A Mathematical Spline-Based Model of Cardiac Left Ventricle Anatomy and Morphology †
Abstract
:1. Introduction
1.1. Examination of Individual Hearts and Empirical Models
1.2. Theoretical Models of Cardiac Anatomy
1.3. Relation to Other Anatomical Ventricular Models
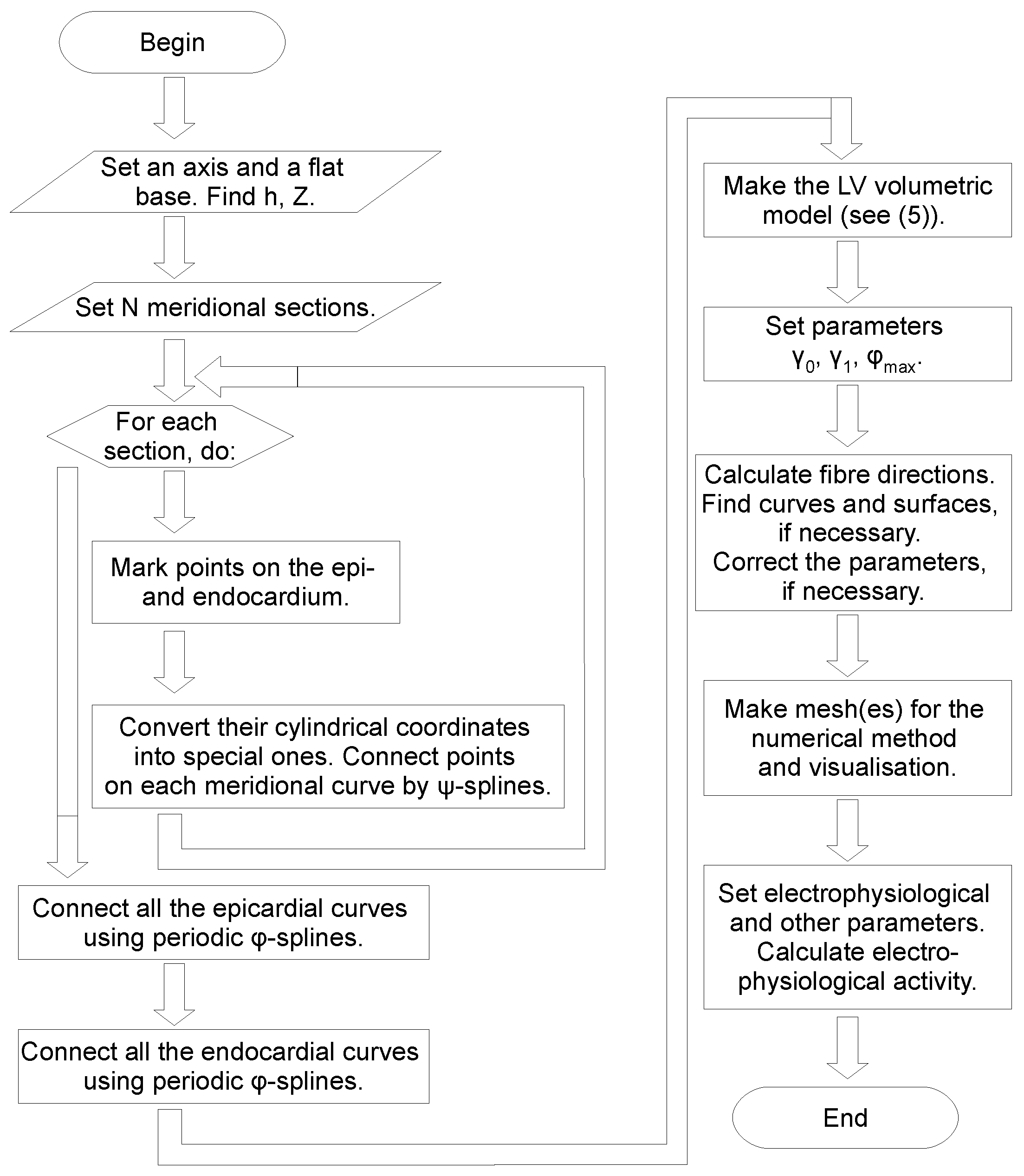
2. Construction of the LV Model
- meridians , where measurements are taken;
- coordinates , , of marked points on the epicardium;
- coordinates , , of marked points on the endocardium.
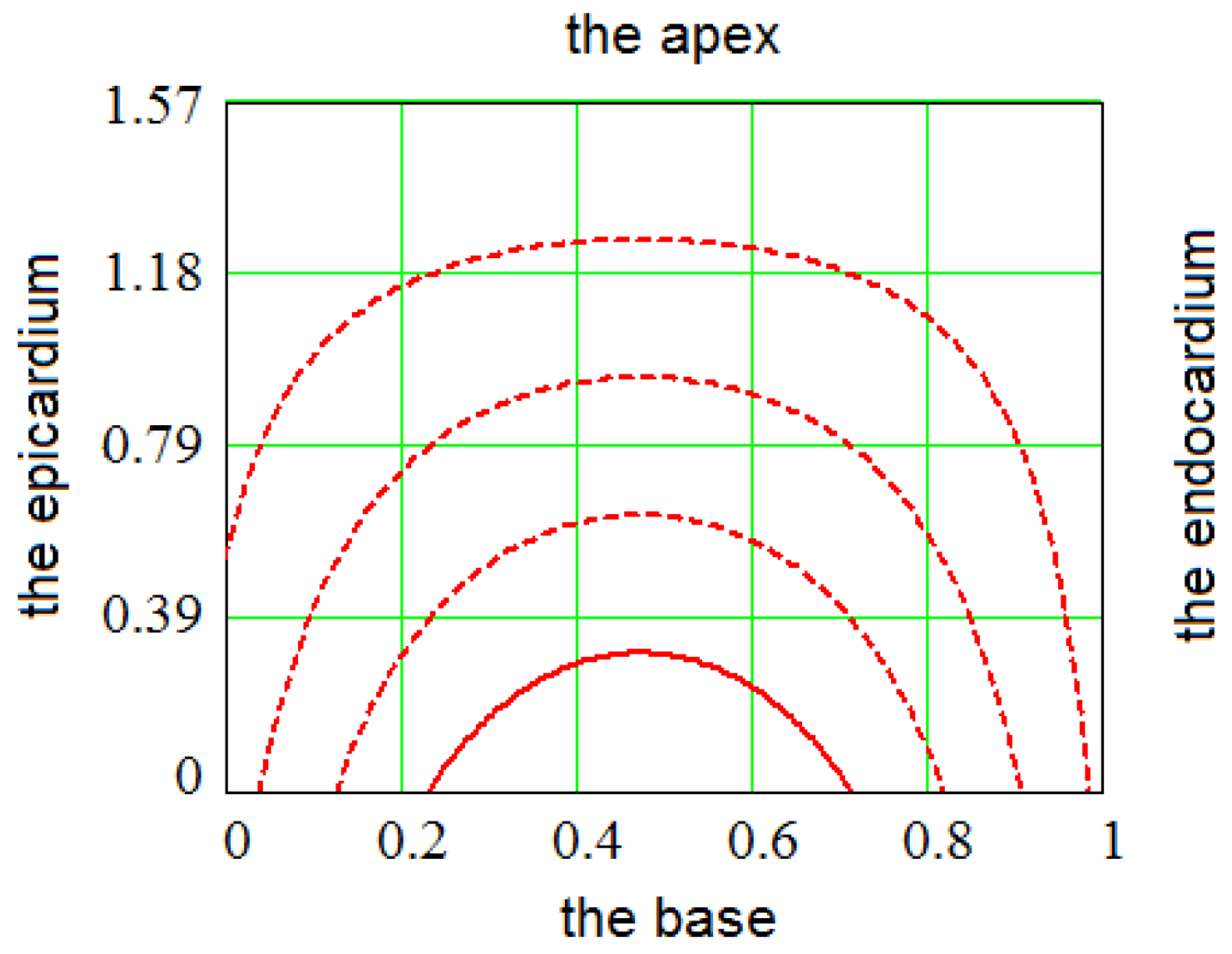
2.1. Spiral Surfaces
2.2. Filling a Spiral Surface with Fibers
2.3. Fitting the LV Form
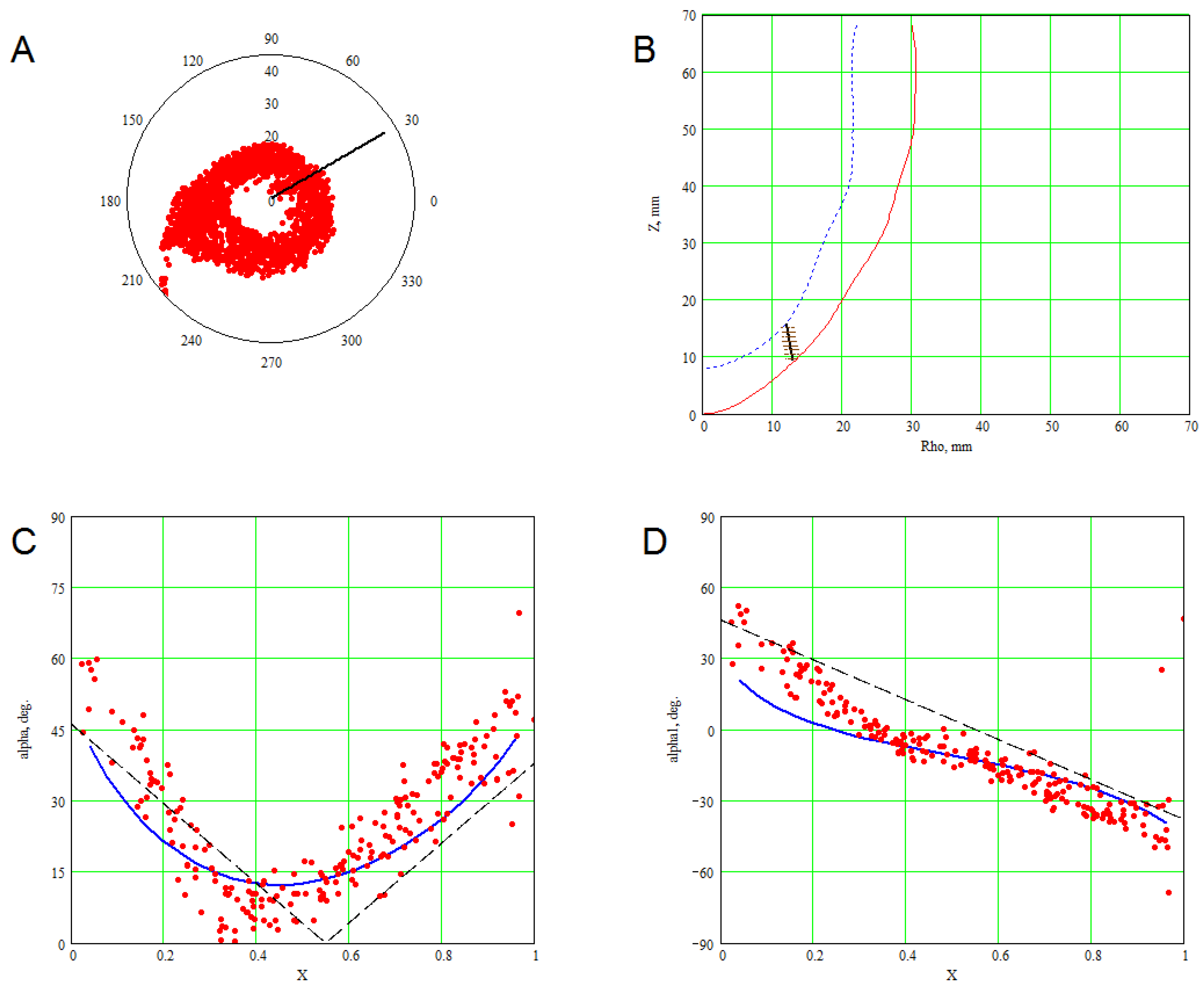
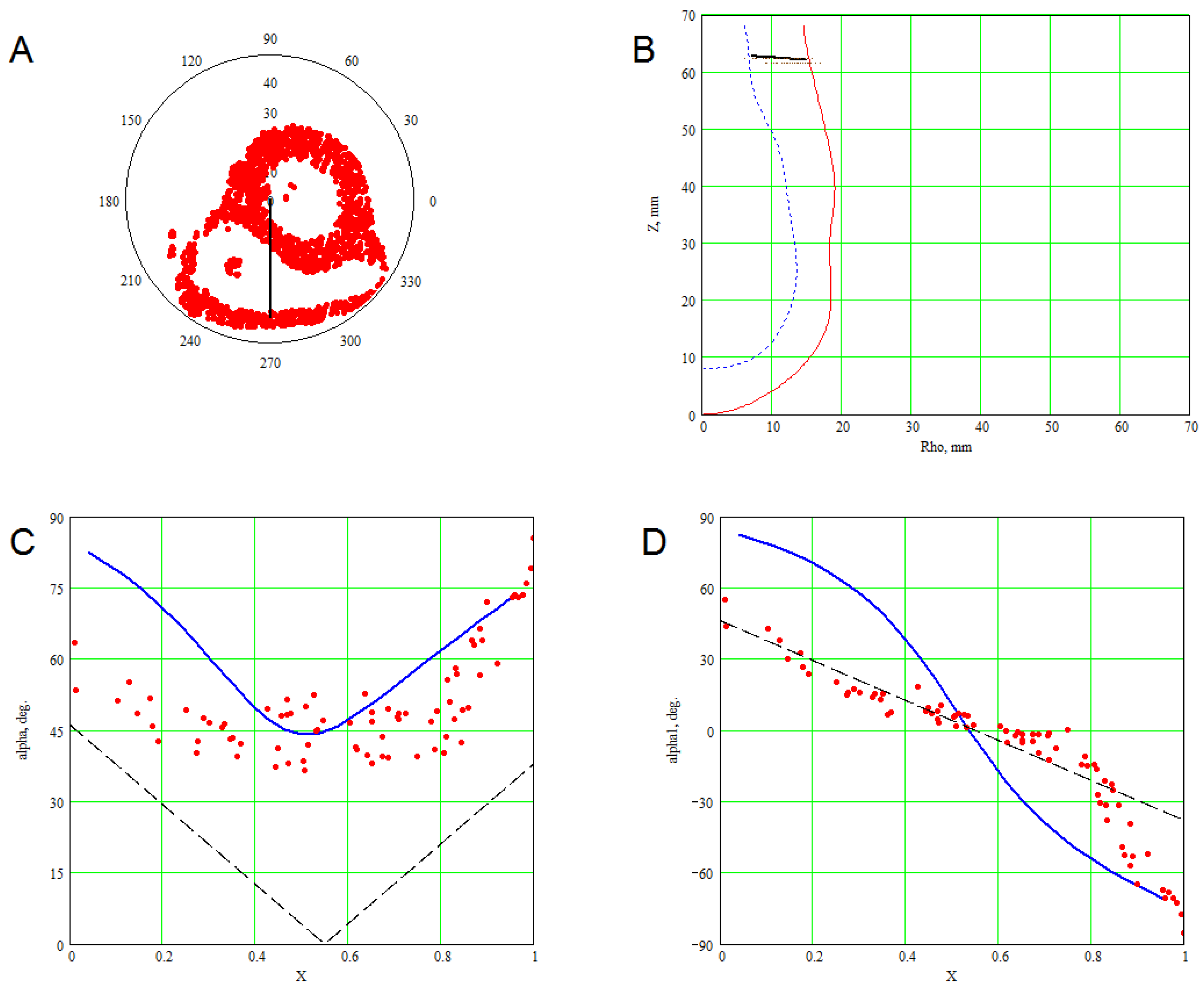
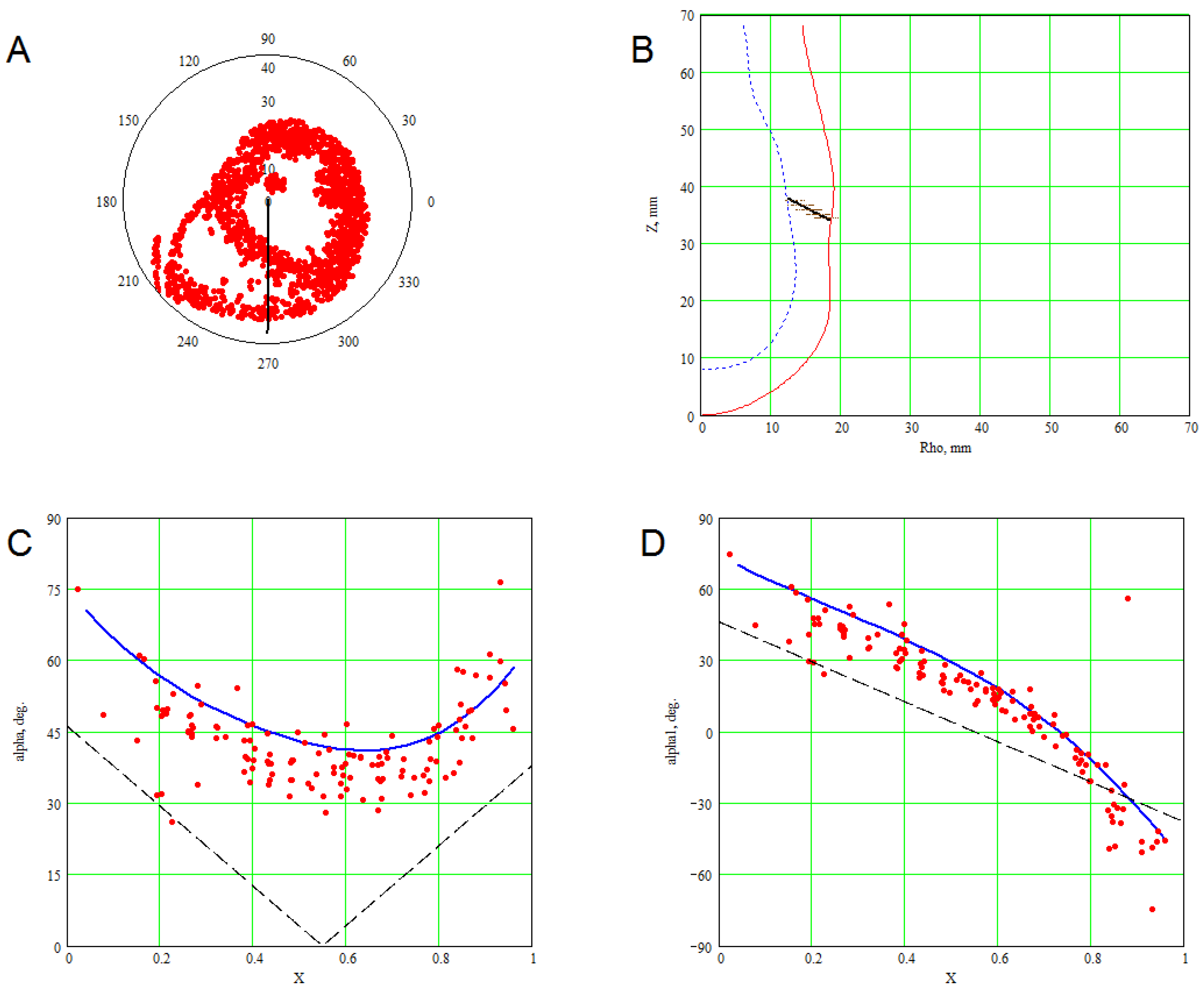
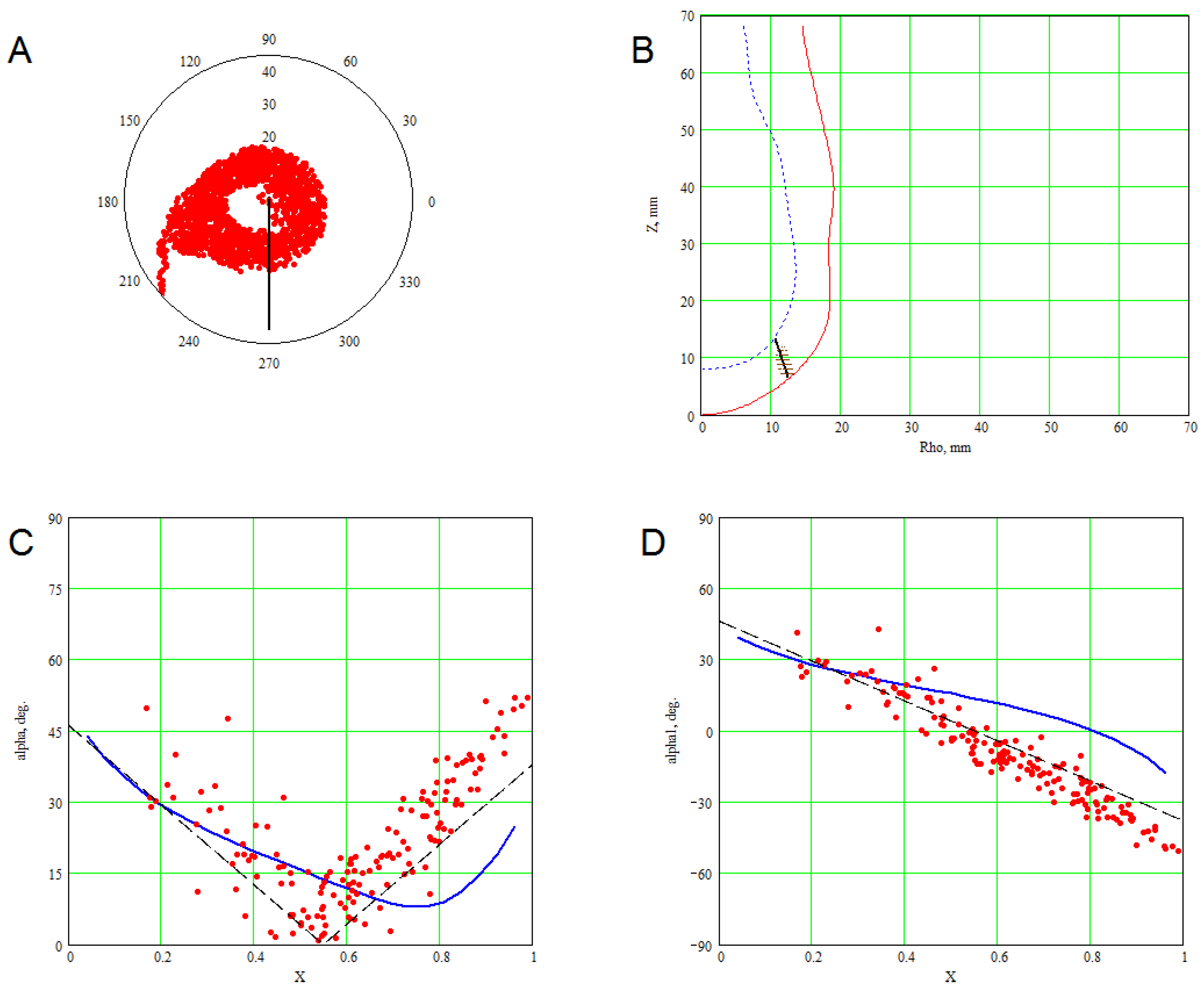
3. Methods for Model and Experimental Data Comparison
4. Results of Comparing the Model with DT-MRI Data
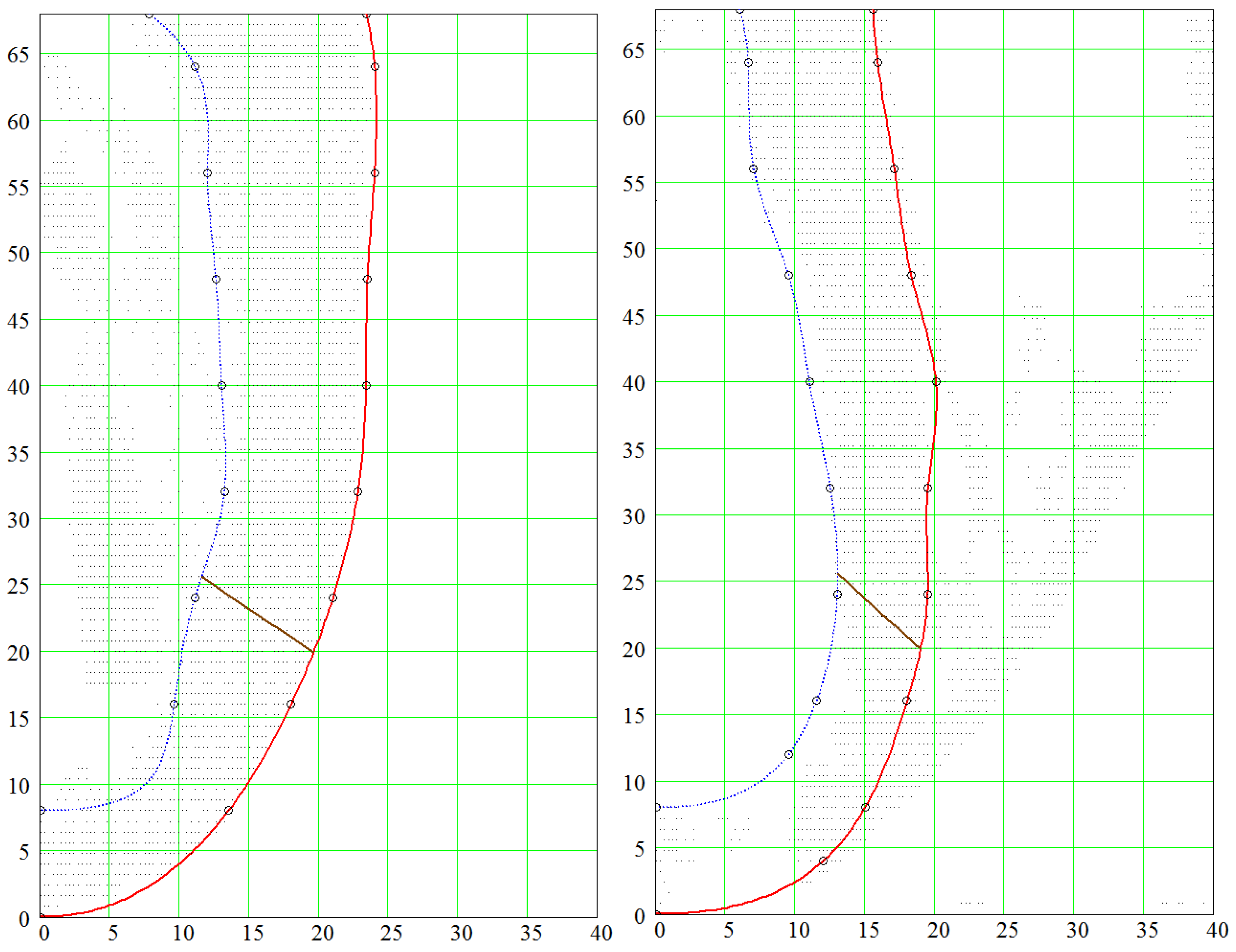
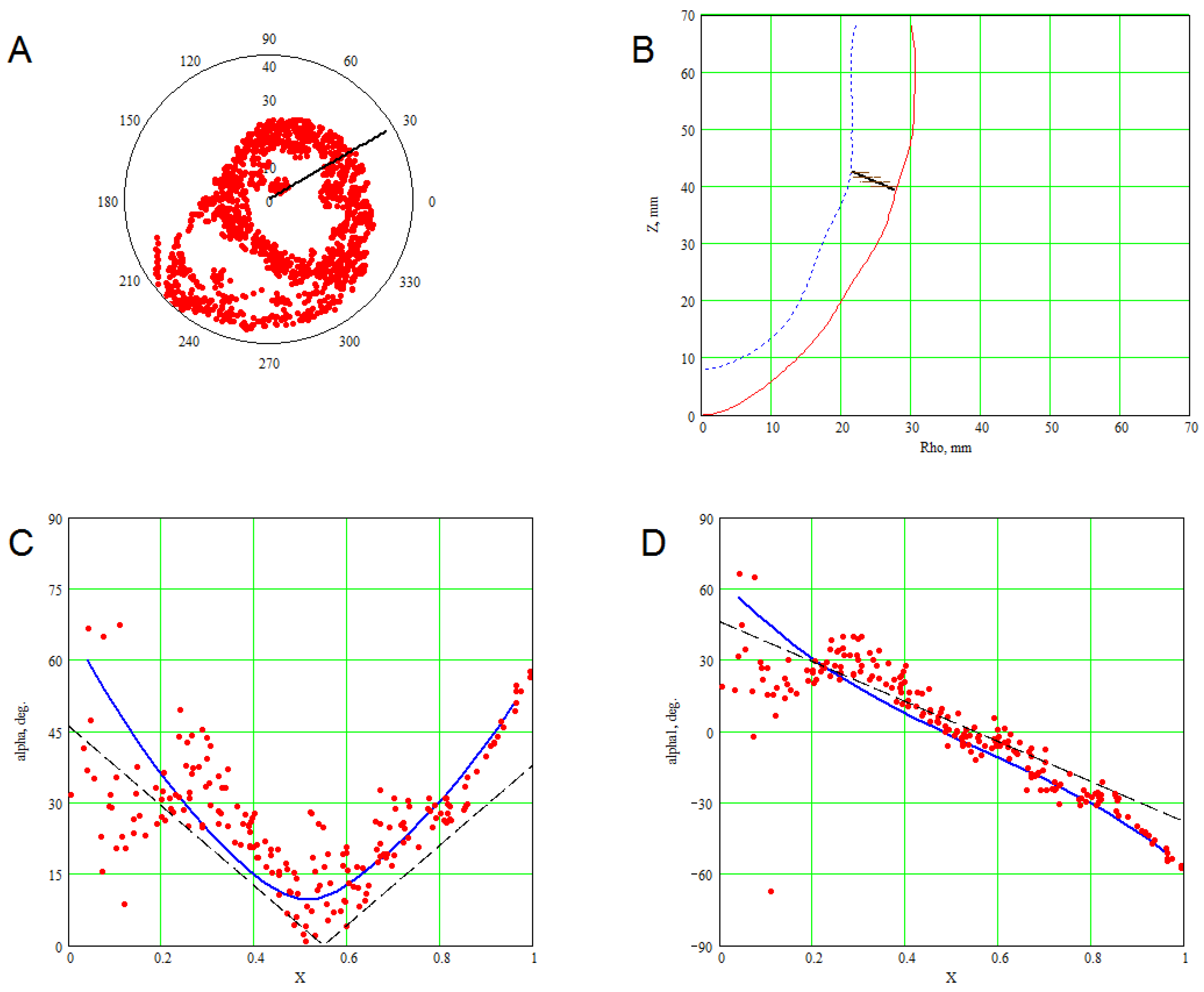
4.1. Comparison of the Model with Human Heart Data along Straight Pinning Lines
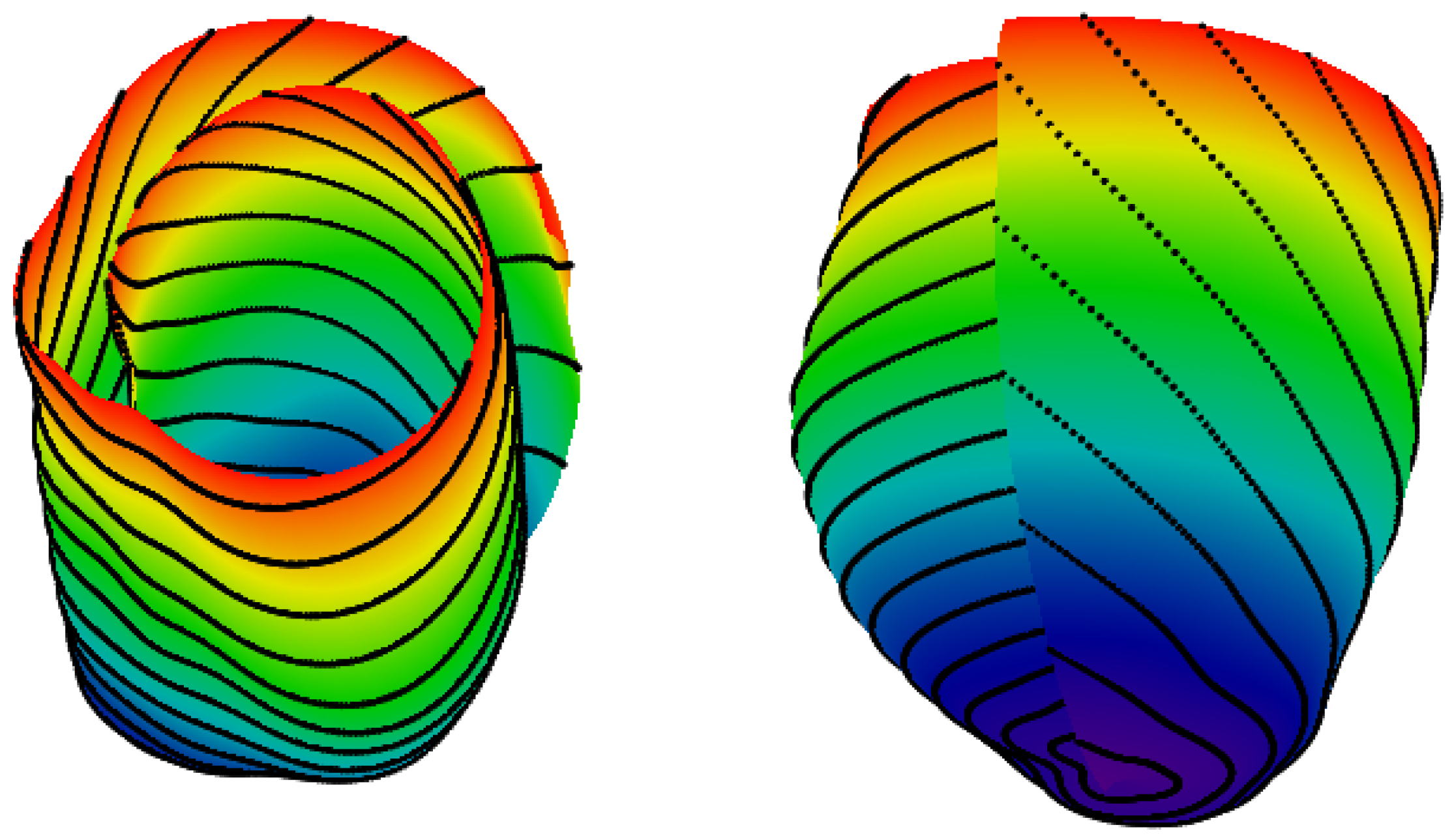
4.2. 3D Verification
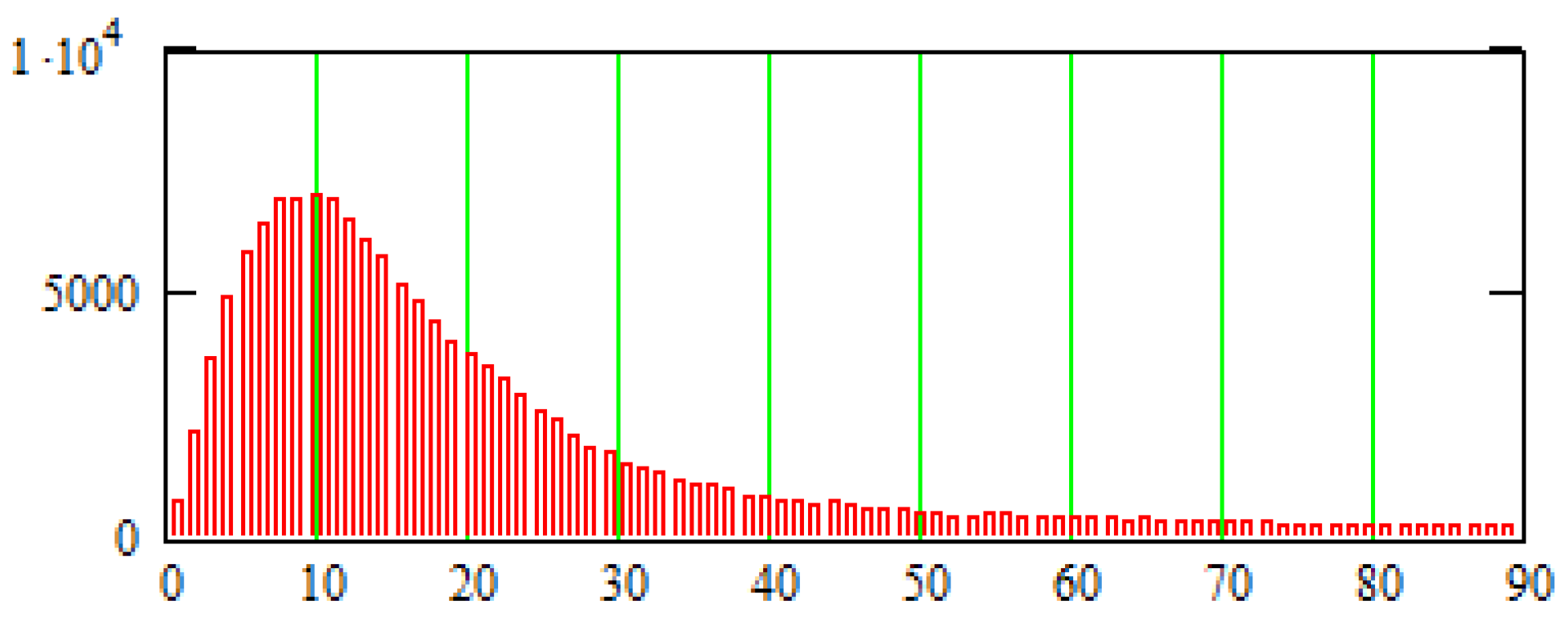
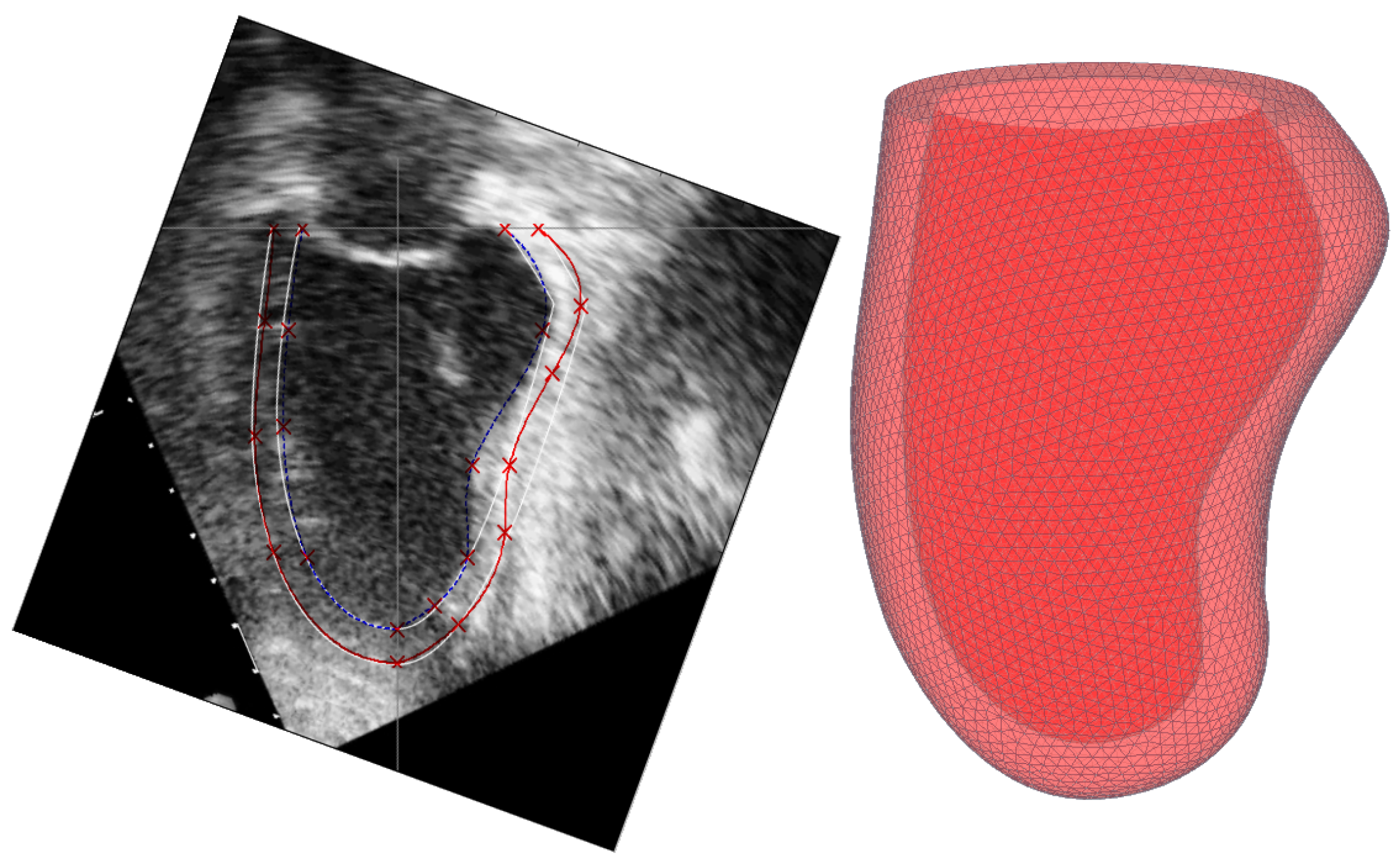
5. Constructing a Model Based on Sonography Data
6. Numerical Method for Solving Reaction-Diffusion Systems on the Model
The Method to Rarefy the Computation Mesh
7. An Example of the Spline-Based Model’s Practical Usage in Electrophysiological Simulations
8. Discussion
8.1. Advantages and Disadvantages of the Proposed Model
- It has relatively few parameters for shape, fibers and sheets.
- The LV apex is smooth thanks to the special choice of function z. This is a useful feature for integration methods sensitive to the smoothness of the boundary.
- It uses only simple one-variable splines, and no two- or three-parametric splines are required.
- The fitting of shape and fiber directions are independent tasks in this model.
- It is flexible enough to fit not only normal, but pathological LVs.
- It yields not only fibers, but also sheets.
- Similar to our previous models, its base is flat, which is not the case in real mammal anatomy.
- The fibers are not geodesic lines in the model sheets. It is difficult to determine if this is a disadvantage of the fibers, the sheets or both.
- Model fibers end at the base. However, this demerit can be amended by combining this model with the toroids-based one described in [62].
- It is unclear whether the model is generalizable for both ventricles.
8.2. The Present Model Compared with Other Qualitative Models
8.3. Comparison with Experimental Data and Quantitative Models
8.4. Further Development and Usage of the Model
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | computed tomography |
| DT-MRI | diffusion tensor magnetic resonance imaging |
| DTI | diffusion tensor imaging |
| IVS | interventricular septum |
| LV | left ventricle |
| RV | right ventricle |
| SS | spiral surface |
Appendix A. Calculation of the Fiber Direction in Points of the LV Model
- Use Formula (5) to find the special coordinates γ and ψ of the point numerically. This problem can be reduced to solving one algebraic equation with one unknown quantity γ on the segment . We utilize the expression for from Formula (1) and substitute it into (5):We solve this equation with respect to γ. Let the root be γ. Hence, the point’s special coordinate
- Differentiate (numerically or analytically) the function with respect to all arguments and obtain three partial derivatives and We can find one derivative analytically:
- The LV model point is an image of a point on the sector . This point, the preimage, has polar coordinates , and Cartesian coordinates , .
- The LV point , parameterized by Φ, has Cartesian coordinates:where , .
- The non-normalized vector of the fiber direction has components:
Appendix B. Formulae for the Laplacian in the Special Coordinates
Appendix C. The No-Flux Boundary Conditions in the Special Coordinates
References
- Trayanova, N.; Constantino, J.; Gurev, V. Electromechanical models of the ventricles. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H279–H286. [Google Scholar] [CrossRef] [PubMed]
- Beyar, R.; Sideman, S. A computer study of the left ventricular performance based on fiber structure, sarcomere dynamics, and transmural electrical propagation velocity. Circ. Res. 1984, 55, 358–375. [Google Scholar] [CrossRef] [PubMed]
- Greenstein, J.; Winslow, R. Integrative systems models of cardiac excitation-contraction coupling. Circ. Res. 2011, 108, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.; McCulloch, A.; Keurs, H.T. Modelling the mechanical properties of cardiac muscle. Prog. Biophys. Mol. Biol. 1998, 69, 289–331. [Google Scholar] [CrossRef]
- Tusscher, K.T.; Noble, D.; Noble, P.; Panfilov, A. A model for human ventricular tissue. Am. J. Physiol. Heart Circ. Physiol. 2004, 286, H1573–H1589. [Google Scholar] [CrossRef] [PubMed]
- Tusscher, K.T.; Panfilov, A. Alternans and spiral breakup in a human ventricular tissue model. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1088–H1100. [Google Scholar] [CrossRef] [PubMed]
- Grandi, E.; Pasqualini, F.; Bers, D. A novel computational model of the human ventricular action potential and Ca transient. J. Mol. Cell. Cardiol. 2010, 48, 112–121. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, T.; Virag, L.; Varro, A.; Rudy, Y. Simulation of the Undiseased Human Cardiac Ventricular Action Potential: Model Formulation and Experimental Validation. PLoS Comput. Biol. 2011, 7, e1002061. [Google Scholar] [CrossRef] [PubMed]
- Sulman, T.; Katsnelson, L.; Solovyova, O.; Markhasin, V. Mathematical modeling of mechanically modulated rhythm disturbances in homogeneous and heterogeneous myocardium with attenuated activity of Na+ – K+ pump. Bull. Math. Biol. 2008, 70, 910–949. [Google Scholar] [CrossRef] [PubMed]
- Niederer, S.; Hunter, P.; Smith, N. A quantitative analysis of cardiac myocyte relaxation: A simulation study. Biophys. J. 2006, 90, 1697–1722. [Google Scholar] [CrossRef] [PubMed]
- Ter Keurs, H.; Shinozaki, T.; Zhang, Y.; Zhang, M.; Wakayama, Y.; Sugai, Y.; Kagaya, Y.; Miura, M.; Boyden, P.; Stuyvers, B.; et al. Sarcomere mechanics in uniform and non-uniform cardiac muscle: A link between pump function and arrhythmias. Prog. Biophys. Mol. Biol. 2008, 97, 312–331. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.; de Tombe, P. Approaches to modeling crossbridges and calcium-dependent activation in cardiac muscle. Prog. Biophys. Mol. Biol. 2004, 85, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Gurev, V.; Lee, T.; Constantino, J.; Arevalo, H.; Trayanova, N. Models of cardiac electromechanics based on individual hearts imaging data: Image-based electromechanical models of the heart. Biomech. Model. Mechanobiol. 2011, 10, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Vetter, F.; McCulloch, A. Three-dimensional analysis of regional cardiac function: A model of rabbit ventricular anatomy. Prog. Biophys. Mol. Biol. 1998, 69, 157–183. [Google Scholar] [CrossRef]
- Nickerson, D.; Smith, N.; Hunter, P. New developments in a strongly coupled cardiac electromechanical model. Europace 2005, 7, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Helm, P.; Tseng, H.; Younes, L.; McVeigh, E.; Winslow, R. Ex Vivo 3D Diffusion Tensor Imaging and Quantification of Cardiac Laminar Structure. Magn. Reson. Med. 2005, 54, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Lunkenheimer, P.; Redmann, K.; Kling, N.; Jiang, X.; Rothaus, K.; Cryer, C.W.; Wübbeling, F.; Niederer, P.; Heitz, P.U.; Ho, S.Y.; et al. Three-Dimensional Architecture of the Left Ventricular Myocardium. Anat. Rec. Part A 2006, 288, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Helm, P.; Beg, M.; Miller, M.; Winslow, R. Measuring and mapping cardiac fiber and laminar architecture using diffusion tensor MR imaging. Ann. N. Y. Acad. Sci. 2005, 1047, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Vadakkumpadan, F.; Arevalo, H.; Prassl, A.; Chen, J.; Kickinger, F.; Kohl, P.; Plank, G.; Trayanova, N. Image-based models of cardiac structure in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Streeter, D. Gross morphology and fiber geometry of the heart. In The Heart; Handbook of Physiology; American Physiology Socienty: Bethesda, MD, USA, 1979; Section 2; Volume I, pp. 61–112. [Google Scholar]
- Nielsen, P.; LeGrice, I.; Smaill, B.; Hunter, P. Mathematical model of the geometry and fibrous structure of the heart. Am. J. Physiol. 1991, 260, H1365–H1378. [Google Scholar] [PubMed]
- LeGrice, I.; Smaill, B.; Chai, L.; Edgar, S.; Gavin, J.; Hunter, P. Laminar structure of the heart: Ventricular myocyte arrangement and connective tissue architecture in the dog. Am. J. Physiol. 1995, 269, H571–H582. [Google Scholar] [PubMed]
- Gilbert, S.H.; Benson, A.P.; Li, P.; Holden, A.V. Regional localisation of left ventricular sheet structure: Integration with current models of cardiac fiber, sheet and band structure. Eur. J. Cardio-Thorac. Surg. 2007, 32, 231–249. [Google Scholar] [CrossRef] [PubMed]
- Frangi, A.; Rueckert, D.; Schnabel, J.; Niessen, W. Automatic construction of multiple-object three-dimensional statistical shape models: Application to cardiac modeling. IEEE Trans. Med. Imaging 2002, 21, 1151–1166. [Google Scholar] [CrossRef] [PubMed]
- Young, A.; Frangi, A. Computational cardiac atlases: From patient to population and back. Exp. Physiol. 2009, 94, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Peyrat, J.; Sermesant, M.; Pennec, X.; Delingette, H.; Xu, C.; McVeigh, E.; Ayache, N. Towards a Statistical Atlas of Cardiac Fiber Structure. Med. Image comput. Comput.-Assist. Interv. 2006, 4190, 297–304. [Google Scholar]
- Peyrat, J.; Sermesant, M.; Pennec, X.; Delingette, H.; Xu, C.; McVeigh, E.; Ayache, N. A computational framework for the statistical analysis of cardiac diffusion tensors: Application to a small database of canine hearts. IEEE Trans. Med. Imaging 2007, 26, 1500–1514. [Google Scholar] [CrossRef] [PubMed]
- Torrent-Guasp, F. The Cardiac Muscle; Fundacion Juan March: Madrid, Spain, 1973. [Google Scholar]
- Rushmer, R.; Crystal, D.; Wagner, C. The functional anatomy of ventricular contraction. Circ. Res. 1953, 1, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Jouk, P.; Usson, Y.; Michalowicz, G.; Grossi, L. Three-dimensional cartography of the pattern of the myofibers in the second trimester fetal human heart. Anat. Embryol. 2000, 202, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Mall, F. On the muscular architecture of the ventricles of the human heart. Am. J. Anat. 1911, 11, 211–266. [Google Scholar] [CrossRef]
- Benninghoff, A. Die Architektur des Herzmuskels. Eine vergleichend anatomische und vergleichend funktionelle Betrachtung. Morph. Jahrb. 1931, 67, 262–317. (In German) [Google Scholar]
- Robb, J.; Robb, R. The normal heart. Anatomy and physiology of the structural units. Am. Heart J. 1942, 23, 455–467. [Google Scholar] [CrossRef]
- Lev, M.; Simkins, C.S. Architecture of the human ventricular myocardium; technic for study using a modification of the Mall–MacCallum method. Lab. Investig. 1956, 5, 396–409. [Google Scholar] [PubMed]
- Torrent-Guasp, F. Organización de la musculatura cardíaca ventricular. In El Fallo Mecánico del Corazón; Ediciones Toray: Barcelona, Spain, 1975; pp. 3–36. (In Spanish) [Google Scholar]
- Lunkenheimer, P.; Redmann, K.; Scheld, H.; Dietl, K.H.; Cryer, C.; Richter, K.D.; Merker, J.; Whimster, W. The heart muscle’s putative “secondary structure”. Functional implications of a band-like anisotropy. Technol. Health Care 1997, 5, 53–64. [Google Scholar] [PubMed]
- Schmid, P.; Niederer, P.; Lunkenheimer, P.; Torrent-Guasp, F. The anisotropic structure of the human left and right ventricles. Technol. Health Care 1997, 5, 29–44. [Google Scholar] [PubMed]
- Torrent-Guasp, F.; Whimster, W.; Redman, K. A silicone rubber mould of the heart. Technol. Health Care 1997, 5, 13–20. [Google Scholar] [PubMed]
- Hort, W. Makroskopische und mikrometrische Untersuchungen am Myokard verschieden stark gefüllter linker Kammern. Virchows Arch. Pathol. Anat. 1960, 333, 523–564. (In German) [Google Scholar] [CrossRef]
- Hort, W. Quantitative morphology and structural dynamics of the myocardium. Methods Achiev. Exp. Pathol. 1971, 5, 3–21. [Google Scholar] [PubMed]
- Fox, C.; Hutchins, G. The architecture of the human ventricular myocardium. Hopkins Med. J. 1972, 130, 289–299. [Google Scholar]
- Krehl, L. Beiträge zur Kenntnis der Füllung und Entleerung des Herzens. Abh Math-Phys Kl Saechs Akad Wiss 1891, 17, 341–362. (In German) [Google Scholar]
- Grant, R. Notes on the muscular architecture of the left ventricle. Circulation 1965, 32, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Seemann, G. Modeling of Electrophysiology and Tension Development in the Human Heart. Ph.D. Thesis, Universitat Karlsruhe, Karlsruhe, Germany, 2005. [Google Scholar]
- Bishop, M.J.; Plank, G.; Burton, R.A.B.; Schneider, J.E.; Gavaghan, D.J.; Grau, V.; Kohl, P. Development of an anatomically detailed MRI-derived rabbit ventricular model and assessment of its impact on simulations of electrophysiological function. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H699–H718. [Google Scholar] [CrossRef] [PubMed]
- Bayer, J.; Blake, R.; Plank, G.; Trayanova, N. A Novel Rule-Based Algorithm for Assigning Myocardial Fiber Orientation to Computational Heart Models. Ann. Biomed. Eng. 2012, 40, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Hren, R. A Realistic Model of the Human Ventricular Myocardium: Application to the Study of Ectopic Activation. Ph.D. Thesis, Dalhousie University, Halifax, NS, Canada, 1996. [Google Scholar]
- Peskin, C. Fiber architecture of the left ventricular wall: An asymptotic analysis. Commun. Pure. Appl. Math. 1989, 42, 79–113. [Google Scholar] [CrossRef]
- Chadwick, R. Mechanics of the left ventricle. Biophys. J. 1982, 39, 279–288. [Google Scholar] [CrossRef]
- Bovendeerd, P.; Arts, T.; Huyghe, J.; Campen, D.V.; Reneman, R. Dependence of local left ventricular wall mechanics on myocardial fiber orientation: A model study. J Biomech. 1992, 25, 1129–1140. [Google Scholar] [CrossRef]
- Arts, T.; Veenstra, P.; Reneman, R. Epicardial deformation and left ventricular wall mechanics during ejection in the dog. Am. J. Physiol. 1982, 243, H379–H390. [Google Scholar] [PubMed]
- Pettigrew, J. On the arrangement of the muscular fibers of the ventricular portion of the heart of the mammal. Proc. Roy. Soc. Lond. 1860, 10, 433–440. [Google Scholar] [CrossRef]
- Pravdin, S.; Berdyshev, V.; Panfilov, A.; Katsnelson, L.; Solovyova, O.; Markhasin, V. Mathematical Model of the Anatomy and Fiber Orientation Field of the Left Ventricle of the Heart. Biomed. Eng. Online 2013, 54, 1–21. [Google Scholar]
- Pravdin, S. Non-axisymmetric mathematical model of the cardiac left ventricle anatomy. Russ. J. Biomech. 2013, 17, 75–94. [Google Scholar]
- Cardiac DT-MRI data. Available online: http://gforge.icm.jhu.edu/gf/project/dtmri_data_sets/docman/ (accessed on 24 October 2016).
- Smoothn. Available online: http://www.biomecardio.com/matlab/smoothn.html (accessed on 24 October 2016).
- Advanced Numerical Instruments 3D-Ani3D. Available online: https://sourceforge.net/projects/ani3d/ (accessed on 24 October 2016).
- Pravdin, S.; Dierckx, H.; Katsnelson, L.; Solovyova, O.; Markhasin, V.; Panfilov, A. Electrical Wave Propagation in an Anisotropic Model of the Left Ventricle Based on Analytical Description of Cardiac Architecture. PLoS ONE 2014, 9, e93617. [Google Scholar] [CrossRef] [PubMed]
- Pravdin, S.; Dierckx, H.; Markhasin, V.; Panfilov, A. Drift of scroll wave filaments in an anisotropic model of the left ventricle of the human heart. BioMed. Res. Int. J. 2015, 2015, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pravdin, S. A method of solving reaction-diffusion problem on a non-symmetrical model of the cardiac left ventricle. In Proceedings of the 47th International Youth School-Conference “Modern Problems in Mathematics and Its Applications”, Ekaterinburg, Russia, 31 January–6 February 2016; pp. 284–296.
- Aliev, R.; Panfilov, A. A simple two-variable model of cardiac excitation. Chaos Solitons Fractals 1996, 7, 293–301. [Google Scholar] [CrossRef]
- Koshelev, A.A.; Bazhutina, A.E.; Pravdin, S.F.; Ushenin, K.S.; Katsnelson, L.B.; Solovyova, O.E. A modified mathematical model of the cardiac left ventricular anatomy. Biofizika 2016, 61, 986–995. (In Russian) [Google Scholar]
- Kocica, M.J.; Corno, A.F.; Carreras-Costa, F.; Ballester-Rodes, M.; Moghbel, M.C.; Cueva, C.N.; Lackovic, V.; Kanjuh, V.I.; Torrent-Guasp, F. The helical ventricular myocardial band: Global, three-dimensional, functional architecture of the ventricular myocardium. Eur. J. Cardio-Thorac. Surg. 2006, 29, S21–S40. [Google Scholar] [CrossRef] [PubMed]
- Rethinking the Cardiac Helix—A Structure/Function Journey: Overview. Eur. J. Cardio-Thorac. Surg. 2006, 29. [CrossRef]
- Anderson, R.H.; Ho, S.Y.; Redmann, K.; Sanchez-Quintana, D.; Lunkenheimer, P.P. The anatomical arrangement of the myocardial cells making up the ventricular mass. Eur. J. Cardio-Thorac. Surg. 2005, 28, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Aslanidi, O.; Nikolaidou, T.; Zhao, J.; Smaill, B.; Gilbert, S.; Holden, A.; Lowe, T.; Withers, P.; Stephenson, R.; Jarvis, J.; et al. Application of Micro-Computed Tomography With Iodine Staining to Cardiac Imaging, Segmentation, and Computational Model Development. IEEE Trans. Med. Imaging 2013, 32, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Trew, M.; Caldwell, B.; Sands, G.; LeGrice, I.; Smaill, B. Three-dimensional cardiac tissue image registration for analysis of in vivo electrical mapping. Ann. Biomed. Eng. 2011, 39, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Vicky, Y.W. Modelling In Vivo Cardiac Mechanics using MRI and FEM. Ph.D. Thesis, Auckland Bioengineering Institute, The University of Auckland, Auckland, New Zealand, 2012. [Google Scholar]
- Zhang, Y.; Liang, X.; Ma, J.; Jing, Y.; Gonzales, M.J.; Villongco, C.; Krishnamurthy, A.; Frank, L.R.; Nigam, V.; Stark, P.; et al. An atlas-based geometry pipeline for cardiac Hermite model construction and diffusion tensor reorientation. Med. Image Anal. 2012, 16, 1130–1141. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Stein, K.M.; Christini, D.J. Critical role of inhomogeneities in pacing termination of cardiac reentry. Chaos Interdiscip. J. Nonlinear Sci. 2002, 12, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Denney, T.S.J. Left ventricular motion reconstruction with a prolate spheroidal B-spline model. Phys. Med. Biol. 2006, 51, 517–537. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Gomez, A.D.; Morgan, J.L.; Hsu, E.W. Parametric modeling of the mouse left ventricular myocardial fiber structure. Ann. Biomed. Eng. 2016, 44, 2661–2673. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.G.; Howard, E.; Aguado-Sierra, J.; Coppola, B.A.; Omens, J.H.; Mulligan, L.J.; McCulloch, A.D.; Kerckhoffs, R.C.P. Effect of transmurally heterogeneous myocyte excitation-contraction coupling on canine left ventricular electromechanics. Exp. Physiol. 2009, 94, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.C.L.; Billet, F.; Mansi, T.; Chabiniok, R.; Sermesant, M.; Delingette, H.; Ayache, N. Cardiac Motion Estimation Using a ProActive Deformable Model: Evaluation and Sensitivity Analysis. In Statistical Atlases and Computational Models of the Heart; Springer: Berlin/Heidelberg, Germany, 2010; pp. 154–163. [Google Scholar]
- Sánchez, R.A. Electromechanical Large Scale Computational Models of the Ventricular Myocardium. Ph.D. Thesis, Universitat Politecnica de Catalunya, Barcelona, Spain, 2014. [Google Scholar]
- Baron, L.; Fritz, T.; Seemann, G.; Dössel, O. Sensitivity study of fiber orientation on stroke volume in the human left ventricle. In Proceedings of the Computing in Cardiology Conference (CinC), Cambridge, MA, USA, 7–10 September 2014; pp. 681–684.






| True Fiber | Canine | Human | ||
|---|---|---|---|---|
| Angle | Raw Data | Averaged | Raw Data | Averaged |
| mean | ||||
| median | ||||
| st. dev. | ||||
| Helix | Canine | Human | ||
|---|---|---|---|---|
| Angle | Raw Data | Averaged | Raw Data | Averaged |
| mean | ||||
| median | ||||
| st. dev. | ||||
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pravdin, S. A Mathematical Spline-Based Model of Cardiac Left Ventricle Anatomy and Morphology. Computation 2016, 4, 42. https://doi.org/10.3390/computation4040042
Pravdin S. A Mathematical Spline-Based Model of Cardiac Left Ventricle Anatomy and Morphology. Computation. 2016; 4(4):42. https://doi.org/10.3390/computation4040042
Chicago/Turabian StylePravdin, Sergei. 2016. "A Mathematical Spline-Based Model of Cardiac Left Ventricle Anatomy and Morphology" Computation 4, no. 4: 42. https://doi.org/10.3390/computation4040042
APA StylePravdin, S. (2016). A Mathematical Spline-Based Model of Cardiac Left Ventricle Anatomy and Morphology. Computation, 4(4), 42. https://doi.org/10.3390/computation4040042






