Functional Analysis of Cotton Leaf Curl Kokhran Virus/Cotton Leaf Curl Multan Betasatellite RNA Silencing Suppressors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of Constructs
2.2. Plants and Plant Transformation
2.3. Agro-Infiltration Patch Assay
2.4. Expression and Purification of Recombinant Proteins
2.5. Electrophoretic Mobility Shift Assay (EMSA)
2.6. SDS-PAGE
2.7. Western Blotting
3. Results
3.1. Suppression of S-PTGS

3.2. Suppression of IR-PTGS
3.3. Effects of Virus Proteins on Systemic Silencing
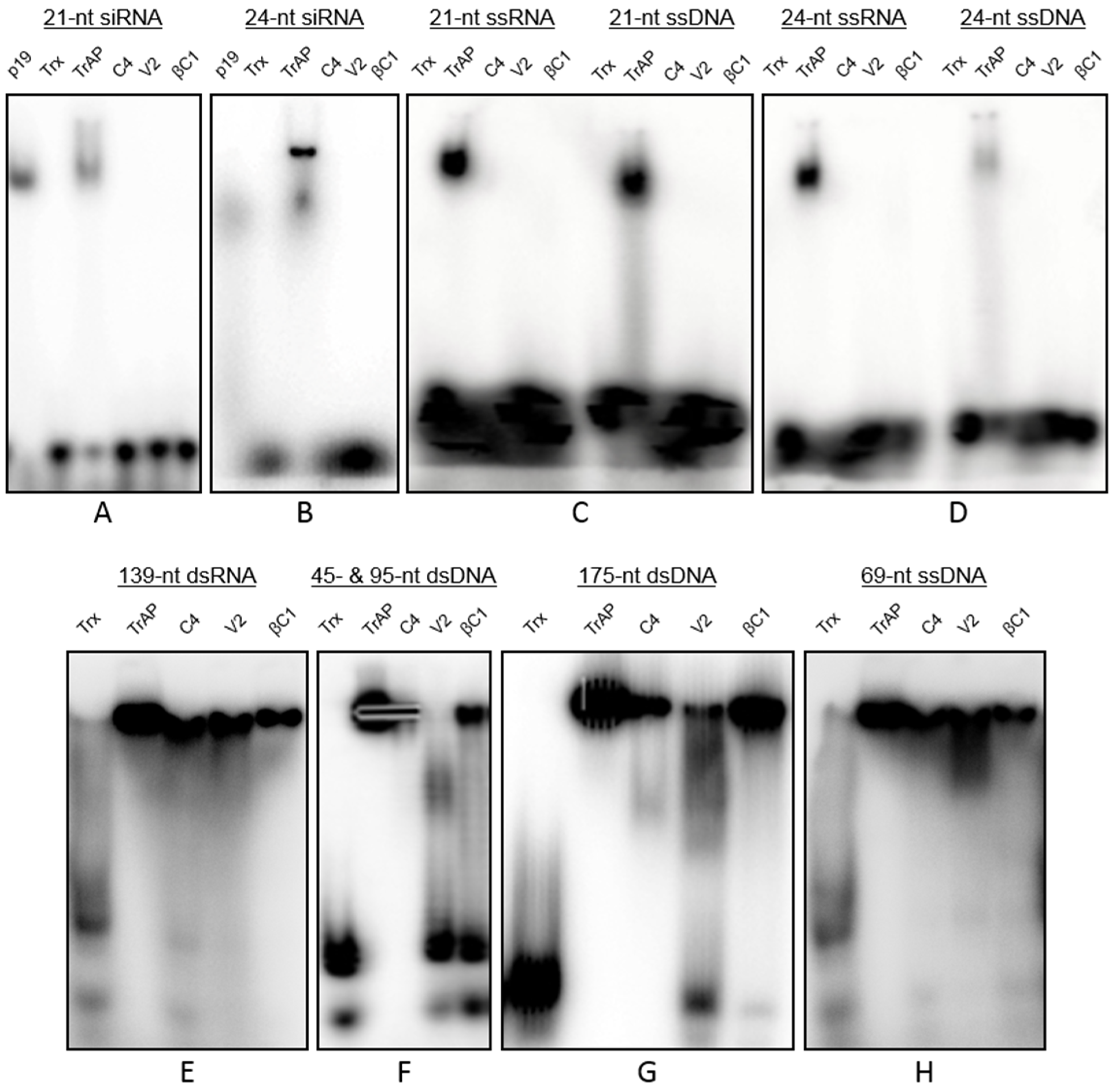
3.4. Interactions of Proteins with Nucleic Acids
3.5. Effects of Transgenic Expression of Protein in Plants

| Inoculum | No. of plant systemically silenced/no. of plants inoculated | Silencing(%) |
|---|---|---|
| GFP+GUS | 27/49 | 55 C |
| GFP+p19 | 2/27 | 7 A |
| GFP+TrAP | 11/27 | 41 B |
| GFP+C4 | 1/29 | 3 A |
| GFP+V2 | 15/27 | 56 B |
| GFP+ βC1 | 30/44 | 68 C |
| IR-GFP+GUS | 28/48 | 58 C |
| IR-GFP+p19 | 2/28 | 7 A |
| IR-GFP+TrAP | 13/27 | 48 B |
| IR-GFP++C4 | 2/30 | 6 A |
| IR-GFP+V2 | 16/26 | 62 B |
| IR-GFP+βC1 | 32/44 | 73 C |
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wassenegger, M.; Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006, 11, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Matthew, L.; Smith, N.A.; Curtin, S.J.; Dedic-Hagan, J.; Ellacott, G.A.; Watson, J.M.; Wang, M.B.; Brosnan, C.; Carroll, B.J.; et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006, 7, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ebright, Y.W.; Yu, B.; Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2' OH of the 3' terminal nucleotide. Nucleic Acids Res. 2006, 34, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N. Sorting out small RNAs. Cell 2008, 133, 25–26. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M.; Heimes, S.; Riedel, L.; Sanger, H.L. RNA-directed de novo methylation of genomic sequences in plants. Cell 1994, 76, 567–576. [Google Scholar] [CrossRef]
- Moissiard, G.; Parizotto, E.A.; Himber, C.; Voinnet, O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 2007, 13, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C. Amplified silencing. Science 2007, 315, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Aregger, M.; Zvereva, A.S.; Borah, B.K.; Gubaeva, E.G.; Pooggin, M.M. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 2012, 40, 6241–6254. [Google Scholar] [CrossRef] [PubMed]
- Rajeswaran, R.; Pooggin, M.M. RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 2011, 40, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Jones, L.; Baulcombe, D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 2002, 14, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Himber, C.; Ruiz-Ferrer, V.; Alioua, A.; Voinnet, O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007, 39, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Schott, G.; Himber, C.; Meyer, D.; Takeda, A.; Carrington, J.C.; Voinnet, O. Small RNA duplexes function as mobile silencing signals between plant cells. Science 2010, 328, 912–916. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, C.A.; Voinnet, O. Cell-to-cell and long-distance siRNA movement in plants: Mechanisms and biological implications. Curr. Opin. Plant Biol. 2011, 14, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Voinnet, O. Mixing and matching: The essence of plant systemic silencing? Trends Genet. 2008, 24, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, C.W.; Molnar, A.; Baulcombe, D.C. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011, 30, 3553–3563. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Melgarejo, T.A.; Kon, T.; Rojas, M.R.; Paz-Carrasco, L.; Zerbini, F.M.; Gilbertson, R.L. Characterization of a new world monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J. Virol. 2013, 87, 5397–5413. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Ikegami, M. RNA-silencing suppressors of geminiviruses. J. Gen. Plant Pathol. 2008, 74, 189–202. [Google Scholar] [CrossRef]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Stanley, J. Sub-viral agents associated with plant single-stranded DNA viruses. Virology 2006, 344, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Advances in understanding begomovirus satellites. Ann. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant-Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Qazi, J.; Amin, I.; Mansoor, S.; Iqbal, J.; Briddon, R.W. Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res. 2007, 128, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease—An emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Briddon, R.W.; Bull, S.E.; Bedford, I.D.; Bashir, A.; Hussain, M.; Saeed, M.; Zafar, M.Y.; Malik, K.A.; Fauquet, C.; et al. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 2003, 148, 1969–1986. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Saunders, K.; Stanley, J.; Zafar, Y.; Malik, K.A.; Markham, P.G. Identification of DNA components required for induction of cotton leaf curl disease. Virology 2001, 285, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Galetzka, D.; Russo, M.; Rubino, L.; Krczal, G. Molecular characterization of a tombusvirus associated with disease of statice (Goniolimon tataricum (L.) boiss.). J. Plant Pathol. 2000, 82, 151–155. [Google Scholar]
- Aoyama, T.; Chua, N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997, 11, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Vogt, U.; Pélissier, T.; Pütz, A.; Razvi, F.; Fischer, R.; Wassenegger, M. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 2004, 38, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Moser, M.; Zwiebel, M.; Krczal, G.; Hell, R.; Wassenegger, M. A hairpin RNA construct residing in an intron efficiently triggered RNA-directed DNA methylation in tobacco. Plant J. 2009, 60, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.T.; Voinnet, O.; Baulcombe, D.C. Initiation and maintenance of virus-induced gene silencing. Plant Cell 1998, 10, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Fuellgrabe, M.W.; Boonrod, K.; Jamous, R.; Moser, M.; Shiboleth, Y.; Krczal, G.; Wassenegger, M. Expression, purification and functional characterization of recombinant Zucchini yellow mosaic virus HC-Pro. Protein Expr. Purif. 2011, 75, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Weinheimer, I.; Boonrod, K.; Moser, M.; Zwiebel, M.; Fullgrabe, M.; Krczal, G.; Wassenegger, M. Analysis of an autoproteolytic activity of Rice yellow mottle virus silencing suppressor P1. Biol. Chem. 2010, 391, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Baulcombe, D.C. Systemic signalling in gene silencing. Nature 1997, 389, 553–553. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, L.; Szittya, G.; Silhavy, D.; Burgyán, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004, 23, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Voinnet, O.; Chappell, L.; Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002, 21, 4671–4679. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Vain, P.; Angell, S.; Baulcombe, D.C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 1998, 95, 177–187. [Google Scholar] [CrossRef]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyán, J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.-P.; Dolja, V.V.; Calvino, L.F.; López-Moya, J.J.; Burgyán, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef] [PubMed]
- Vanitharani, R.; Chellappan, P.; Pita, J.S.; Fauquet, C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004, 78, 9487–9498. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Fauquet, C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA 2005, 102, 10381–10386. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Patil, B.L.; Briddon, R.W.; Mansoor, S.; Fauquet, C.M. Comparison of phenotypes produced in response to transient expression of genes encoded by four distinct begomoviruses in Nicotiana benthamiana and their correlation with the levels of developmental miRNAs. Virol. J. 2011. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Krczal, G.; Wassenegger, M. Three gene products of a begomovirus-betasatellite complex restore expression of a transcriptionally silenced green fluorescent protein transgene in Nicotiana benthamiana. Virus Genes 2015, 50, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Noris, E.; Jupin, I.; Accotto, G.P.; Gronenborn, B. DNA-binding activity of the AC2 protein of tomato yellow leaf curl geminivirus. Virology 1996, 217, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.K.; Coutts, R.H.A. Potato yellow mosaic geminivirus AC2 protein is a sequence non-specific DNA binding protein. FEBS Lett. 1996, 383, 51–54. [Google Scholar] [CrossRef]
- Hartitz, M.D.; Sunter, G.; Bisaro, D.M. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 1999, 263, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Trinks, D.; Rajeswaran, R.; Shivaprasad, P.V.; Akbergenov, R.; Oakeley, E.J.; Veluthambi, K.; Hohn, T.; Pooggin, M.M. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005, 79, 2517–2527. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.Y.; Sunter, G. Interaction between the transcription factor AtTIFY4B and begomovirus AL2 protein impacts pathogenicity. Plant Mol. Biol. 2014, 86, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cázares, B.; Sudarshana, M.R.; Lucas, W.J.; Gilbertson, R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 2001, 291, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Hak, H.; Levy, Y.; Chandran, S.A.; Belausov, E.; Loyter, A.; Lapidot, M.; Gafni, Y. TYLCV-IS movement in planta does not require V2 protein. Virology 2015, 477c, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Sharma, P.K.; Malathi, V.G. Functional characterization of βC1 gene of Cotton leaf curl multan betasatellite. Virus Genes 2013, 46, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Zafar, Y.; Randles, J.W.; Rezaian, M.A. A monopartite begomovirus-associated DNA B satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007, 88, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Dogra, S.C.; Dry, I.B.; Randles, J.W. Silencing suppressor activity of a begomovirus DNA β encoded protein and its effect on heterologous helper virus replication. Virus Res. 2012, 167, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Canizares, M.C.; Lozano-Duran, R.; Canto, T.; Bejarano, E.R.; Bisaro, D.M.; Navas-Castillo, J.; Moriones, E. Effects of the crinivirus coat protein-interacting plant protein SAHH on post-transcriptional RNA silencing and its suppression. Mol. Plant Microbe Interact. 2013, 26, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Jackel, J.N.; Buchmann, R.C.; Singhal, U.; Bisaro, D.M. Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J. Virol. 2015, 89, 3176–3187. [Google Scholar] [CrossRef] [PubMed]
- Zrachya, A.; Glick, E.; Levy, Y.; Arazi, T.; Citovsky, V.; Gafni, Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 2007, 358, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Himber, C.; Dunoyer, P.; Moissiard, G.; Ritzenthaler, C.; Voinnet, O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003, 22, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Negrete, E.; Lozano-Durán, R.; Piedra-Aguilera, A.; Cruzado, L.; Bejarano, E.R.; Castillo, A.G. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013, 199, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dang, M.; Hou, H.; Mei, Y.; Qian, Y.; Zhou, X. Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 2014, 95, 2082–2088. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeed, M.; Briddon, R.W.; Dalakouras, A.; Krczal, G.; Wassenegger, M. Functional Analysis of Cotton Leaf Curl Kokhran Virus/Cotton Leaf Curl Multan Betasatellite RNA Silencing Suppressors. Biology 2015, 4, 697-714. https://doi.org/10.3390/biology4040697
Saeed M, Briddon RW, Dalakouras A, Krczal G, Wassenegger M. Functional Analysis of Cotton Leaf Curl Kokhran Virus/Cotton Leaf Curl Multan Betasatellite RNA Silencing Suppressors. Biology. 2015; 4(4):697-714. https://doi.org/10.3390/biology4040697
Chicago/Turabian StyleSaeed, Muhammad, Rob W. Briddon, Athanasios Dalakouras, Gabi Krczal, and Michael Wassenegger. 2015. "Functional Analysis of Cotton Leaf Curl Kokhran Virus/Cotton Leaf Curl Multan Betasatellite RNA Silencing Suppressors" Biology 4, no. 4: 697-714. https://doi.org/10.3390/biology4040697





