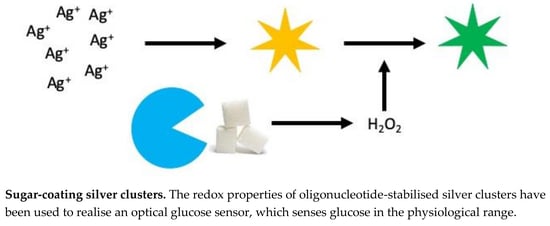
Glucose Sensor Using Redox Active Oligonucleotide-Templated Silver Nanoclusters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Silver Nanocluster Synthesis
2.3. Characterisation
2.4. Protocol for Glucose Sensing
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Driehorst, T.; O’Neill, P.; Goodwin, P.M.; Pennathur, S.; Fygenson, D.K. Distinct Conformations of DNA-Stabilized Fluorescent Silver Nanoclusters Revealed by Electrophoretic Mobility and Diffusivity Measurements. Langmuir 2011, 27, 8923–8933. [Google Scholar] [PubMed]
- O’Neill, P.R.; Velazquez, L.R.; Dunn, D.G.; Gwinn, E.G.; Fygenson, D.K. Hairpins with Poly-C Loops Stabilize Four Types of Fluorescent Agn:DNA. J. Phys. Chem. C 2009, 113, 4229–4233. [Google Scholar]
- Ritchie, C.M.; Johnsen, K.R.; Kiser, J.R.; Antoku, Y.; Dickson, R.M.; Petty, J.T. Ag Nanocluster Formation Using a Cytosine Oligonucleotide Template. J. Phys. Chem. C 2007, 111, 175–181. [Google Scholar]
- Richards, C.I.; Choi, S.; Hsiang, J.C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y.L.; Dickson, R.M. Oligonucleotide-Stabilized Ag Nanocluster Fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039. [Google Scholar] [PubMed] [Green Version]
- Petty, J.T.; Story, S.P.; Hsiang, J.C.; Dickson, R.M. DNA-Templated Molecular Silver Fluorophores. J. Phys. Chem. Lett. 2013, 4, 1148–1155. [Google Scholar] [PubMed] [Green Version]
- Lan, G.Y.; Chen, W.Y.; Chang, H.T. Control of synthesis and optical properties of DNA templated silver nanoclusters by varying DNA length and sequence. RSC Adv. 2011, 1, 802. [Google Scholar]
- Enkin, N.; Wang, F.; Sharon, E.; Albada, B.; Willner, I. Multiplexed Analysis of Genes Using Nucleic Acid-Stabilized Silver-Nanocluster Quantum Dots. ACS Nano 2014, 8, 11666–11673. [Google Scholar] [PubMed]
- Schultz, D.; Gardner, K.; Oemrawsingh, S.S.R.; Markešević, N.; Olsson, K.; Debord, M.; Bouwmeester, D.; Gwinn, E. Evidence for Rod-Shaped DNA-Stabilized Silver Nanocluster Emitters. Adv. Mater. 2013, 25, 2797–2803. [Google Scholar]
- Park, J.; Lee, J.; Ban, C.; Kim, W.J. An approach toward SNP detection by modulating the fluorescence of DNA-templated silver nanoclusters. Biosens. Bioelectron. 2013, 43, 419–424. [Google Scholar]
- Sengupta, B.; Corley, C.; Cobb, K.; Saracino, A.; Jockusch, S. DNA Scaffolded Silver Clusters: A Critical Study. Molecules 2016, 21, 216. [Google Scholar] [Green Version]
- Javani, S.; Lorca, R.; Latorre, A.; Flors, C.; Cortajarena, A.L.; Somoza, Á. Antibacterial Activity of DNA-Stabilized Silver Nanoclusters Tuned by Oligonucleotide Sequence. ACS Appl. Mater. Interfaces 2016, 8, 10147–10154. [Google Scholar] [PubMed]
- Sharma, J.; Yeh, H.C.; Yoo, H.; Werner, J.H.; Martinez, J.S. A complementary palette of fluorescent silver nanoclusters. Chem. Commun. 2010, 46, 3280–3282. [Google Scholar]
- Lentner, C. Wissenschaftliche Tabellen Geigy, 8th ed.; Ciba-Geigy: Basel, Switzerland, 1985. [Google Scholar]
- Zhao, Q.; Chen, S.; Huang, H.; Zhang, L.; Wang, L.; Liu, F.; Chen, J.; Zeng, Y.; Chu, P.K. Colorimetric and ultra-sensitive fluorescence resonance energy transfer determination of H2O2 and glucose by multi-functional Au nanoclusters. Analyst 2014, 139, 1498–1503. [Google Scholar] [PubMed]
- Bellary, S.; Cameron, H.; Macleod, K.; Malecha, M.; Koria, K.; Raja, P.; Cabezudo, J.D.; Ellison, J. Clinical evaluation of a novel test strip technology for blood glucose monitoring: Accuracy at hypoglycaemic glucose levels. Diabetes Res. Clin. Pract. 2012, 98, 430–435. [Google Scholar] [PubMed]
- Mayo Clinic Staff. Hyperglycemia in Diabetes. Symptoms and Causes. In Mayo Clinic. Available online: http://www.mayoclinic.org/diseases-conditions/hyperglycemia/symptoms-causes/syc-20373631 (accessed on 28 August 2018).
- Yoo, E.H.; Lee, S.Y. Glucose Biosensors: An Overview of Use in Clinical Practice. Sensors 2010, 10, 4558–4576. [Google Scholar] [PubMed] [Green Version]
- Wang, H.C.; Lee, A.R. Recent developments in blood glucose sensors. J. Food Drug Anal. 2015, 23, 191–200. [Google Scholar] [PubMed] [Green Version]
- Raba, J.; Mottola, H.A. Glucose oxidase as an analytical reagent. Crit. Rev. Anal. Chem. 1995, 25, 1–42. [Google Scholar]
- Zong, C.; Li, B.; Wang, J.; Liu, X.; Zhao, W.; Zhang, Q.; Nie, X.; Yu, Y. Visual and colorimetric determination of H2O2 and glucose based on citrate-promoted H2O2 sculpturing of silver nanoparticles. Microchim. Acta 2018, 185, 199. [Google Scholar]
- Endo, T.; Ikeda, R.; Yanagida, Y.; Hatsuzawa, T. Stimuli-responsive hydrogel–silver nanoparticles composite for development of localized surface plasmon resonance-based optical biosensor. Anal. Chim. Acta 2008, 611, 205–211. [Google Scholar]
- Tashkhourian, J.; Hormozi-Nezhad, M.R.; Khodaveisi, J.; Dashti, R. A novel photometric glucose biosensor based on decolorizing of silver nanoparticles. Sens. Actuators B Chem. 2011, 158, 185–189. [Google Scholar]
- Radhakumary, C.; Sreenivasan, K. Naked Eye Detection of Glucose in Urine Using Glucose Oxidase Immobilized Gold Nanoparticles. Anal. Chem. 2011, 83, 2829–2833. [Google Scholar] [PubMed]
- Cowart, S.L.; Stachura, M.E. Glucosuria. In Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd ed.; Walker, H.K., Hall, W.D., Hurst, J.W., Eds.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- He, D.; Garg, S.; Waite, T.D. H2O2-Mediated Oxidation of Zero-Valent Silver and Resultant Interactions among Silver Nanoparticles, Silver Ions, and Reactive Oxygen Species. Langmuir 2012, 28, 10266–10275. [Google Scholar]
- Christiansen, M.P.; Klaff, L.J.; Brazg, R.; Chang, A.R.; Levy, C.J.; Lam, D.; Denham, D.S.; Atiee, G.; Bode, B.W.; Walters, S.J.; et al. A Prospective Multicenter Evaluation of the Accuracy of a Novel Implanted Continuous Glucose Sensor: PRECISE II. Diabetes Technol. Ther. 2018, 20, 197–206. [Google Scholar]
- Colvin, A.E.; Jiang, H. Increased in vivo stability and functional lifetime of an implantable glucose sensor through platinum catalysis. J. Biomed. Mater. Res. A 2013, 101, 1274–1282. [Google Scholar] [PubMed]
- Choi, S.; Park, S.; Lee, K.; Yu, J. Oxidant-resistant imaging and ratiometric luminescence detection by selective oxidation of silver nanodots. Chem. Commun. 2013, 49, 10908–10910. [Google Scholar]
- Liu, X.; Wang, F.; Niazov-Elkan, A.; Guo, W.; Willner, I. Probing Biocatalytic Transformations with Luminescent DNA/Silver Nanoclusters. Nano Lett. 2013, 13, 309–314. [Google Scholar] [PubMed]
- Liu, C.; Ding, Y.; Li, Q.; Lin, Y. Photochemical synthesis of glutathione-stabilized silver nanoclusters for fluorometric determination of hydrogen peroxide. Microchim. Acta 2017, 184, 2497–2503. [Google Scholar]
- Ren, X.; Liu, J.; Ren, J.; Meng, X.; Fang, Z.; Tang, F. Fluorescent Ag Nanoprobes for the Highly Selective Detection of Mercury (II) Ions and Colorimetric Detection of Hydrogen Peroxide. J. Nanosci. Nanotechnol. 2016, 16, 6719–6725. [Google Scholar]
- Sharma, J.; Rocha, R.C.; Phipps, M.L.; Yeh, H.C.; Balatsky, K.A.; Vu, D.M.; Shreve, A.P.; Werner, J.H.; Martinez, J.S. A DNA-templated fluorescent silver nanocluster with enhanced stability. Nanoscale 2012, 4, 4107. [Google Scholar]
- Nakamura, S.; Ogura, Y. Mode of Inhibition of Glucose Oxidase by Metal Ions. J. Biochem. 1968, 64, 439–447. [Google Scholar]
- Walla, P.J. Optical Properties of Biomolecules. In Modern Biophysical Chemistry; Wiley-Blackwell: Hoboken, NJ, USA, 2014; pp. 41–60. [Google Scholar]
- Copp, S.M.; Schultz, D.; Swasey, S.; Pavlovich, J.; Debord, M.; Chiu, A.; Olsson, K.; Gwinn, E. Magic Numbers in DNA-Stabilized Fluorescent Silver Clusters Lead to Magic Colors. J. Phys. Chem. Lett. 2014, 5, 959–963. [Google Scholar] [PubMed]
- Ramazanov, R.R.; Sych, T.S.; Reveguk, Z.V.; Maksimov, D.A.; Vdovichev, A.A.; Kononov, A.I. Ag–DNA Emitter: Metal Nanorod or Supramolecular Complex? J. Phys. Chem. Lett. 2016, 7, 3560–3566. [Google Scholar] [PubMed]
- Huard, D.J.E.; Demissie, A.; Kim, D.; Lewis, D.; Dickson, R.M.; Petty, J.T.; Lieberman, R.L. Atomic Structure of a Fluorescent Ag8 Cluster Templated by a Multistranded DNA Scaffold. J. Am. Chem. Soc. 2018. [Google Scholar] [CrossRef]
- Manura, J.J.; Manura, D.J. Isotope Distribution Calculator and Mass Spec Plotter. In Scientific Instrument Services. Available online: http://www.sisweb.com/mstools/isotope.htm (accessed on 7 September 2016).
- Li, T.; He, N.; Wang, J.; Deng, Y. Effects of the i-motif DNA loop on the fluorescence of silver nanoclusters. RSC Adv. 2016, 6, 22839–22844. [Google Scholar]
- Xu, J.; Wei, C. The aptamer DNA-templated fluorescence silver nanoclusters: ATP detection and preliminary mechanism investigation. Biosens. Bioelectron. 2017, 87, 422–427. [Google Scholar] [PubMed]
- Borghei, Y.S.; Hosseini, M.; Ganjali, M.R. Detection of large deletion in human BRCA1 gene in human breast carcinoma MCF-7 cells by using DNA-Silver Nanoclusters. Methods Appl. Fluoresc. 2018, 6, 015001. [Google Scholar]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schroeder, K.L.; Goreham, R.V.; Nann, T. Glucose Sensor Using Redox Active Oligonucleotide-Templated Silver Nanoclusters. Nanomaterials 2019, 9, 1065. https://doi.org/10.3390/nano9081065
Schroeder KL, Goreham RV, Nann T. Glucose Sensor Using Redox Active Oligonucleotide-Templated Silver Nanoclusters. Nanomaterials. 2019; 9(8):1065. https://doi.org/10.3390/nano9081065
Chicago/Turabian StyleSchroeder, Kathryn L., Renee V. Goreham, and Thomas Nann. 2019. "Glucose Sensor Using Redox Active Oligonucleotide-Templated Silver Nanoclusters" Nanomaterials 9, no. 8: 1065. https://doi.org/10.3390/nano9081065