Recent Advances in Bioprinting and Applications for Biosensing
Abstract
:1. Introduction
2. Transduction and Detection Methods for Biosensing
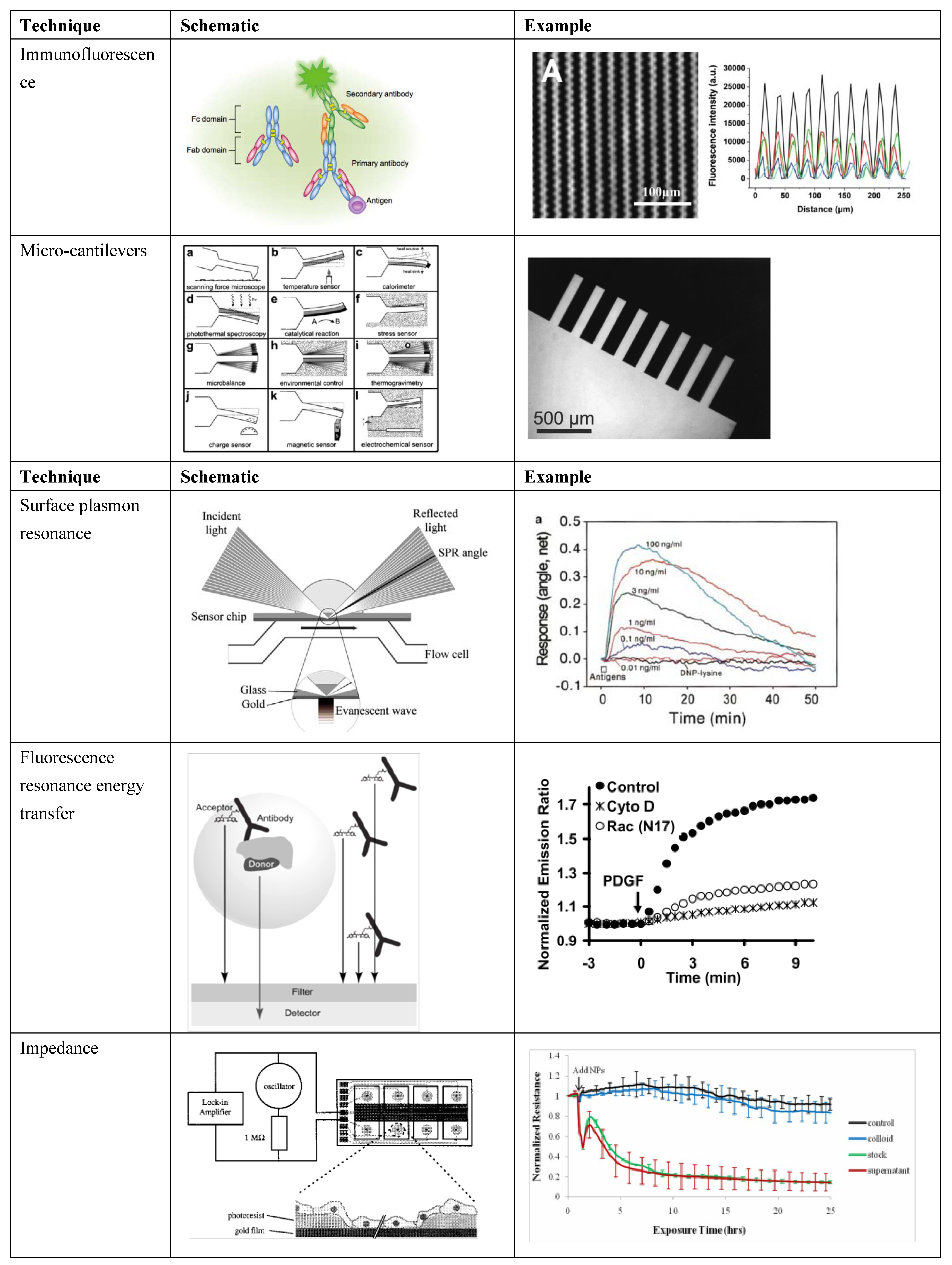
| Transduction method | Transduction mechanism | Simultaneous multiple analyte detection
+++ | Real-time capability
+++ | Fabrication speed and customizability
+++ |
|---|---|---|---|---|
| Immunofluorescence | Fluorescent molecule specific binding | ++ | + | +++ |
| Micro-cantilevers | Actuation during binding event | +++ | + | + |
| Surface plasmon resonance | Adsorption or binding changes local index of refraction and resonance of surface plasmon waves | - | +++ | +++ |
| Resonance energy transfer | Specific binding or interaction enables emission from and detection of target | ++ | +++ | + |
| Impedance | Impedance of cell membrane measured | - | +++ | ++ |
3. Methods for Bioprinting and Applications to Biosensing
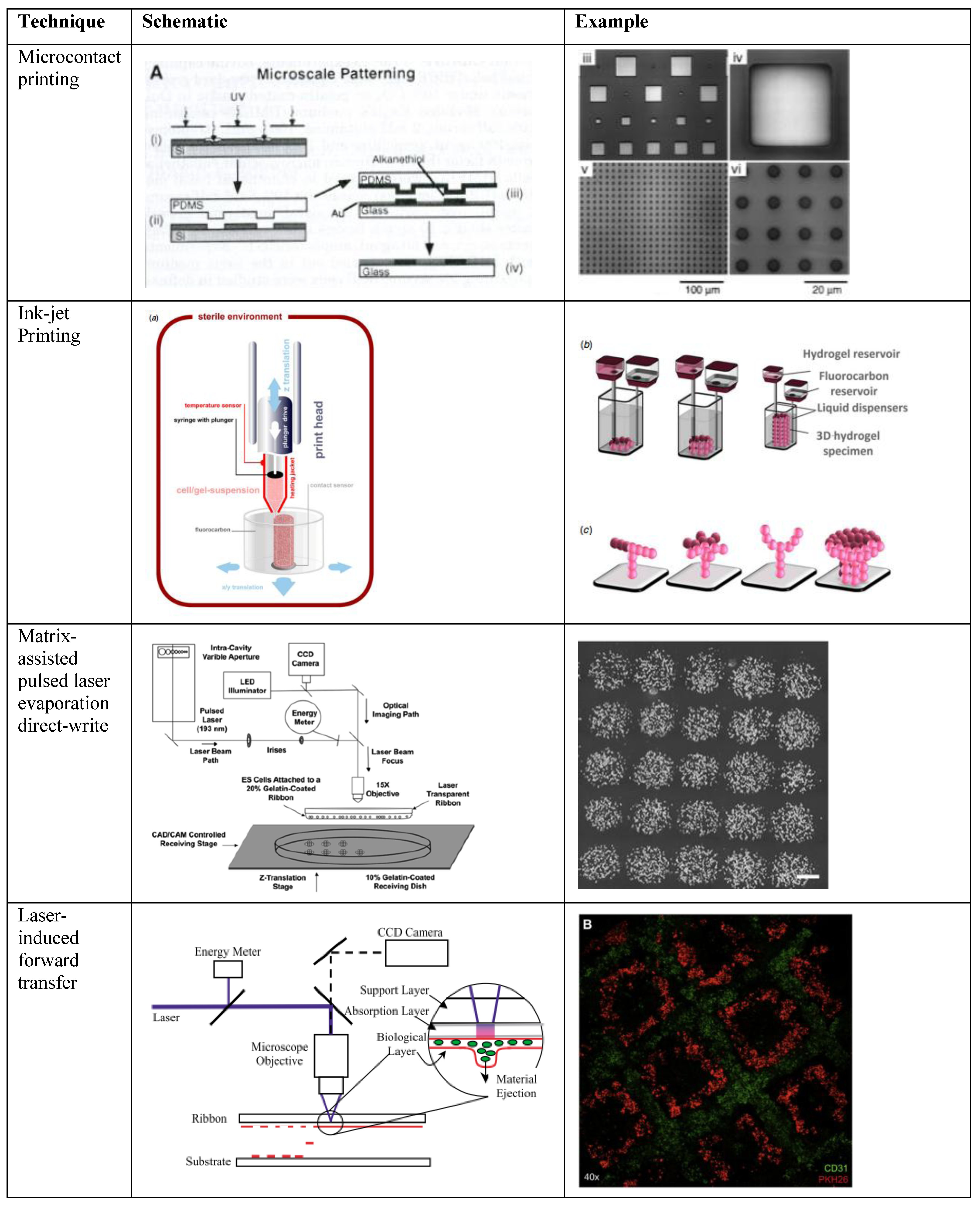
| Patterning Technique | Technique mechanism | Resolution +++ | Throughput +++ | Printable materials library +++ |
| Microcontact printing | Contact-based method, typically microstamp. | +++ | ++ | ++ |
| Ink-jet printing | Droplet ejection from nozzle invovling thermal piezoelectric or pressure | + | +++ | ++ |
| Matrix-assisted pulsed laser evaporation direct-write | Non-contact deposition via pulsed laser directly onto gel with cell suspension | +++ | + | +++ |
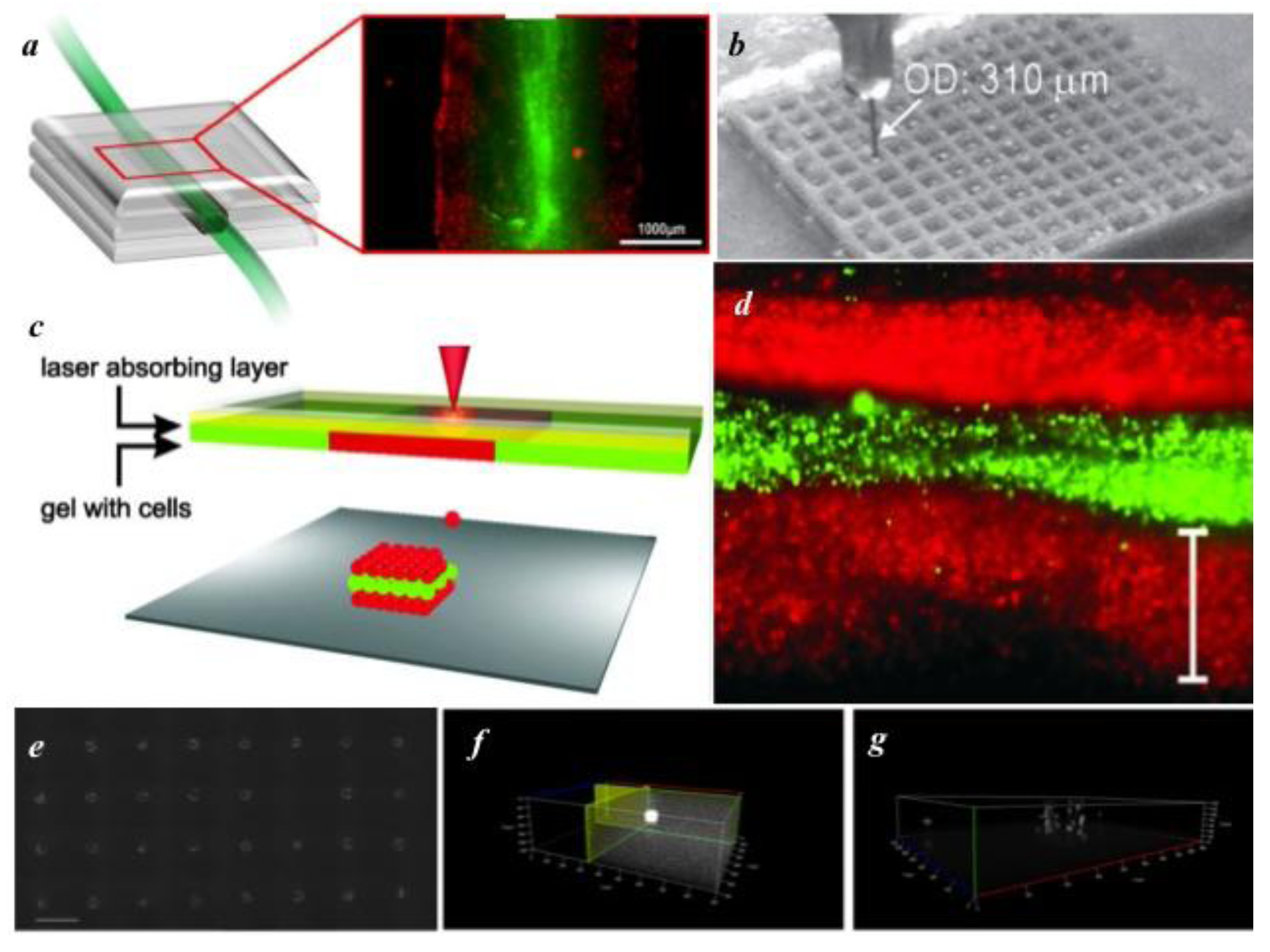
| Laser-induced forward transfer | Non-contact deposition via pulsed laser on sacrificial layer | +++ | ++ | +++ |
3.1. Contact-Based Bioprinting
3.2. Non-Contact Printing
4. In Situ Crosslinking for 3D Bioprinting
5. Conclusions and Future Direction of the Field
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Soper, S.A.; Brown, K.; Ellington, A.; Frazier, B.; Garcia-Manero, G.; Gau, V.; Gutman, S.I.; Hayes, D.F.; Korte, B.; Landers, J.L.; et al. Point-of-care biosensor systems for cancer diagnostics/prognostics. Biosens. Bioelectron. 2006, 21, 1932–1942. [Google Scholar] [CrossRef]
- Tothill, I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar]
- Taitt, C.R.; Golden, J.P.; Shubin, Y.S.; Shriver-Lake, L.C.; Sapsford, K.E.; Rasooly, A.; Ligler, F.S. A portable array biosensor for detecting multiple analytes in complex samples. Microb. Ecol. 2004, 47, 175–185. [Google Scholar] [CrossRef]
- Curtis, T.M.; Widder, M.W.; Brennan, L.M.; Schwager, S.J.; van der Schalie, W.; Fey, J.; Salazar, N. A portable cell-based impedance sensor for toxicity testing of drinking water. Lab Chip 2009, 9, 2176–2183. [Google Scholar] [CrossRef]
- Heeres, J.T.; Hergenrother, P.J. High-throughput screening for modulators of protein-protein interactions: Use of photonic crystal biosensors and complementary technologies. Chem. Soc. Rev. 2011, 40, 4398–4410. [Google Scholar] [CrossRef]
- Lang, H.P.; Baller, M.K.; Berger, R.; Gerber, C.; Gimzewski, J.K.; Battiston, F.M.; Fornaro, P.; Ramseyer, J.P.; Meyer, E.; Guntherodt, H.J. An artificial nose based on a micromechanical cantilever array. Anal. Chim. Acta 1999, 393, 59–65. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, J.; Shi, M.; Zhao, S.; Chen, Z.-F.; Liang, H. A gold nanoparticle-enhanced fluorescence polarization biosensor for amplified detection of T4 polynucleotide kinase activity and inhibition. J. Mater. Chem. B 2013, 1, 2018–2021. [Google Scholar] [CrossRef]
- Aristotelous, T.; Ahn, S. Discovery of β2 adrenergic receptor ligands using biosensor fragment screening of tagged wild-type receptor. ACS Med. Chem. Lett. 2013, 4, 1005–1010. [Google Scholar] [CrossRef]
- Hartati, Y.W.; Topkaya, S.N.; Maksum, I.P.; Ozsoz, M. Sensitive detection of mitochondrial DNA A3243G tRNALeu mutation via an electrochemical biosensor using meldola’s blue as a hybridization indicator. Adv. Anal. Chem. 2013, 3, 20–27. [Google Scholar]
- Lee, S.; Kim, G.-Y.; Moon, J.-H. Detection of 6-benzylaminopurine plant growth regulator in bean sprouts using OFRR biosensor and QuEChERS method. Anal. Methods 2013, 5, 961–966. [Google Scholar] [CrossRef]
- Loh, Q.L.; Choong, C. Three-dimensional scaffolds for tissue engineering applications: Role of porosity and pore size. Tissue Eng. B 2013, 19, 485–502. [Google Scholar] [CrossRef]
- Gruene, M.; Pflaum, M.; Hess, C.; Diamantouros, S.; Schlie, S.; Deiwick, A.; Koch, L.; Wilhelmi, M.; Jockenhoevel, S.; Haverich, A.; Chichkov, B. Laser printing of three-dimensional multicellular arrays for studies of cell–cell and cell–environment interactions. Tissue Eng. C Meth. 2011, 17, 973–982. [Google Scholar] [CrossRef]
- Seyama, T.; Suh, E.Y.; Kondo, T. Three-dimensional culture of epidermal cells on ordered cellulose scaffolds. Biofabrication 2013, 5. [Google Scholar] [CrossRef]
- Terzaki, K.; Kalloudi, E.; Mossou, E.; Mitchell, E.P.; Forsyth, V.T.; Rosseeva, E.; Simon, P.; Vamvakaki, M.; Chatzinikolaidou, M.; Mitraki, A.; et al. Mineralized self-assembled peptides on 3D laser-made scaffolds: A new route toward “scaffold on scaffold” hard tissue engineering. Biofabrication 2013, 5. [Google Scholar] [CrossRef]
- Morimoto, Y.; Tsuda, Y.; Takeuchi, S. Reconstruction of 3D Hierarchic Micro-Tissues Using Monodisperse Collagen Microbeads. In Proceedings of IEEE 22nd International Conference on Micro Electro Mechanical Systems (MEMS 2009), Sorrento, Italy, 25–29 January 2009; pp. 56–59.
- Yu, J.; Du, K.T.; Fang, Q.; Gu, Y.; Mihardja, S.S.; Sievers, R.E.; Wu, J.C.; Lee, R.J. The use of human mesenchymal stem cells encapsulated in RGD modified alginate microspheres in the repair of myocardial infarction in the rat. Biomaterials 2010, 31, 7012–7020. [Google Scholar] [CrossRef]
- Solorio, L.D.; Vieregge, E.L.; Dhami, C.D.; Alsberg, E. High-density cell systems incorporating polymer microspheres as microenvironmental regulators in engineered cartilage tissues. Tissue Eng. B 2013, 19, 209–220. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Choi, B.; Wu, B.; Lee, M. Customized biomimetic scaffolds created by indirect three-dimensional printing for tissue engineering. Biofabrication 2013, 5. [Google Scholar] [CrossRef]
- Payne, G.F.; Kim, E.; Cheng, Y.; Wu, H.-C.; Ghodssi, R.; Rubloff, G.W.; Raghavan, S.R.; Culver, J.N.; Bentley, W.E. Accessing biology’s toolbox for the mesoscale biofabrication of soft matter. Soft Matter 2013, 9, 6019–6032. [Google Scholar] [CrossRef]
- Song, W.; Yu, X.; Markel, D.C.; Shi, T.; Ren, W. Coaxial PCL/PVA electrospun nanofibers: Osseointegration enhancer and controlled drug release device. Biofabrication 2013, 5. [Google Scholar] [CrossRef]
- Squires, T.M.; Messinger, R.J.; Manalis, S.R. Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 2008, 26, 417–426. [Google Scholar] [CrossRef]
- Situ, C.; Buijs, J.; Mooney, M.H.; Elliott, C.T. Advances in surface plasmon resonance biosensor technology towards high-throughput, food-safety analysis. Trends Anal. Chem. 2010, 29, 1305–1315. [Google Scholar] [CrossRef]
- Ligler, F.S.; Taitt, C.R.; Shriver-Lake, L.C.; Sapsford, K.E.; Shubin, Y.; Golden, J.P. Array biosensor for detection of toxins. Anal. Bioanal. Chem. 2003, 377, 469–477. [Google Scholar] [CrossRef]
- Volcke, C.; Gandhiraman, R.P.; Basabe-Desmonts, L.; Iacono, M.; Gubala, V.; Cecchet, F.; Cafolla, A.A.; Williams, D.E. Protein pattern transfer for biosensor applications. Biosens. Bioelectron. 2010, 25, 1295–1300. [Google Scholar] [CrossRef]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Annu. Rev. Mater. Sci. 1998, 37, 550–575. [Google Scholar]
- Khademhosseini, A.; Langer, R.; Borenstein, J.; Vacanti, J.P. Microscale technologies for tissue engineering and biology. Proc. Natl. Acad. Sci. USA 2006, 103, 2480–2487. [Google Scholar] [CrossRef]
- Wilson, G.S.; Hu, Y. Enzyme-based biosensors for in vivo measurements. Chem. Rev. 2000, 100, 2693–2704. [Google Scholar] [CrossRef]
- Plaxco, K.W.; Soh, H.T. Switch-based biosensors: A new approach towards real-time, in vivo molecular detection. Trends Biotechnol. 2011, 29, 1–5. [Google Scholar] [CrossRef]
- Kellenberger, C.A.; Wilson, S.C.; Sales-Lee, J.; Hammond, M.C. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135, 4906–4909. [Google Scholar] [CrossRef]
- Odell, I.D.; Cook, D. Immunofluorescence techniques. J. Invest. Dermatol. 2013, 133. [Google Scholar] [CrossRef]
- Hide, M.; Tsutsui, T.; Sato, H.; Nishimura, T.; Morimoto, K.; Yamamoto, S.; Yoshizato, K. Real-time analysis of ligand-induced cell surface and intracellular reactions of living mast cells using a surface plasmon resonance-based biosensor. Anal. Biochem. 2002, 302, 28–37. [Google Scholar] [CrossRef]
- Bastiaens, P.I.; Squire, A. Fluorescence lifetime imaging microscopy: Spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999, 9, 48–52. [Google Scholar] [CrossRef]
- Ouyang, M.; Sun, J.; Chien, S.; Wang, Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. USA 2008, 105, 14353–14358. [Google Scholar] [CrossRef]
- Wegener, J.; Keese, C.R.; Giaever, I. Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp. Cell Res. 2000, 259, 158–166. [Google Scholar] [CrossRef]
- Mcauley, E.; Plopper, G.E.; Mohanraj, B.; Phamduy, T.; Corr, D.T.; Chrisey, D.B. Evaluation of electric cell-substrate impedance sensing for the detection of nanomaterial toxicity. Int. J. Biomed. Nanosci. Nanotechnol. 2011, 2, 136–151. [Google Scholar] [CrossRef]
- Baird, C.L.; Myszka, D.G. Current and emerging commercial optical biosensors. J. Mol. Recognit. 2001, 14, 261–268. [Google Scholar] [CrossRef]
- Quinn, J.G.; O’Neill, S.; Doyle, A.; McAtamney, C.; Diamond, D.; MacCraith, B.D.; O’Kennedy, R. Development and application of surface plasmon resonance-based biosensors for the detection of cell-ligand interactions. Anal. Biochem. 2000, 281, 135–143. [Google Scholar] [CrossRef]
- Charych, D.H.; Nagy, J.O.; Spevak, W.; Bednarski, M.D. Direct colorimetric detection of a receptor-ligand interaction by a polymerized bilayer assembly. Science 1993, 261, 585–588. [Google Scholar]
- Bai, S.; Li, S.; Yao, T.; Hu, Y.; Bao, F.; Zhang, J.; Zhang, Y.; Zhu, S.; He, Y. Rapid detection of eight vegetable oils on optical thin-film biosensor chips. Food Control 2011, 22, 1624–1628. [Google Scholar] [CrossRef]
- Wang, W.; Han, J.; Wu, Y.; Yuan, F.; Chen, Y.; Ge, Y. Simultaneous detection of eight food allergens using optical thin-film biosensor chips. J. Agric. Food Chem. 2011, 59, 6889–6894. [Google Scholar] [CrossRef]
- Chen, C.; Xie, Q.; Yang, D.; Xiao, H.; Fu, Y.; Tan, Y.; Yao, S. Recent advances in electrochemical glucose biosensors: A review. RSC Adv. 2013, 3, 4473–4491. [Google Scholar] [CrossRef]
- Hamidi-Asl, E.; Palchetti, I.; Hasheminejad, E.; Mascini, M. A review on the electrochemical biosensors for determination of microRNAs. Talanta 2013, 115, 74–83. [Google Scholar] [CrossRef]
- Homola, J.; Yee, S.S.; Gauglitz, G. Surface plasmon resonance sensors: Review. Sens. Actuator B Chem. 1999, 54, 3–15. [Google Scholar] [CrossRef]
- Mayer, K.M.; Hafner, J.H. Localized surface plasmon resonance sensors. Chem. Rev. 2011, 111, 3828–3857. [Google Scholar] [CrossRef]
- Tiefenthaler, K.; Lukosz, W. Sensitivity of grating couplers as integrated-optical chemical sensors. J. Opt. Soc. Am. B 1989, 6, 209–220. [Google Scholar] [CrossRef]
- Fang, Y. Non-invasive optical biosensor for probing cell signaling. Sensors 2007, 7, 2316–2329. [Google Scholar] [CrossRef]
- Goral, V.; Wu, Q.; Sun, H.; Fang, Y. Label-free optical biosensor with microfluidics for sensing ligand-directed functional selectivity on trafficking of thrombin receptor. FEBS Lett. 2011, 585, 1054–1060. [Google Scholar] [CrossRef]
- Dong, Y.; Wilkop, T.; Xu, D.; Wang, Z.; Cheng, Q. Microchannel chips for the multiplexed analysis of human immunoglobulin G-antibody interactions by surface plasmon resonance imaging. Anal. Bioanal. Chem. 2008, 390, 1575–1583. [Google Scholar] [CrossRef]
- Cetin, A.E.; Coskun, A.F.; Galarreta, B.C.; Huang, M.; Herman, D.; Ozcan, A.; Altug, H. Handheld high-throughput plasmonic biosensor using computational on-chip imaging. Light Sci. Appl. 2014, 3. [Google Scholar] [CrossRef]
- Raiteri, R.; Grattarola, M.; Butt, H.-J.; Skládal, P. Micromechanical cantilever-based biosensors. Sens. Actuator B Chem. 2001, 79, 115–126. [Google Scholar] [CrossRef]
- Battiston, F.; Ramseyer, J.-P.; Lang, H.; Baller, M.; Gerber, C.; Gimzewski, J.; Meyer, E.; Güntherodt, H.-J. A chemical sensor based on a microfabricated cantilever array with simultaneous resonance-frequency and bending readout. Sens. Actuator B Chem. 2001, 77, 122–131. [Google Scholar] [CrossRef]
- Wee, K.W.; Kang, G.Y.; Park, J.; Kang, J.Y.; Yoon, D.S.; Park, J.H.; Kim, T.S. Novel electrical detection of label-free disease marker proteins using piezoresistive self-sensing micro-cantilevers. Biosens. Bioelectron. 2005, 20, 1932–1938. [Google Scholar] [CrossRef]
- Carrascosa, L.G.; Moreno, M.; Álvarez, M.; Lechuga, L.M. Nanomechanical biosensors: A new sensing tool. TrAC Trends Anal. Chem. 2006, 25, 196–206. [Google Scholar] [CrossRef]
- Zhang, H.; Shepherd, J.N.H.; Nuzzo, R.G. Microfluidic contact printing: A versatile printing platform for patterning biomolecules on hydrogel substrates. Soft Matter 2010, 6, 2238–2245. [Google Scholar] [CrossRef]
- Zheng, C.; Wang, J.; Pang, Y.; Wang, J.; Li, W.; Ge, Z.; Huang, Y. High-throughput immunoassay through in-channel microfluidic patterning. Lab Chip 2012, 12, 2487–2490. [Google Scholar] [CrossRef]
- Lo, C.M.; Keese, C.R.; Giaever, I. Impedance analysis of MDCK cells measured by electric cell-substrate impedance sensing. Biophys. J. 1995, 69, 2800–2807. [Google Scholar] [CrossRef]
- Xiao, C.; Luong, J.H.T. On-line monitoring of cell growth and cytotoxicity using electric cell-substrate impedance sensing (ECIS). Biotechnol. Prog. 2003, 19, 1000–1005. [Google Scholar] [CrossRef]
- Hynes, A.; Mohanraj, B.; Schiele, N.; Corr, D.T.; Plopper, G.E.; Dinu, C.Z.; Chrisey, D.B. Cell-based detection of bacillus cereus anthrax simulant using cell impedance sensing. Sens. Lett. 2010, 8, 528–533. [Google Scholar] [CrossRef]
- Ferentinos, K.P.; Yialouris, C.P.; Blouchos, P.; Moschopoulou, G.; Tsourou, V.; Kintzios, S. The use of artificial neural networks as a component of a cell-based biosensor device for the detection of pesticides. Procedia Eng. 2012, 47, 989–992. [Google Scholar] [CrossRef]
- McConnell, H.M.; Owicki, J.C.; Parce, J.W.; Miller, D.L.; Baxter, G.S.; Wada, H.G.; Pitchford, S. The cytosensor microphysiometer: Biological applications of silicon technology. Science 1992, 257, 1906–1912. [Google Scholar]
- Salimi, A.; Noorbakhsh, A.; Rafiee-Pour, H.-A.; Ghourchian, H. Direct voltammetry of copper, zinc-superoxide dismutase immobilized onto electrodeposited nickel oxide nanoparticles: Fabrication of amperometric superoxide biosensor. Electroanalysis 2011, 23, 683–691. [Google Scholar]
- Xiang, C.; Kung, S.; Taggart, D.K.; Yang, F.; Thompson, M.A.; Gu, A.G.; Yang, Y.; Penner, R.M. Lithographically patterned nanowire patterning electrically continuous metal nanowires on dielectrics. ACS Nano 2008, 2, 1939–1949. [Google Scholar] [CrossRef]
- Peng, S.; Zhang, X. Electrodeposition of CdSe quantum dots and its application to an electrochemiluminescence immunoassay for α-fetoprotein. Microchim. Acta 2012, 178, 323–330. [Google Scholar] [CrossRef]
- Loget, G.; Wood, J.B.; Cho, K.; Halpern, A.R.; Corn, R.M. Electrodeposition of polydopamine thin films for DNA patterning and microarrays. Anal. Chem. 2013, 85, 9991–9995. [Google Scholar] [CrossRef]
- Novak, S.; Maver, U.; Peternel, Š.; Venturini, P.; Bele, M.; Gaberšček, M. Electrophoretic deposition as a tool for separation of protein inclusion bodies from host bacteria in suspension. Colloid Surf. A Physicochem. Eng. Asp. 2009, 340, 155–160. [Google Scholar] [CrossRef]
- Ammam, M.; Fransaer, J. Two-enzyme lactose biosensor based on β-galactosidase and glucose oxidase deposited by AC-electrophoresis: Characteristics and performance for lactose determination in milk. Sens. Actuator B Chem. 2010, 148, 583–589. [Google Scholar] [CrossRef]
- Poortinga, A.T.; Bos, R.; Busscher, H.J. Controlled electrophoretic deposition of bacteria to surfaces for the design of biofilms. Biotechnol. Bioeng. 2000, 67, 117–120. [Google Scholar] [CrossRef]
- Suginta, W.; Khunkaewla, P.; Schulte, A. Electrochemical biosensor applications of polysaccharides chitin and chitosan. Chem. Rev. 2013, 113, 5458–5479. [Google Scholar] [CrossRef]
- Moffat, T.P.; Yang, H. Patterned metal electrodeposition using an alkanethiolate mask. J. Electrochem. Soc. 1995, 142, 220–222. [Google Scholar] [CrossRef]
- Fernandes, R.; Wu, L.; Chen, T.; Yi, H.; Rubloff, G.W.; Ghodssi, R.; Bentley, W.E.; Payne, G.F. Electrochemically induced deposition of a polysaccharide hydrogel onto a patterned surface. Langmuir 2003, 19, 4058–4062. [Google Scholar] [CrossRef]
- Chen, P.-C.; Chen, R.L. C.; Cheng, T.-J.; Wittstock, G. Localized deposition of chitosan as matrix for enzyme immobilization. Electroanalysis 2009, 21, 804–810. [Google Scholar]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Micropatterned surfaces for control of cell shape, position, and function. Biotechnol. Prog. 1998, 14, 356–363. [Google Scholar] [CrossRef]
- Duarte Campos, D.F.; Blaeser, A.; Weber, M.; Jäkel, J.; Neuss, S.; Jahnen-Dechent, W.; Fischer, H. Three-dimensional printing of stem cell-laden hydrogels submerged in a hydrophobic high-density fluid. Biofabrication 2013, 5. [Google Scholar] [CrossRef]
- Raof, N.A.; Schiele, N.R.; Xie, Y.; Chrisey, D.B.; Corr, D.T. The maintenance of pluripotency following laser direct-write of mouse embryonic stem cells. Biomaterials 2011, 32, 1802–1808. [Google Scholar] [CrossRef]
- Barron, J.A.; Rosen, R.; Jones-Meehan, J.; Spargo, B.J.; Belkin, S.; Ringeisen, B.R. Biological laser printing of genetically modified Escherichia coli for biosensor applications. Biosens. Bioelectron. 2004, 20, 246–252. [Google Scholar] [CrossRef]
- Gaebel, R.; Ma, N.; Liu, J.; Guan, J.; Koch, L.; Klopsch, C.; Gruene, M.; Toelk, A.; Wang, W.; Mark, P.; et al. Patterning human stem cells and endothelial cells with laser printing for cardiac regeneration. Biomaterials 2011, 32, 9218–9230. [Google Scholar] [CrossRef]
- Britland, S.; Perez-Arnaud, E.; Clark, P.; Mcginn, B.; Connolly, P.; Moores, G. Micropatterning proteins and synthetic peptides on solid supports: A novel application for microelectronics fabrication technology. Biotechnol. Prog. 1992, 8, 155–160. [Google Scholar] [CrossRef]
- Folch, A.; Jo, B.H.; Hurtado, O.; Beebe, D.J.; Toner, M. Microfabricated elastomeric stencils for micropatterning cell cultures. J. Biomed. Mater. Res. 2000, 52, 346–353. [Google Scholar] [CrossRef]
- Ostuni, E.; Kane, R.; Chen, C.; Ingber, D.E.; Whitesides, G.M. Patterning mammalian cells using elastomeric membranes. Langmuir 2000, 7811–7819. [Google Scholar]
- Selvarasah, S.; Chao, S.H.; Chen, C.-L.; Sridhar, S.; Busnaina, A.; Khademhosseini, A.; Dokmeci, M.R. A reusable high aspect ratio parylene-C shadow mask technology for diverse micropatterning applications. Sens. Actuator A Phys. 2008, 145, 306–315. [Google Scholar]
- Jackman, R.J.; Wilbur, J.L.; Whitesides, G.M. Fabrication of submicrometer features on curved substrates by microcontact printing. Science 1995, 269, 664–666. [Google Scholar]
- Tien, J.; Nelson, C.M.; Chen, C.S. Fabrication of aligned microstructures with a single elastomeric stamp. Proc. Natl. Acad. Sci. USA 2002, 99, 1758–1762. [Google Scholar] [CrossRef]
- Beebe, D.J.; Moore, J.S.; Yu, Q.; Liu, R.H.; Kraft, M.L.; Jo, B.H.; Devadoss, C. Microfluidic tectonics: A comprehensive construction platform for microfluidic systems. Proc. Natl. Acad. Sci. USA 2000, 97, 13488–13493. [Google Scholar] [CrossRef]
- Gu, W.; Zhu, X.; Futai, N.; Cho, B.S.; Takayama, S. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proc. Natl. Acad. Sci. USA 2004, 101, 15861–15866. [Google Scholar] [CrossRef]
- Unger, M.A.; Chou, H.P.; Thorsen, T.; Scherer, A.; Quake, S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 2000, 288, 113–116. [Google Scholar] [CrossRef]
- Jeon, N.L.; Dertinger, S.K. W.; Chiu, D.T.; Choi, I.S.; Stroock, A.D.; Whitesides, G.M. Generation of solution and surface gradients using microfluidic systems. Langmuir 2000, 16, 8311–8316. [Google Scholar] [CrossRef]
- Chiu, D.T.; Jeon, N.L.; Huang, S.; Kane, R.S.; Wargo, C.J.; Choi, I.S.; Ingber, D.E.; Whitesides, G.M. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. Proc. Natl. Acad. Sci. USA 2000, 97, 2408–2413. [Google Scholar] [CrossRef]
- King, K.R.; Wang, S.; Irimia, D.; Jayaraman, A.; Toner, M.; Yarmush, M.L. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip 2007, 7, 77–85. [Google Scholar] [CrossRef]
- Pitaval, A.; Tseng, Q.; Bornens, M.; Thery, M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J. Cell Biol. 2010, 191, 303–312. [Google Scholar]
- Théry, M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 2010, 123, 4201–4213. [Google Scholar] [CrossRef]
- Guilak, F.; Cohen, D.M.; Estes, B.T.; Gimble, J.M.; Liedtke, W.; Chen, C.S. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 2009, 5, 17–26. [Google Scholar] [CrossRef]
- Clark, P.; Britland, S.; Connolly, P. Growth cone guidance and neuron morphology on micropatterned laminin surfaces. J. Cell Sci. 1993, 105, 203–212. [Google Scholar]
- Lee, L.H.; Peerani, R.; Ungrin, M.; Joshi, C.; Kumacheva, E.; Zandstra, P.W. Micropatterning of human embryonic stem cells dissects the mesoderm and endoderm lineages. Stem Cell Res. 2009, 2, 155–162. [Google Scholar] [CrossRef]
- Tang, J.; Peng, R.; Ding, J. The regulation of stem cell differentiation by cell-cell contact on micropatterned material surfaces. Biomaterials 2010, 31, 2470–2476. [Google Scholar] [CrossRef]
- Pal, R.; Yang, M.; Lin, R.; Johnson, B.N.; Srivastava, N.; Razzacki, S.Z.; Chomistek, K.J.; Heldsinger, D.C.; Haque, R.M.; Ugaz, V.M.; et al. An integrated microfluidic device for influenza and other genetic analyses. Lab Chip 2005, 5, 1024–1032. [Google Scholar] [CrossRef]
- Irimia, D.; Tompkins, R.G.; Toner, M. Single-cell chemical lysis in picoliter-scale closed volumes using a microfabricated device. Anal. Chem. 2004, 76, 6137–6143. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–82. [Google Scholar] [CrossRef]
- Lai, N.; Sims, J.K.; Jeon, N.L.; Lee, K. Adipocyte induction of preadipocyte differentiation in a gradient chamber. Tissue Eng. C Meth. 2012, 18, 958–967. [Google Scholar] [CrossRef]
- Sung, J.H.; Shuler, M.L. In vitro microscale systems for systematic drug toxicity study. Bioprocess Biosyst. Eng. 2010, 33, 5–19. [Google Scholar] [CrossRef]
- Jeon, B.O.; Alsberg, E. Regulation of stem cell fate in a three-dimensional micropatterned dual-crosslinked hydrogel system. Adv. Funct. Mater. 2013, 23, 4765–4775. [Google Scholar]
- Xie, X.N.; Chung, H.J.; Sow, C.H.; Wee, A.T.S. Nanoscale materials patterning and engineering by atomic force microscopy nanolithography. Mater. Sci. Eng. R Rep. 2006, 54, 1–48. [Google Scholar]
- Piner, R.; Zhu, J.; Xu, F.; Hong, S.; Mirkin, C. “Dip-pen” nanolithography. Science 1999, 283, 661–663. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Hirtz, M.; Deng, X.; Laue, T.; Fuchs, H.; Lahann, J. Substrate-independent dip-pen nanolithography based on reactive coatings. J. Am. Chem. Soc. 2010, 132, 18023–18025. [Google Scholar] [CrossRef]
- Wheeldon, I.; Farhadi, A.; Bick, A.G.; Jabbari, E.; Khademhosseini, A. Nanoscale tissue engineering: Spatial control over cell-materials interactions. Nanotechnology 2011, 22. [Google Scholar] [CrossRef]
- Wang, X.; Yan, C.; Ye, K.; He, Y.; Li, Z.; Ding, J. Effect of RGD nanospacing on differentiation of stem cells. Biomaterials 2013, 34, 2865–2874. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Tenney, R.M.; Discher, D.E. Stem cells, microenvironment mechanics, and growth factor activation. Curr. Opin. Cell Biol. 2009, 21, 630–635. [Google Scholar] [CrossRef]
- Boland, T.; Xu, T.; Damon, B.; Cui, X. Application of inkjet printing to tissue engineering. Biotechnol. J. 2006, 1, 910–917. [Google Scholar] [CrossRef]
- Saunders, R.E.; Gough, J.E.; Derby, B. Delivery of human fibroblast cells by piezoelectric drop-on-demand inkjet printing. Biomaterials 2008, 29, 193–203. [Google Scholar] [CrossRef]
- Lee, W.; Debasitis, J.C.; Lee, V.K.; Lee, J.-H.; Fischer, K.; Edminster, K.; Park, J.-K.; Yoo, S.-S. Multi-layered culture of human skin fibroblasts and keratinocytes through three-dimensional freeform fabrication. Biomaterials 2009, 30, 1587–1595. [Google Scholar] [CrossRef]
- Gonzalez-Macia, L.; Morrin, A.; Smyth, M.R.; Killard, A.J. Advanced printing and deposition methodologies for the fabrication of biosensors and biodevices. Analyst 2010, 135, 845–867. [Google Scholar] [CrossRef]
- Cui, X.; Dean, D.; Ruggeri, Z.M.; Boland, T. Cell damage evaluation of thermal inkjet printed Chinese hamster ovary cells. Biotechnol. Bioeng. 2010, 106, 963–969. [Google Scholar] [CrossRef]
- Roth, E.A.; Xu, T.; Das, M.; Gregory, C.; Hickman, J.J.; Boland, T. Inkjet printing for high-throughput cell patterning. Biomaterials 2004, 25, 3707–3715. [Google Scholar] [CrossRef]
- Pardo, L.; Wilson, W.C.; Boland, T. Characterization of patterned self-assembled monolayers and protein arrays generated by the ink-jet method. Langmuir 2003, 19, 1462–1466. [Google Scholar] [CrossRef]
- Xia, Y.; Mrksich, M.; Kim, E.; Whitesides, G.M. Microcontact printing of octadecylsiloxane on the surface of silicon dioxide and its application in microfabrication. J. Am. Chem. Soc. 1995, 117, 9576–9577. [Google Scholar] [CrossRef]
- Chrisey, D.B.; Pique, A.; Fitz-Gerald, J.; Auyeung, R.C. Y.; McGill, R.A.; Wu, H.D.; Duignan, M. New approach to laser direct writing active and passive mesoscopic circuit elements. Appl. Surf. Sci. 2000, 154–155, 593–600. [Google Scholar] [CrossRef]
- Unger, C.; Gruene, M.; Koch, L.; Koch, J.; Chichkov, B.N. Time-resolved imaging of hydrogel printing via laser-induced forward transfer. Appl. Phys. A 2011, 103, 271–277. [Google Scholar]
- Khetan, S.; Burdick, J. Patterning hydrogels in three dimensions towards controlling cellular interactions. Soft Matter 2011, 7, 830–838. [Google Scholar] [CrossRef]
- Jeon, O.; Alsberg, E. Photofunctionalization of alginate hydrogels to promote adhesion and proliferation of human mesenchymal stem cells. Tissue Eng. A 2013, 19, 1424–1432. [Google Scholar] [CrossRef]
- Ovsianikov, A.; Schlie, S.; Ngezahayo, A.; Haverich, A.; Chichkov, B.N. Two-photon polymerization technique for microfabrication of CAD-designed 3D scaffolds from commercially available photosensitive materials. J. Tissue Eng. Regen. Med. 2007, 1, 443–449. [Google Scholar] [CrossRef]
- Schiele, N.R.; Chrisey, D.B.; Corr, D.T. Gelatin-based laser direct-write technique for the precise spatial patterning of cells. Tissue Eng. C Meth. 2011, 17, 289–298. [Google Scholar] [CrossRef]
- Arnold, C.B.; Serra, P.; Piqué, A. Laser Direct-write techniques for printing of complex materials. MRS Bull. 2007, 32, 23–31. [Google Scholar] [CrossRef]
- Kingsley, D.M.; Dias, A.D.; Chrisey, D.B.; Corr, D.T. Single-step laser-based fabrication and patterning of cell-encapsulated alginate microbeads. Biofabrication 2013, 5. [Google Scholar] [CrossRef]
- Barron, J.A.; Ringeisen, B.R.; Kim, H.; Spargo, B.J.; Chrisey, D.B. Application of laser printing to mammalian cells. Thin Solid Films 2004, 453, 383–387. [Google Scholar]
- Ringeisen, B.R.; Kim, H.; Barron, J.A.; Krizman, D.B.; Chrisey, D.B.; Jackman, S.; Auyeung, R.Y.C.; Spargo, B.J. Laser printing of pluripotent embryonal carcinoma cells. Tissue Eng. 2004, 10, 483–491. [Google Scholar] [CrossRef]
- Schiele, N.R.; Koppes, R.A.; Corr, D.T.; Ellison, K.S.; Thompson, D.M.; Ligon, L.A.; Lippert, T.K. M.; Chrisey, D.B. Laser direct writing of combinatorial libraries of idealized cellular constructs: Biomedical applications. Appl. Surf. Sci. 2009, 255, 5444–5447. [Google Scholar] [CrossRef]
- Wu, P.; Ringeisen, B.R.; Callahan, J.; Brooks, M.; Hubb, D.M.; Wu, H.D.; Pique, A.; Spargo, B.; McGill, R.A.; Chrisey, D.B. The deposition, structure, pattern deposition, and activity of biomaterial thin-films by matrix-assisted pulsed-laser evaporation (MAPLE) and MAPLE direct write. Thin Solid Films 2001, 398–399, 607–614. [Google Scholar]
- Koch, L.; Kuhn, S.; Sorg, H.; Gruene, M.; Schlie, S.; Gaebel, R.; Polchow, B.; Reimers, K.; Stoelting, S.; Ma, N.; Vogt, P.M.; Steinhoff, G.; Chichkov, B. Laser printing of skin cells and human stem cells. Tissue Eng. C. Meth. 2010, 16, 847–854. [Google Scholar] [CrossRef]
- Serra, P.; Fernández-Pradas, J.M.; Colina, M.; Duocastella, M.; Dominguez, J.; Morenza, J.L. Laser-induced forward transfer: A direct-writing technique for biosensors preparation. JLMN 2006, 1, 236–242. [Google Scholar] [CrossRef]
- Gruene, M.; Pflaum, M.; Deiwick, A.; Koch, L.; Schlie, S.; Unger, C.; Wilhelmi, M.; Haverich, A.; Chichkov, B. Adipogenic differentiation of laser-printed 3D tissue grafts consisting of human adipose-derived stem cells. Biofabrication 2011, 3. [Google Scholar] [CrossRef]
- Zaman, M.; Trapani, L.; Sieminski, A.; D, M.; Gong, H.; Kamm, R.; Wells, A.; Lauffenburger, D.; Matsudaira, P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 10889–10894. [Google Scholar] [CrossRef]
- Cukierman, E.; Pankov, R.; Stevens, D.; Yamada, K. Taking cell-matrix adhesions to the third dimension. Am. Assoc. Adv. Sci. 2001, 294, 1708–1712. [Google Scholar]
- Wang, F.; Weaver, V.M.; Petersen, O.W.; Larabell, C.A.; Dedhar, S.; Briands, P.; Lupu, R.; Bissell, M.J. Reciprocal interactions between β1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Cell Biol. 1998, 95, 14821–14826. [Google Scholar]
- Gombotz, W.R.; Wee, S.F. Protein release from alginate matrices. Adv. Drug Deliv. Rev. 1998, 31, 267–285. [Google Scholar] [CrossRef]
- Lee, T.K.; Sokoloski, T.D.; Royer, G.P. Serum albumin beads: An injectable, biodegradable system for the sustained release of drugs. Science 2014, 213, 233–235. [Google Scholar]
- Tan, W.-H.; Takeuchi, S. Monodisperse alginate hydrogel microbeads for cell encapsulation. Adv. Mater. 2007, 19, 2696–2701. [Google Scholar] [CrossRef]
- Ballyns, J.J.; Gleghorn, J.P.; Niebrzydowski, V.; Rawlinson, J.J.; Potter, H.G.; Maher, S.A.; Wright, T.M.; Bonassar, L.J. Image-guided tissue engineering of anatomically shaped implants via MRI and micro-CT using injection molding. Tissue Eng. A 2008, 14, 1195–1202. [Google Scholar] [CrossRef]
- Brown, B.N.; Siebenlist, N.J.; Cheetham, J.; Ducharme, N.G.; Rawlinson, J.J.; Bonassar, L.J. Computed tomography-guided tissue engineering of upper airway cartilage. Tissue Eng. C Meth. 2013. [Google Scholar] [CrossRef]
- Hung, C.T.; Lima, E.G.; Mauck, R.L.; Taki, E.; LeRoux, M.A.; Lu, H.H.; Stark, R.G.; Guo, X.E.; Ateshian, G.A. Anatomically shaped osteochondral constructs for articular cartilage repair. J. Biomech. 2003, 36, 1853–1864. [Google Scholar] [CrossRef]
- Wu, L.; Jing, D.; Ding, J. A “room-temperature” injection molding/particulate leaching approach for fabrication of biodegradable three-dimensional porous scaffolds. Biomaterials 2006, 27, 185–191. [Google Scholar] [CrossRef]
- Chang, R.; Nam, J.; Sun, W. Effects of dispensing pressure and nozzle diameter on cell survival from solid freeform fabrication-based direct cell writing. Tissue Eng. A 2008, 14, 41–48. [Google Scholar]
- Lee, W.; Lee, V.; Polio, S.; Keegan, P.; Lee, J.-H.; Fischer, K.; Park, J.-K.; Yoo, S.-S. On-demand three-dimensional freeform fabrication of multi-layered hydrogel scaffold with fluidic channels. Biotechnol. Bioeng. 2010, 105, 1178–1186. [Google Scholar]
- Koch, L.; Deiwick, A.; Schlie, S.; Michael, S.; Gruene, M.; Coger, V.; Zychlinski, D.; Schambach, A.; Reimers, K.; Vogt, P.M.; Chichkov, B. Skin tissue generation by laser cell printing. Biotechnol. Bioeng. 2012, 109, 1855–1863. [Google Scholar] [CrossRef]
- Zhao, L.; Lee, V.K.; Yoo, S.-S.; Dai, G.; Intes, X. The integration of 3-D cell printing and mesoscopic fluorescence molecular tomography of vascular constructs within thick hydrogel scaffolds. Biomaterials 2012, 33, 5325–5332. [Google Scholar] [CrossRef]
- Lee, H.; Ahn, S.; Bonassar, L.J.; Kim, G. Cell(MC3T3-E1)-printed poly(ε-caprolactone)/alginate hybrid scaffolds for tissue regeneration. Macromol. Rapid Commun. 2013, 34, 142–149. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dias, A.D.; Kingsley, D.M.; Corr, D.T. Recent Advances in Bioprinting and Applications for Biosensing. Biosensors 2014, 4, 111-136. https://doi.org/10.3390/bios4020111
Dias AD, Kingsley DM, Corr DT. Recent Advances in Bioprinting and Applications for Biosensing. Biosensors. 2014; 4(2):111-136. https://doi.org/10.3390/bios4020111
Chicago/Turabian StyleDias, Andrew D., David M. Kingsley, and David T. Corr. 2014. "Recent Advances in Bioprinting and Applications for Biosensing" Biosensors 4, no. 2: 111-136. https://doi.org/10.3390/bios4020111
APA StyleDias, A. D., Kingsley, D. M., & Corr, D. T. (2014). Recent Advances in Bioprinting and Applications for Biosensing. Biosensors, 4(2), 111-136. https://doi.org/10.3390/bios4020111




