Exploratory Spatial Mapping of the Occurrence of Antimicrobial Resistance in E. coli in the Community
Abstract
:1. Introduction
2. Results
2.1. Descriptives
| N | % | |
|---|---|---|
| Female | 682 | 91% |
| Nursing home residents | 70 | 9% |
| mean | SD | |
| Age | 56 | (±21) |
2.2. Distribution of UTI Cases
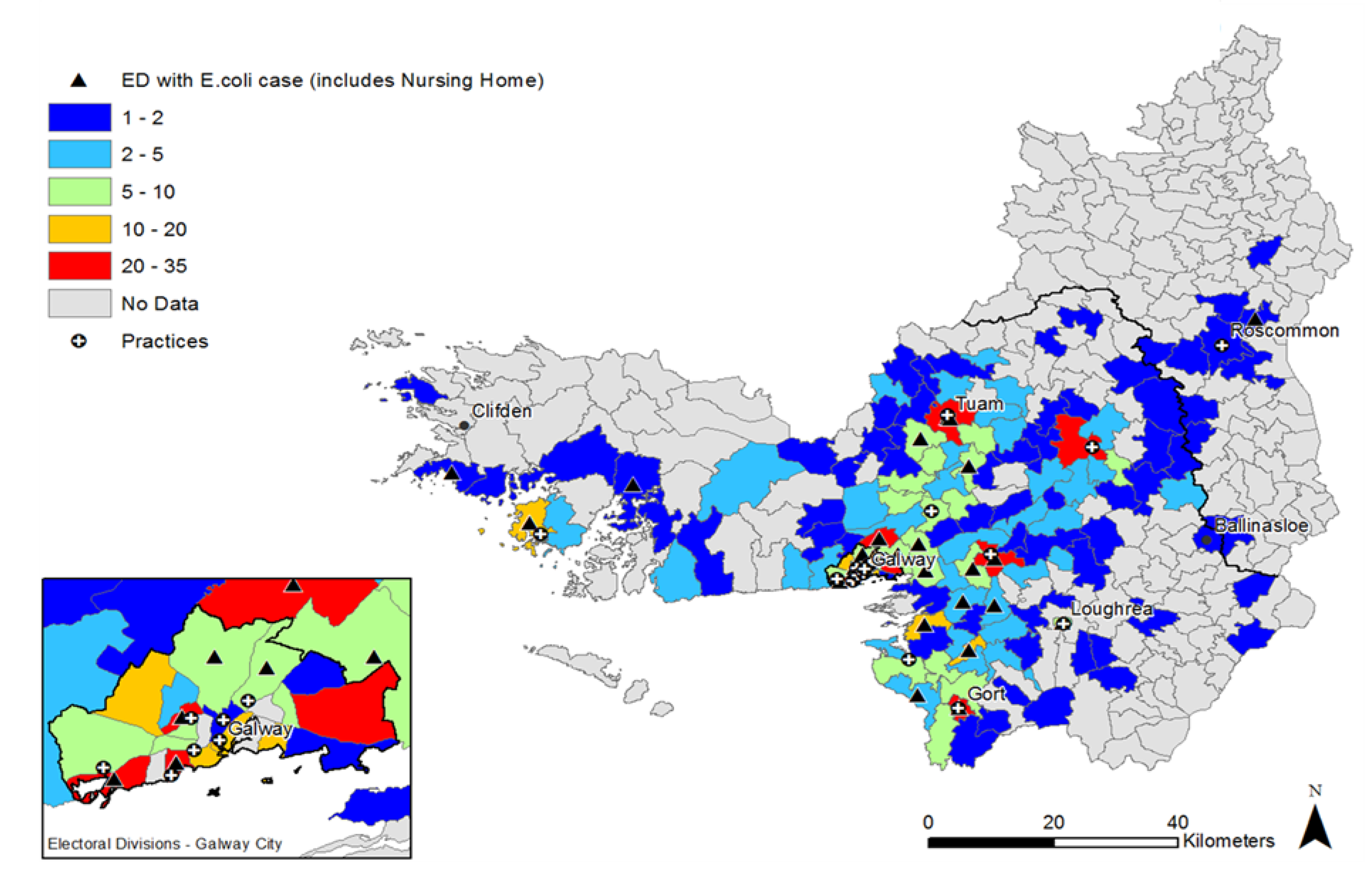
2.3. Antimicrobial Resistance Patterns
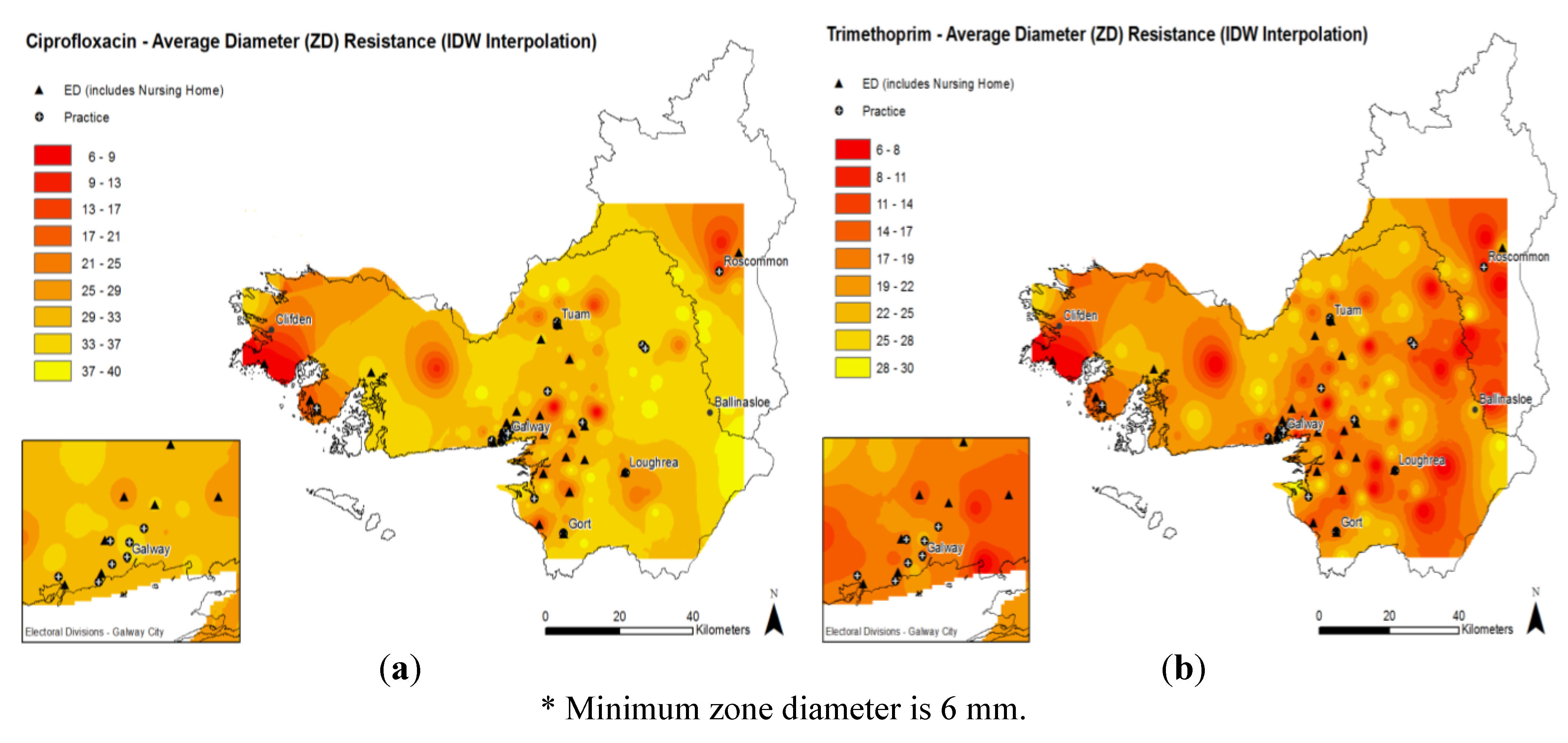

3. Discussion
4. Experimental
4.1. Patient Selection
4.2. Data Collection
4.3. Geocoding, Mapping and Cartographic Displays
5. Conclusions
Acknowledgements
Conflict of Interest
Ethical Approval
References
- Wise, R.; Hart, T.; Cars, O.; Streulens, M.; Helmuth, R.; Huovinen, P.; Sprenger, M. Antimicrobial resistance is a major threat to public health. Br. Med. J. 1998, 317, 609–610. [Google Scholar] [CrossRef]
- Cormican, M.; Vellinga, A. Existing classes of antibiotics are probably the best we will ever have. Br. Med. J. 2012, 344, e3369. [Google Scholar] [CrossRef]
- Vellinga, A.; Tansey, S.; Hanahoe, B.; Bennett, K.; Murphy, A.W.; Cormican, M. Trimethoprim and ciprofloxacin resistance and prescribing in urinary tract infection associated with Escherichia coli: A multilevel model. J. Antimicrob. Chemother. 2012, 67, 2523–2530. [Google Scholar]
- Sannes, M.R.; Kuskowski, M.A.; Johnson, J.R. Geographical distribution of antimicrobial resistance among Escherichia coli causing acute uncomplicated pyelonephritis in the united states. FEMS Immunol. Med. Microbiol. 2004, 42, 213–218. [Google Scholar] [CrossRef]
- McCormick, A.W.; Whitney, C.G.; Farley, M.M.; Lynfield, R.; Harrison, L.H.; Bennett, N.M.; Schaffner, W.; Reingold, A.; Hadler, J.; Cieslak, P.; et al. Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the united states. Nat. Med. 2003, 9, 424–430. [Google Scholar]
- Kalter, H.D.; Gilman, R.H.; Moulton, L.H.; Cullotta, A.R.; Cabrera, L.; Velapatino, B. Risk factors for antibiotic-resistant Escherichia coli carriage in young children in Peru: Community-based cross-sectional prevalence study. Am. J. Trop. Med. Hyg. 2010, 82, 879–888. [Google Scholar]
- Magee, J.T.; Heginbothom, M.L.; Mason, B.W. Finding a strategy: The case for co-operative research on resistance epidemiology. J. Antimicrob. Chemother. 2005, 55, 628–633. [Google Scholar] [CrossRef]
- Lipsitch, M.; Samore, M.H. Antimicrobial use and antimicrobial resistance: A population perspective. Emerg. Infect. Dis. 2002, 8, 347–354. [Google Scholar]
- ECDC Ears-net Database. Available online: http://ecdc.europa.eu/en/activities/surveillance/EARS-net/database/Pages/database.aspx (accessed on 5 April 2013).
- European Surveillance of Antimicrobial Consumption (ESAC) database. Available online: http://app.esac.ua.ac.be/public/ (accessed on 5 April 2013).
- Carey, B.; Murphy, M.; Bradley, C.; Cunney, R.; Byrne, S.; O’Connor, N.; Sheehan, A. Guidelines for Antimicrobial Prescribing in Primary Care in Ireland; Health Protection Surveillance Centre: Dublin, Ireland, 2011; p. 24. [Google Scholar]
- Perez-Trallero, E.; Garcia-de-la-Fuente, C.; Garcia-Rey, C.; Baquero, F.; Aguilar, L.; Dal-Re, R.; Garcia-de-Lomas, J. Spanish surveillance group for respiratory, geographical and ecological analysis of resistance, coresistance, and coupled resistance to antimicrobials in respiratory pathogenic bacteria in spain. Antimicrob. Agents Chemother. 2005, 49, 1965–1972. [Google Scholar] [CrossRef]
- Samore, M.H.; Magill, M.K.; Alder, S.C.; Severina, E.; Morrison-De Boer, L.; Lyon, J.L.; Carroll, K.; Leary, J.; Stone, M.B.; Bradford, D.; et al. High rates of multiple antibiotic resistance in Streptococcus pneumoniae from healthy children living in isolated rural communities: Association with cephalosporin use and intrafamilial transmission. Pediatrics 2001, 108, 856–865. [Google Scholar] [CrossRef]
- Carmeli, Y.; Castro, J.; Eliopoulos, G.M.; Samore, M.H. Clinical isolation and resistance patterns of and superinfection with 10 nosocomial pathogens after treatment with ceftriaxone versus ampicillin-sulbactam. Antimicrob. Agents Chemother. 2001, 45, 275–279. [Google Scholar] [CrossRef]
- Masiero, G.; Filippini, M.; Ferech, M.; Goossens, H. Socioeconomic determinants of outpatient antibiotic use in Europe. Int. J. Public Health. 2010, 55, 469–478. [Google Scholar] [CrossRef]
- Samarasundera, E.; Walsh, T.; Cheng, T.; Koenig, A.; Jattansingh, K.; Dawe, A.; Soljak, M. Methods and tools for geographical mapping and analysis in primary health care. Prim. Health Care Res. Dev. 2012, 13, 10–21. [Google Scholar] [CrossRef]
- Noble, D.; Smith, D.; Mathur, R.; Robson, J.; Greenhalgh, T. Feasibility study of geospatial mapping of chronic disease risk to inform public health commissioning. Br. Med. J. Open 2012, 2, e000711. [Google Scholar]
- Callaghan, M.; Cormican, M.; Prendergast, M.; Pelly, H.; Cloughley, R.; Hanahoe, B.; O'Donovan, D. Temporal and spatial distribution of human cryptosporidiosis in the west of ireland 2004–2007. Int. J. Health Geogr. 2009, 8, e64. [Google Scholar] [CrossRef]
- McGrath, G.; Abernethy, D.; Stringer, L.; More, S. An all-island approach to mapping bovine tuberculosis in ireland. Irish Vet. J. 2009, 62, 192–197. [Google Scholar]
- Tirabassi, M.V.; Wadie, G.; Moriarty, K.P.; Garb, J.; Konefal, S.H.; Courtney, R.A.; Sachs, B.F.; Wait, R. Geographic information system localization of community-acquired MRSA soft tissue abscesses. J. Ped. Surg. 2005, 40, 962–965; discussion 965–966. [Google Scholar] [CrossRef]
- Mouro, A.; Kiffer, C.; Koga, P.C.; Monteiro, A.M.; Camargo, E.C.; Pignatari, A.C. Spatial exploration of Streptococcus pneumoniae clonal clustering in Sao Paulo, Brazil. Braz. J. Infect. Dis. 2011, 15, 462–466. [Google Scholar] [CrossRef]
- Kiffer, C.R.; Camargo, E.C.; Shimakura, S.E.; Ribeiro, P.J., Jr.; Bailey, T.C.; Pignatari, A.C.; Monteiro, A.M. A spatial approach for the epidemiology of antibiotic use and resistance in community-based studies: The emergence of urban clusters of Escherichia coli quinolone resistance in Sao Paulo, Brasil. Int. J. Health Geogr. 2011, 10, e17. [Google Scholar]
- Vellinga, A.; Cormican, M.; Hanahoe, B.; Bennett, K.; Murphy, A.W. Antimicrobial management and appropriateness of treatment of urinary tract infection in general practice in ireland. BMC Fam Pract. 2011, 12, e108. [Google Scholar] [CrossRef]
- Vellinga, A.; Cormican, M.; Hanahoe, B.; Bennett, K.; Murphy, A.W. Opt-out as an acceptable method of obtaining consent in medical research: A short report. BMC Med. Res. Meth. 2011, 11, e40. [Google Scholar] [CrossRef]
- Central Statistics Office. Available online: http://www.cso.ie/en/index.html (accessed on 6 February 2013).
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) Clinical breakpoints. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 15 September 2012).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Galvin, S.; Bergin, N.; Hennessy, R.; Hanahoe, B.; Murphy, A.W.; Cormican, M.; Vellinga, A. Exploratory Spatial Mapping of the Occurrence of Antimicrobial Resistance in E. coli in the Community. Antibiotics 2013, 2, 328-338. https://doi.org/10.3390/antibiotics2030328
Galvin S, Bergin N, Hennessy R, Hanahoe B, Murphy AW, Cormican M, Vellinga A. Exploratory Spatial Mapping of the Occurrence of Antimicrobial Resistance in E. coli in the Community. Antibiotics. 2013; 2(3):328-338. https://doi.org/10.3390/antibiotics2030328
Chicago/Turabian StyleGalvin, Sandra, Niall Bergin, Ronan Hennessy, Belinda Hanahoe, Andrew W. Murphy, Martin Cormican, and Akke Vellinga. 2013. "Exploratory Spatial Mapping of the Occurrence of Antimicrobial Resistance in E. coli in the Community" Antibiotics 2, no. 3: 328-338. https://doi.org/10.3390/antibiotics2030328
APA StyleGalvin, S., Bergin, N., Hennessy, R., Hanahoe, B., Murphy, A. W., Cormican, M., & Vellinga, A. (2013). Exploratory Spatial Mapping of the Occurrence of Antimicrobial Resistance in E. coli in the Community. Antibiotics, 2(3), 328-338. https://doi.org/10.3390/antibiotics2030328






