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Vasquez, P.A.; Falvo, M.R.; Taylor, R.M., 2nd; Vicci, L.; Yeh, E.; Forest, M.G.; Bloom, K. DNA loops generate intracentromere tension in mitosis. J. Cell Biol. 2015, 210, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Quammen, C.W.; Chang, B.; Haase, J.; Taylor, R.M., 2nd; Bloom, K. The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol. Biol. Cell 2013, 24, 3909–3919. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Haase, J.; Vicci, L.; Taylor, R.M., 2nd; Bloom, K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J. Cell Biol. 2011, 193, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, R.; Rechtsteiner, A.; Yuen, K.W.; Muroyama, A.; Egelhofer, T.; Gaydos, L.; Barron, F.; Maddox, P.; Essex, A.; Monen, J.; et al. An inverse relationship to germline transcription defines centromeric chromatin in c. Elegans. Nature 2012, 484, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Steiner, F.A.; Henikoff, S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife 2014, 3, e02025. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.S.; Portier, N.; Desai, A.; Oegema, K. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc. Natl. Acad. Sci. USA 2006, 103, 15097–15102. [Google Scholar] [CrossRef] [PubMed]
- Grenfell, A.W.; Heald, R.; Strzelecka, M. Mitotic noncoding rna processing promotes kinetochore and spindle assembly in xenopus. J. Cell Biol. 2016, 214, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Qu, Q.; Warrington, R.; Rice, A.; Cheng, N.; Yu, H. Mitotic transcription installs sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell 2015, 59, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Gent, J.I.; Dawe, R.K. Rna as a structural and regulatory component of the centromere. Annu. Rev. Genet. 2012, 46, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.L.; Marshall, O.J.; Saffery, R.; Kim, B.W.; Earle, E.; Choo, K.H.; Wong, L.H. Active transcription and essential role of rna polymerase ii at the centromere during mitosis. Proc. Natl. Acad. Sci. USA 2012, 109, 1979–1984. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, L.; Muller, S.; Liu, Y.; Huang, H.; Dingli, F.; Loew, D.; Vassias, I.; Patel, D.J.; Sullivan, K.F.; Almouzni, G. The cenp-t/-w complex is a binding partner of the histone chaperone fact. Genes Dev. 2016, 30, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Quenet, D.; Dalal, Y. A long non-coding rna is required for targeting centromeric protein a to the human centromere. eLife 2014, 3, e03254. [Google Scholar] [CrossRef] [PubMed]
- Rosic, S.; Kohler, F.; Erhardt, S. Repetitive centromeric satellite rna is essential for kinetochore formation and cell division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Catania, S.; Pidoux, A.L.; Allshire, R.C. Sequence features and transcriptional stalling within centromere DNA promote establishment of cenp-a chromatin. PLoS Genet. 2015, 11, e1004986. [Google Scholar] [CrossRef] [PubMed]
- Schuh, M.; Lehner, C.F.; Heidmann, S. Incorporation of drosophila cid/cenp-a and cenp-c into centromeres during early embryonic anaphase. Curr. Biol. 2007, 17, 237–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunleavy, E.M.; Almouzni, G.; Karpen, G.H. H3.3 is deposited at centromeres in s phase as a placeholder for newly assembled cenp-a in g(1) phase. Nucleus 2011, 2, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Pearson, C.G.; Yeh, E.; Gardner, M.; Odde, D.; Salmon, E.D.; Bloom, K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004, 14, 1962–1967. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Fujita, Y.; Iwasaki, O.; Adachi, Y.; Takahashi, K.; Yanagida, M. Mis16 and mis18 are required for cenp-a loading and histone deacetylation at centromeres. Cell 2004, 118, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Hayashi, T.; Kiyomitsu, T.; Toyoda, Y.; Kokubu, A.; Obuse, C.; Yanagida, M. Priming of centromere for cenp-a recruitment by human hmis18alpha, hmis18beta, and m18bp1. Dev. Cell 2007, 12, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Shiroiwa, Y.; Hayashi, T.; Fujita, Y.; Villar-Briones, A.; Ikai, N.; Takeda, K.; Ebe, M.; Yanagida, M. Mis17 is a regulatory module of the mis6-mal2-sim4 centromere complex that is required for the recruitment of cenh3/cenp-a in fission yeast. PLoS ONE 2011, 6, e17761. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.C.; Lin, Z.; Yuen, K.W. Rbap46/48(lin-53) is required for holocentromere assembly in caenorhabditis elegans. Cell Rep. 2016, 14, 1819–1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, L.; Medina-Pritchard, B.; Barton, R.; Spiller, F.; Kulasegaran-Shylini, R.; Radaviciute, G.; Allshire, R.C.; Arockia Jeyaprakash, A. Centromere localization and function of mis18 requires yippee-like domain-mediated oligomerization. EMBO Rep. 2016, 17, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Nardi, I.K.; Zasadzinska, E.; Stellfox, M.E.; Knippler, C.M.; Foltz, D.R. Licensing of centromeric chromatin assembly through the mis18alpha-mis18beta heterotetramer. Mol. Cell 2016, 61, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.S.; Hyndman, F.; Monen, J.; Oegema, K.; Desai, A. Functional genomics identifies a myb domain-containing protein family required for assembly of cenp-a chromatin. J. Cell Biol. 2007, 176, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Klare, K.; Petrovic, A.; Take, A.; Walstein, K.; Singh, P.; Rondelet, A.; Bird, A.W.; Musacchio, A. Cdk-regulated dimerization of m18bp1 on a mis18 hexamer is necessary for cenp-a loading. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Ebe, M.; Nagao, K.; Kokubu, A.; Sajiki, K.; Yanagida, M. Schizosaccharomyces pombe centromere protein mis19 links mis16 and mis18 to recruit cenp-a through interacting with nmd factors and the swi/snf complex. Genes Cells 2014, 19, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, L.; Toda, N.R.; Rappsilber, J.; Allshire, R.C. Eic1 links mis18 with the ccan/mis6/ctf19 complex to promote cenp-a assembly. Open Biol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, E.M.; Roche, D.; Tagami, H.; Lacoste, N.; Ray-Gallet, D.; Nakamura, Y.; Daigo, Y.; Nakatani, Y.; Almouzni-Pettinotti, G. Hjurp is a cell-cycle-dependent maintenance and deposition factor of cenp-a at centromeres. Cell 2009, 137, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Foltz, D.R.; Jansen, L.E.; Bailey, A.O.; Yates, J.R., 3rd; Bassett, E.A.; Wood, S.; Black, B.E.; Cleveland, D.W. Centromere-specific assembly of cenp-a nucleosomes is mediated by hjurp. Cell 2009, 137, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; Sanchez, P.; Rivera, T.; Rodriguez-Corsino, M.; Boyarchuk, E.; Vassias, I.; Ray-Gallet, D.; Arnaoutov, A.; Dasso, M.; Almouzni, G.; et al. Xenopus hjurp and condensin ii are required for cenp-a assembly. J. Cell Biol. 2011, 192, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Iyer, L.M.; Wu, C. Domain architectures of the scm3p protein provide insights into centromere function and evolution. Cell Cycle 2007, 6, 2511–2515. [Google Scholar] [CrossRef] [PubMed]
- Stoler, S.; Rogers, K.; Weitze, S.; Morey, L.; Fitzgerald-Hayes, M.; Baker, R.E. Scm3, an essential saccharomyces cerevisiae centromere protein required for g2/m progression and cse4 localization. Proc. Natl. Acad. Sci. USA 2007, 104, 10571–10576. [Google Scholar] [CrossRef] [PubMed]
- Camahort, R.; Li, B.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Gerton, J.L. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 2007, 26, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, A.L.; Choi, E.S.; Abbott, J.K.; Liu, X.; Kagansky, A.; Castillo, A.G.; Hamilton, G.L.; Richardson, W.; Rappsilber, J.; He, X.; et al. Fission yeast scm3: A cenp-a receptor required for integrity of subkinetochore chromatin. Mol. Cell 2009, 33, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, G.; Xiao, H.; Wisniewski, J.; Smith, M.M.; Wu, C. Nonhistone scm3 and histones cenh3-h4 assemble the core of centromere-specific nucleosomes. Cell 2007, 129, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Dechassa, M.L.; Bettini, E.; Ledoux, M.B.; Belisario, C.; Heun, P.; Luger, K.; Mellone, B.G. Cal1 is the drosophila cenp-a assembly factor. J. Cell Biol. 2014, 204, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Hayashi, T.; Yanagida, M.; Russell, P. Fission yeast scm3 mediates stable assembly of cnp1/cenp-a into centromeric chromatin. Mol. Cell 2009, 33, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pulido, L.; Pidoux, A.L.; Ponting, C.P.; Allshire, R.C. Common ancestry of the cenp-a chaperones scm3 and hjurp. Cell 2009, 137, 1173–1174. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, S.; Mellone, B.G.; Betts, C.M.; Zhang, W.; Karpen, G.H.; Straight, A.F. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008, 183, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Schittenhelm, R.B.; Althoff, F.; Heidmann, S.; Lehner, C.F. Detrimental incorporation of excess cenp-a/cid and cenp-c into drosophila centromeres is prevented by limiting amounts of the bridging factor cal1. J. Cell Sci. 2010, 123, 3768–3779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosin, L.; Mellone, B.G. Co-evolving cenp-a and cal1 domains mediate centromeric cenp-a deposition across drosophila species. Dev. Cell 2016, 37, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Zasadzinska, E.; Barnhart-Dailey, M.C.; Kuich, P.H.; Foltz, D.R. Dimerization of the cenp-a assembly factor hjurp is required for centromeric nucleosome deposition. EMBO J. 2013, 32, 2113–2124. [Google Scholar] [CrossRef] [PubMed]
- Shuaib, M.; Ouararhni, K.; Dimitrov, S.; Hamiche, A. Hjurp binds cenp-a via a highly conserved n-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. USA 2010, 107, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Cho, U.S.; Harrison, S.C. Recognition of the centromere-specific histone cse4 by the chaperone scm3. Proc. Natl. Acad. Sci. USA 2011, 108, 9367–9371. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Liu, Y.; Wang, M.; Fang, J.; Huang, H.; Yang, N.; Li, Y.; Wang, J.; Yao, X.; Shi, Y.; et al. Structure of a cenp-a-histone h4 heterodimer in complex with chaperone hjurp. Genes Dev. 2011, 25, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Feng, H.; Zhou, B.R.; Ghirlando, R.; Hu, K.; Zwolak, A.; Miller Jenkins, L.M.; Xiao, H.; Tjandra, N.; Wu, C.; et al. Structural basis for recognition of centromere histone variant cenh3 by the chaperone scm3. Nature 2011, 472, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Perpelescu, M.; Hori, T.; Toyoda, A.; Misu, S.; Monma, N.; Ikeo, K.; Obuse, C.; Fujiyama, A.; Fukagawa, T. Hjurp is involved in the expansion of centromeric chromatin. Mol. Biol. Cell 2015, 26, 2742–2754. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Dou, Z.; Chen, L.; Jiang, H.; Fu, C.; Fu, G.; Liu, D.; Zhang, J.; Zhu, T.; et al. Mitotic regulator mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (hjurp). J. Biol. Chem. 2014, 289, 8326–8336. [Google Scholar] [CrossRef] [PubMed]
- Moree, B.; Meyer, C.B.; Fuller, C.J.; Straight, A.F. Cenp-c recruits m18bp1 to centromeres to promote cenp-a chromatin assembly. J. Cell Biol. 2011, 194, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Stellfox, M.E.; Nardi, I.K.; Knippler, C.M.; Foltz, D.R. Differential binding partners of the mis18alpha/beta yippee domains regulate mis18 complex recruitment to centromeres. Cell Rep. 2016, 15, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Dambacher, S.; Deng, W.; Hahn, M.; Sadic, D.; Frohlich, J.; Nuber, A.; Hoischen, C.; Diekmann, S.; Leonhardt, H.; Schotta, G. Cenp-c facilitates the recruitment of m18bp1 to centromeric chromatin. Nucleus 2012, 3, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Chen, E.S.; Yanagida, M. Requirement of mis6 centromere connector for localizing a cenp-a-like protein in fission yeast. Science 2000, 288, 2215–2219. [Google Scholar] [CrossRef] [PubMed]
- Shono, N.; Ohzeki, J.; Otake, K.; Martins, N.M.; Nagase, T.; Kimura, H.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. Cenp-c and cenp-i are key connecting factors for kinetochore and cenp-a assembly. J. Cell Sci. 2015, 128, 4572–4587. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Bodor, D.L.; Stellfox, M.E.; Martins, N.M.; Hochegger, H.; Foltz, D.R.; Jansen, L.E. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 2012, 22, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhou, X.; Wang, W.; Deng, W.; Fang, J.; Hu, H.; Wang, Z.; Li, S.; Cui, L.; Shen, J.; et al. Dynamic phosphorylation of cenp-a at ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell 2015, 32, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Montes de Oca, R.; Lacoste, N.; Dingli, F.; Loew, D.; Almouzni, G. Phosphorylation and DNA binding of hjurp determine its centromeric recruitment and function in cenh3(cenp-a) loading. Cell Rep. 2014, 8, 190–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKinley, K.L.; Cheeseman, I.M. Polo-like kinase 1 licenses cenp-a deposition at centromeres. Cell 2014, 158, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Stankovic, A.; Guo, L.Y.; Mata, J.F.; Bodor, D.L.; Cao, X.J.; Bailey, A.O.; Shabanowitz, J.; Hunt, D.F.; Garcia, B.A.; Black, B.E.; et al. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted cenp-a assembly. Mol. Cell 2016. [Google Scholar] [CrossRef] [PubMed]
- Miell, M.D.; Straight, A.F. Regulating the timing of cenp-a nucleosome assembly by phosphorylation. Dev. Cell 2015, 32, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Deyter, G.M.; Biggins, S. The fact complex interacts with the e3 ubiquitin ligase psh1 to prevent ectopic localization of cenp-a. Genes Dev. 2014, 28, 1815–1826. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Bowers, S.; Lipinszki, Z.; Palladino, J.; Trusiak, S.; Bettini, E.; Rosin, L.; Przewloka, M.R.; Glover, D.M.; O’Neill, R.J.; et al. Establishment of centromeric chromatin by the cenp-a assembly factor cal1 requires fact-mediated transcription. Dev. Cell 2015, 34, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Lagana, A.; Dorn, J.F.; De Rop, V.; Ladouceur, A.M.; Maddox, A.S.; Maddox, P.S. A small gtpase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated cenp-a. Nat. Cell Biol. 2010, 12, 1186–1193. [Google Scholar] [CrossRef] [PubMed]
- Perpelescu, M.; Nozaki, N.; Obuse, C.; Yang, H.; Yoda, K. Active establishment of centromeric cenp-a chromatin by rsf complex. J. Cell Biol. 2009, 185, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Boltengagen, M.; Huang, A.; Boltengagen, A.; Trixl, L.; Lindner, H.; Kremser, L.; Offterdinger, M.; Lusser, A. A novel role for the histone acetyltransferase hat1 in the cenp-a/cid assembly pathway in Drosophila melanogaster. Nucleic Acids Res. 2016, 44, 2145–2159. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo-Lambert, A.; Hao, J.; Xiao, H.; Li, Y.; Han, Z.; Zhu, W. Acidic nucleoplasmic DNA-binding protein (and-1) controls chromosome congression by regulating the assembly of centromere protein a (cenp-a) at centromeres. J. Biol. Chem. 2013, 288, 1480–1488. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.S.; Stralfors, A.; Catania, S.; Castillo, A.G.; Svensson, J.P.; Pidoux, A.L.; Ekwall, K.; Allshire, R.C. Factors that promote h3 chromatin integrity during transcription prevent promiscuous deposition of cenp-a(cnp1) in fission yeast. PLoS Genet. 2012, 8, e1002985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamnun, Y.M.; Katayama, S.; Toda, T. Fission yeast mcl1 interacts with scf(pof3) and is required for centromere formation. Biochem. Biophys. Res. Commun. 2006, 350, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Sullivan, B.A.; Trazzi, S.; Della Valle, G.; Robertson, K.D. Dnmt3b interacts with constitutive centromere protein cenp-c to modulate DNA methylation and the histone code at centromeric regions. Hum. Mol. Genet. 2009, 18, 3178–3193. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Lee, M.; Park, K.C.; Jeon, Y.; Park, J.H.; Hwang, E.J.; Jeon, T.I.; Ko, S.; Lee, H.; Baek, S.H.; et al. Roles of mis18alpha in epigenetic regulation of centromeric chromatin and cenp-a loading. Mol. Cell 2012, 46, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Vernarecci, S.; Ornaghi, P.; Bagu, A.; Cundari, E.; Ballario, P.; Filetici, P. Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol. Cell. Biol. 2008, 28, 988–996. [Google Scholar] [CrossRef] [PubMed]
- Ohzeki, J.; Shono, N.; Otake, K.; Martins, N.M.; Kugou, K.; Kimura, H.; Nagase, T.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. Kat7/hbo1/myst2 regulates cenp-a chromatin assembly by antagonizing suv39h1-mediated centromere inactivation. Dev. Cell 2016, 37, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Rodriguez, M.G.; Martins, N.M.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.; Earnshaw, W.C. Epigenetic engineering shows h3k4me2 is required for hjurp targeting and cenp-a assembly on a synthetic human kinetochore. EMBO J. 2011, 30, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Ohzeki, J.; Bergmann, J.H.; Kouprina, N.; Noskov, V.N.; Nakano, M.; Kimura, H.; Earnshaw, W.C.; Larionov, V.; Masumoto, H. Breaking the hac barrier: Histone h3k9 acetyl/methyl balance regulates cenp-a assembly. EMBO J. 2012, 31, 2391–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raychaudhuri, N.; Dubruille, R.; Orsi, G.A.; Bagheri, H.C.; Loppin, B.; Lehner, C.F. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on cid/cenp-a presence in drosophila sperm. PLoS Biol. 2012, 10, e1001434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, J.E.; Woodlief, K.S.; Sullivan, B.A. Inheritance of the cenp-a chromatin domain is spatially and temporally constrained at human centromeres. Epigenet. Chromatin 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. Atp-driven exchange of histone h2az variant catalyzed by swr1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Podhraski, V.; Campo-Fernandez, B.; Worle, H.; Piatti, P.; Niederegger, H.; Bock, G.; Fyodorov, D.V.; Lusser, A. Cenh3/cid incorporation is not dependent on the chromatin assembly factor chd1 in drosophila. PLoS ONE 2010, 5, e10120. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Sylvester, J.E.; Gonzalez, I.L.; Costanzi, C.C.; Gillespie, D. Definition of a second dimeric subfamily of human alpha satellite DNA. Nucleic Acids Res. 1989, 17, 2769–2782. [Google Scholar] [CrossRef] [PubMed]
- Cellamare, A.; Catacchio, C.R.; Alkan, C.; Giannuzzi, G.; Antonacci, F.; Cardone, M.F.; Della Valle, G.; Malig, M.; Rocchi, M.; Eichler, E.E.; et al. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol. Biol. Evol. 2009, 26, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Camahort, R.; Shivaraju, M.; Mattingly, M.; Li, B.; Nakanishi, S.; Zhu, D.; Shilatifard, A.; Workman, J.L.; Gerton, J.L. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 2009, 35, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, B.; Gull, K. Discovery of unconventional kinetochores in kinetoplastids. Cell 2014, 156, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- D’Archivio, S.; Wickstead, B. Trypanosome outer kinetochore proteins suggest conservation of chromosome segregation machinery across eukaryotes. J. Cell Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Monen, J.; Maddox, P.S.; Hyndman, F.; Oegema, K.; Desai, A. Differential role of cenp-a in the segregation of holocentric c. Elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 2005, 7, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
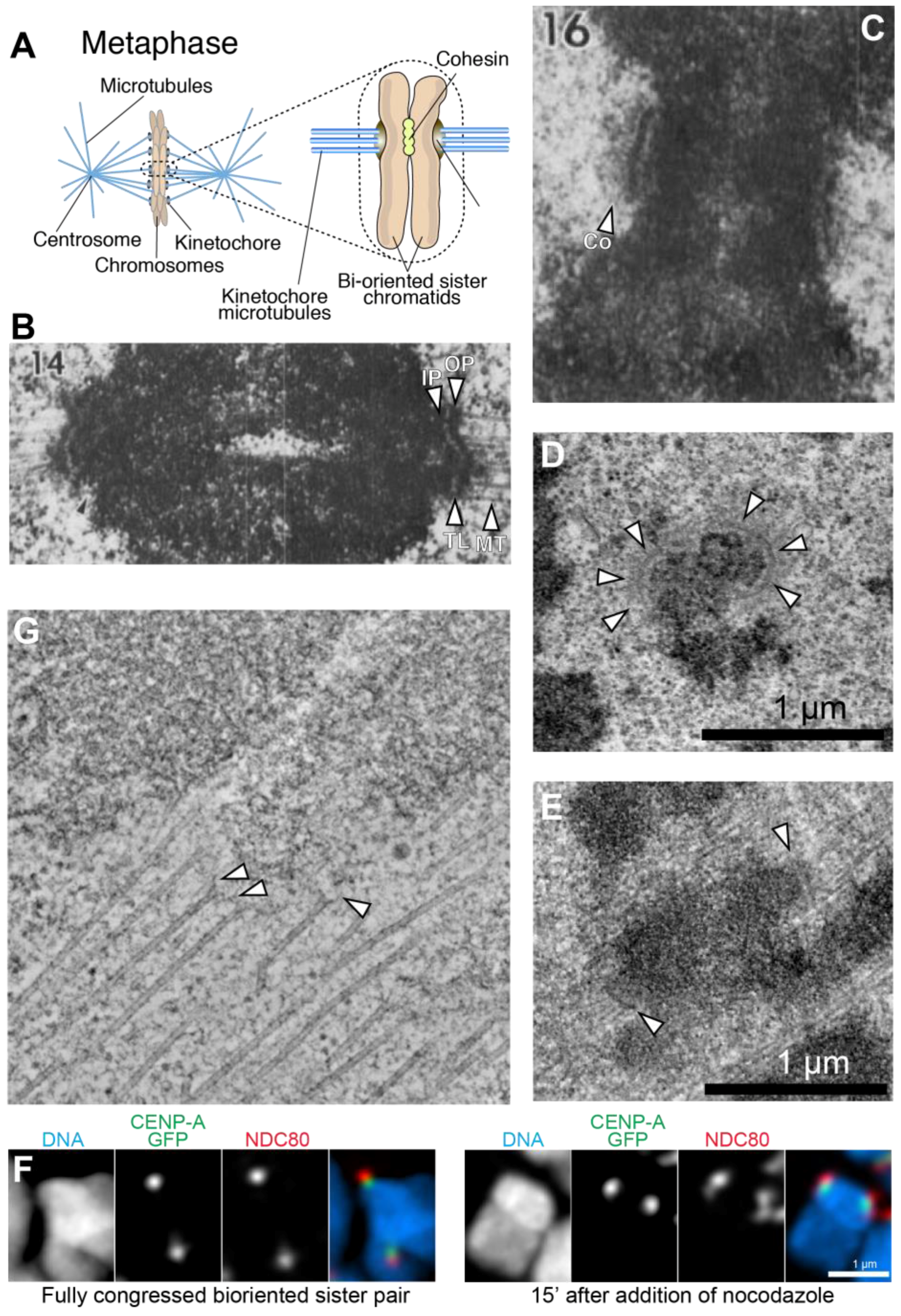

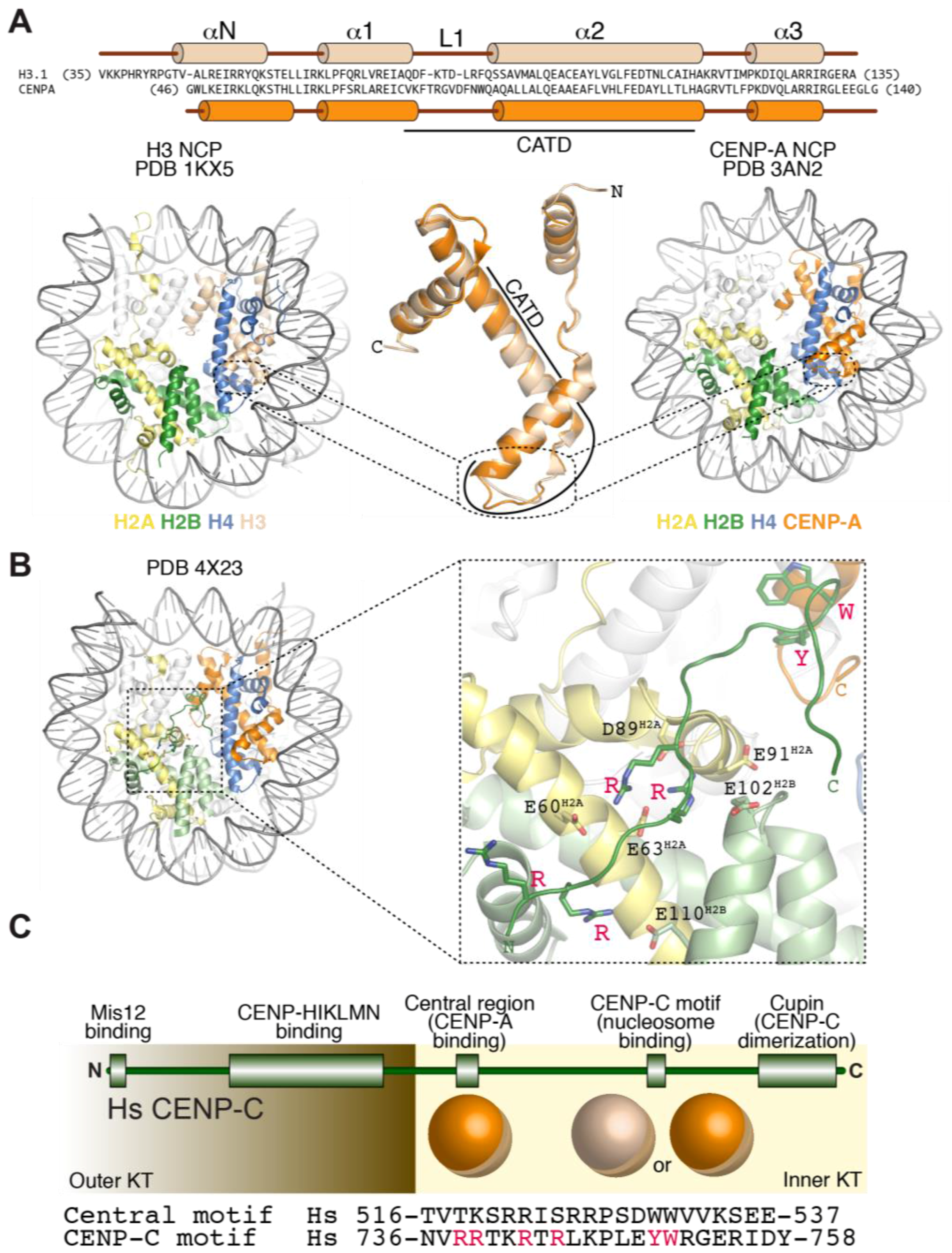
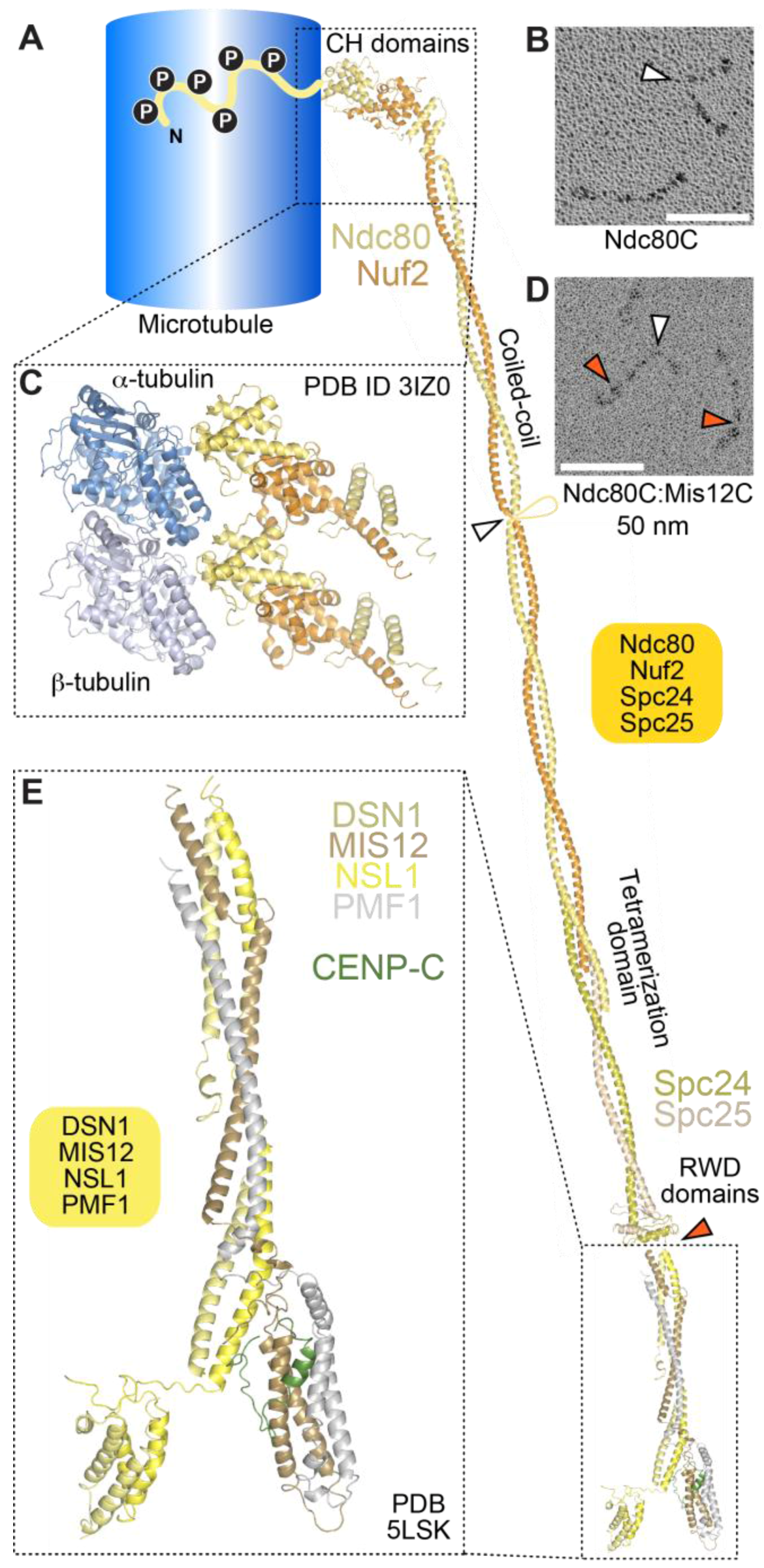
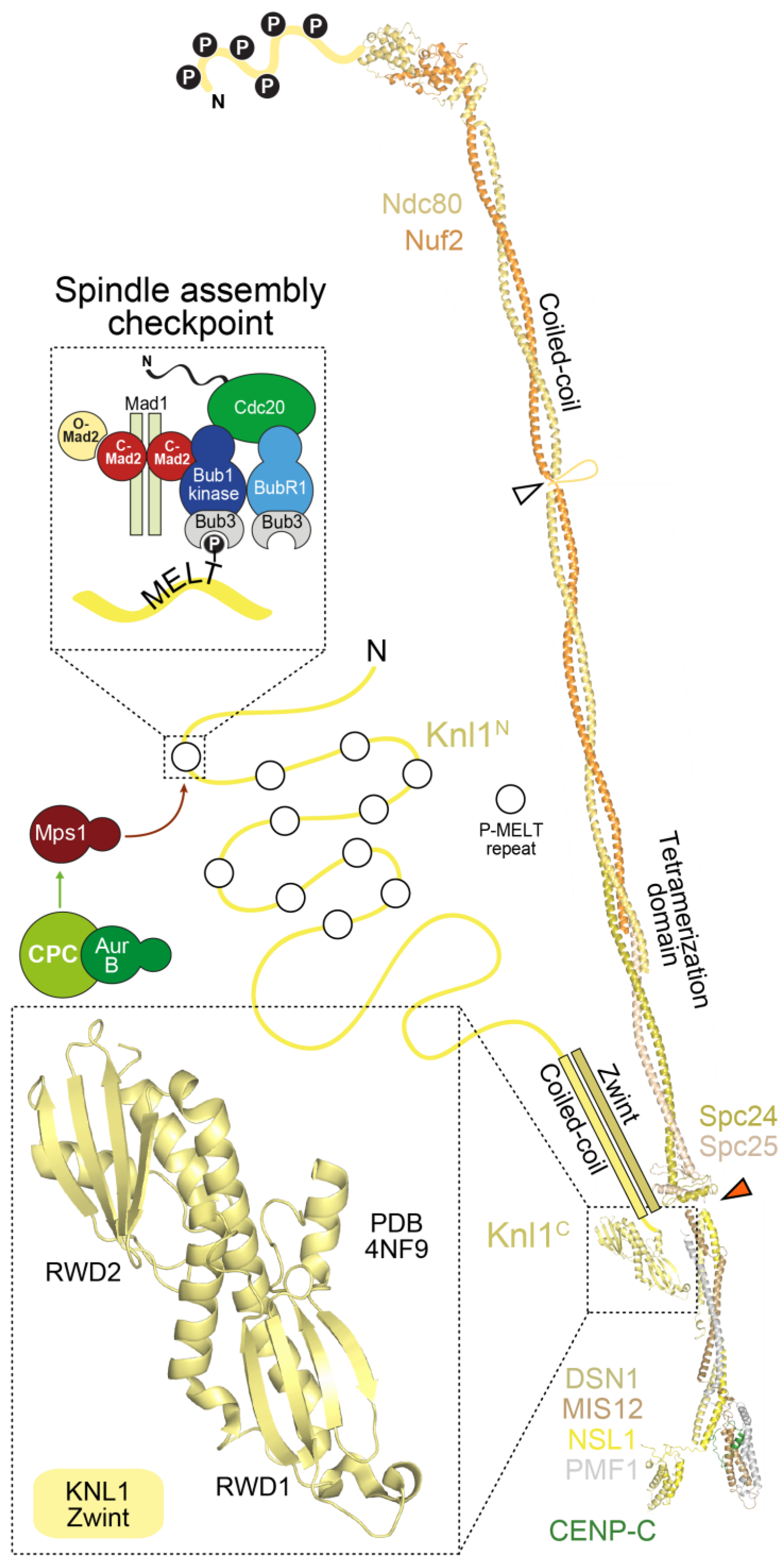



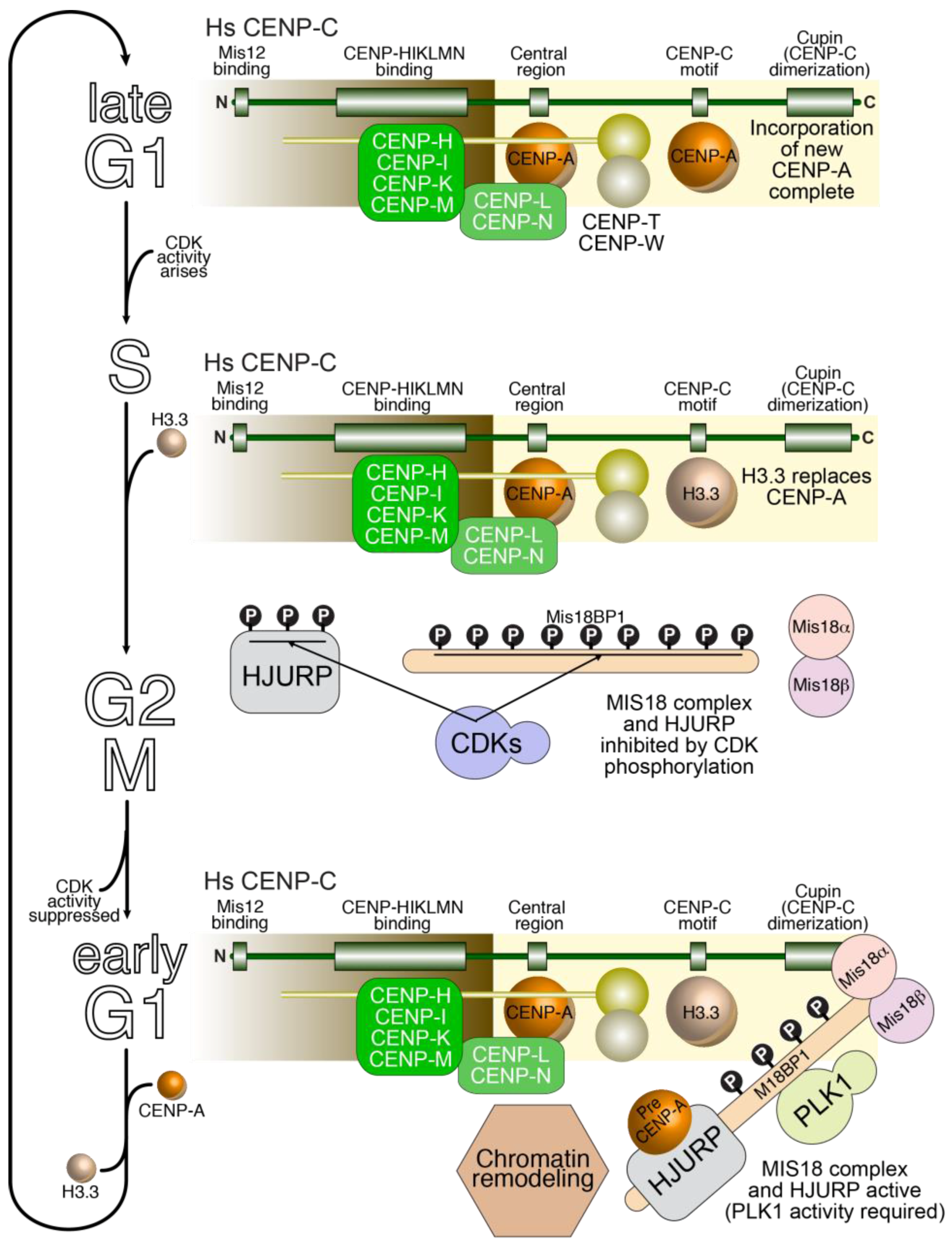
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musacchio, A.; Desai, A. A Molecular View of Kinetochore Assembly and Function. Biology 2017, 6, 5. https://doi.org/10.3390/biology6010005
Musacchio A, Desai A. A Molecular View of Kinetochore Assembly and Function. Biology. 2017; 6(1):5. https://doi.org/10.3390/biology6010005
Chicago/Turabian StyleMusacchio, Andrea, and Arshad Desai. 2017. "A Molecular View of Kinetochore Assembly and Function" Biology 6, no. 1: 5. https://doi.org/10.3390/biology6010005





