Designing Microfluidic PCR Chip Device Using CFD Software for the Detection of Malaria
Abstract
1. Introduction
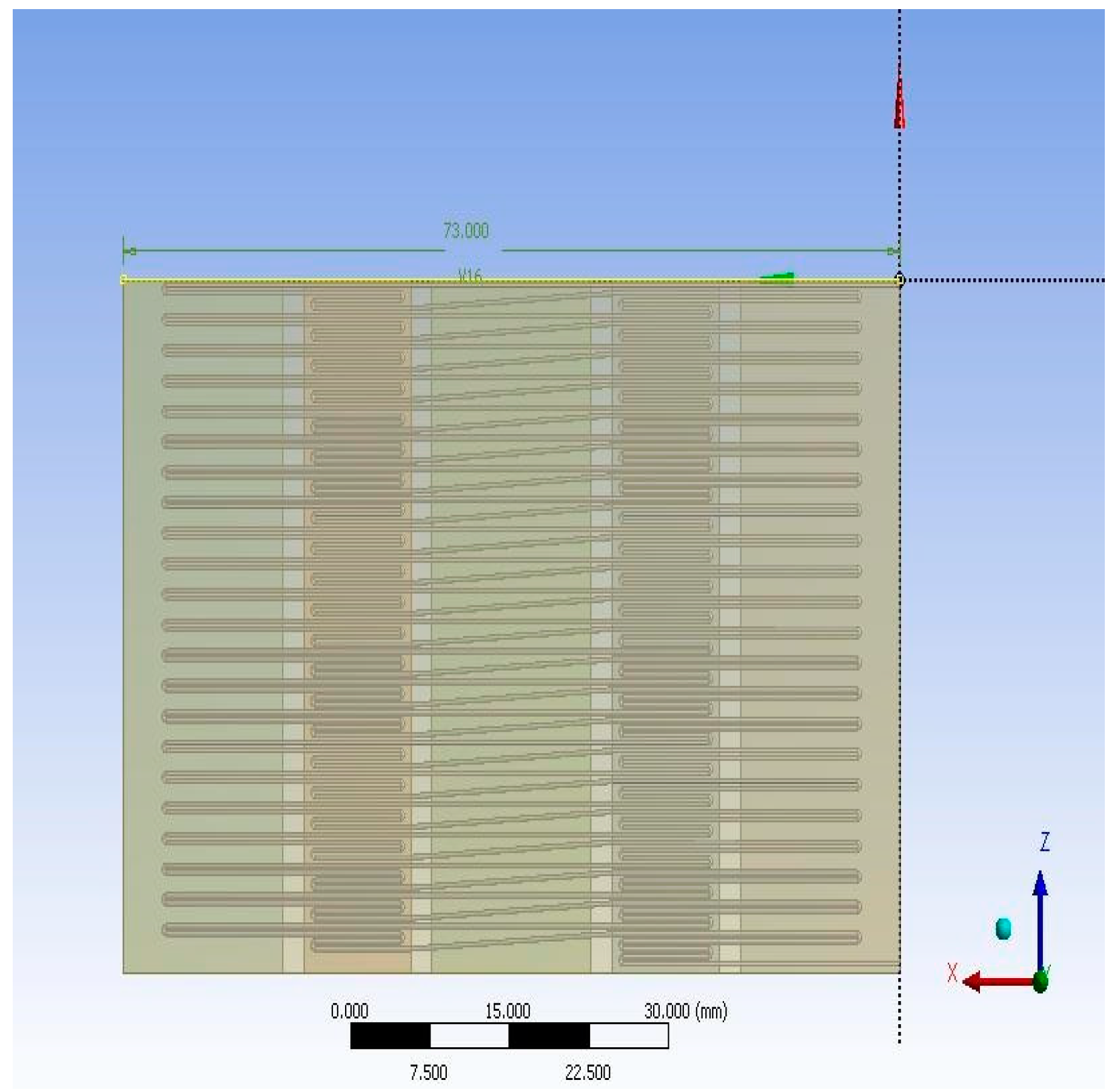
2. Materials and Methods
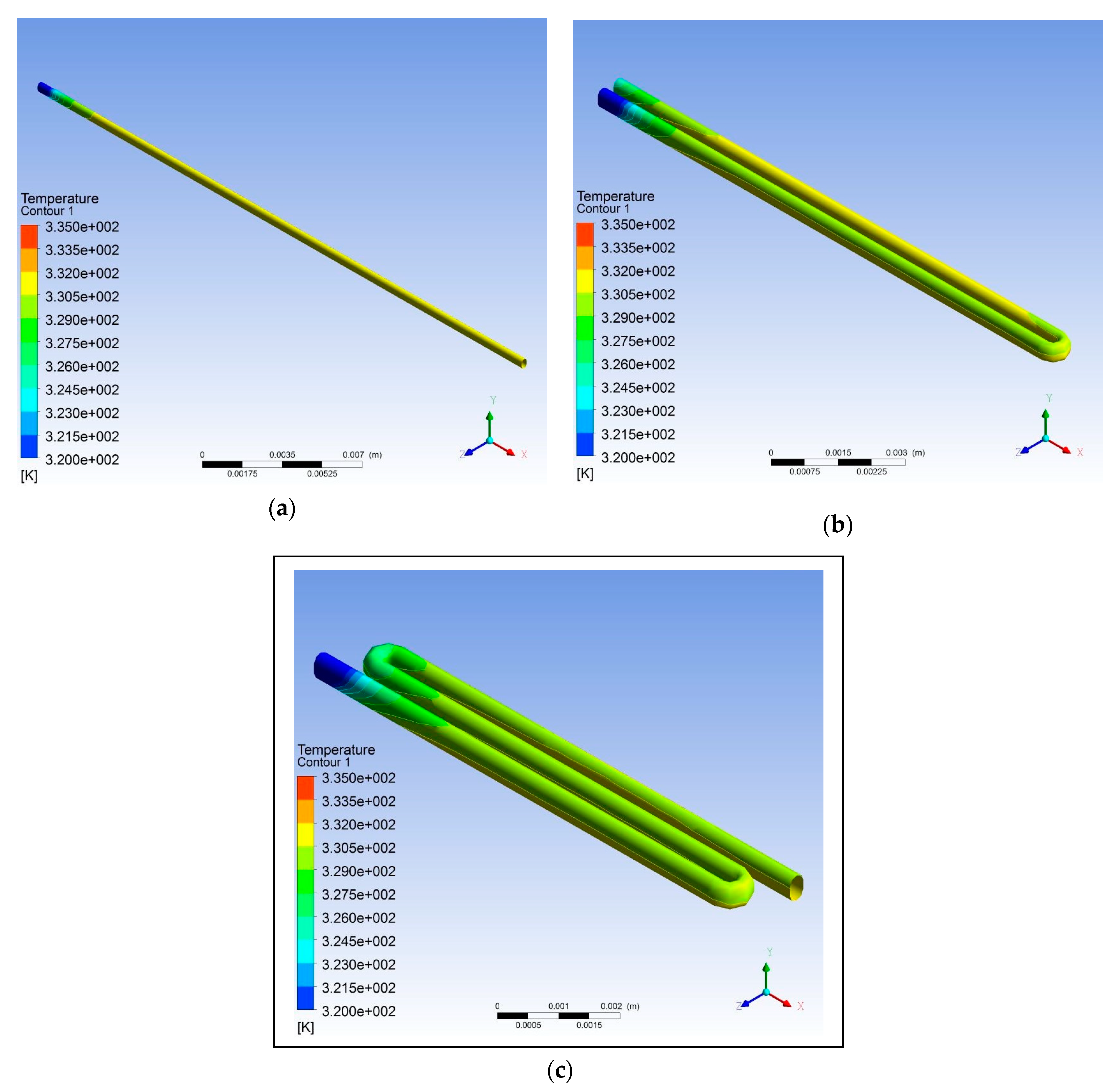
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sakihama, N.; Mitamura, T.; Kaneko, A.; Horii, T.; Tanabe, K. Long PCR Amplification of Plasmodium falciparum DNA Extracted from Filter Paper Blots. Exp. Parasitol. 2001, 97, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Ansah, F.; Suurbaar, J.; Darko, D.; Anabire, N.G.; Blankson, S.O.; Domson, B.K.S.; Soulama, A.; Kpasra, P.; Chirawurah, J.D.; Amenga-Etego, L.; et al. Development of Cooperative Primer-Based Real-Time PCR Assays for the Detection of Plasmodium malariae and Plasmodium ovale. J. Mol. Diagn. 2021, 23, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Azmi, W.A.; Rizki, A.F.M.; Djuardi, Y.; Artika, I.M.; Siregar, J.E. Molecular insights into artemisinin resistance in Plasmodium falciparum: An updated review. Infect. Genet. Evol. 2023, 112, 105460. [Google Scholar] [CrossRef] [PubMed]
- Torkashvand, H.; Dehdast, S.A.; Nateghpour, M.; Haghi, A.M.; Fard, G.C.; Elmi, T.; Shabani, M.; Tabatabaie, F. Antimalarial nano-drug delivery system based on graphene quantum dot on Plasmodium falciparum: Preparation, characterization, toxicological evaluation. Diam. Relat. Mater. 2023, 132, 109670. [Google Scholar] [CrossRef]
- Chemwor, G.C.; Andagalu, B.M.; Onyango, I.A.; Opot, B.H.; Okoth, R.O.; Yedah, R.A.; Juma, J.A.; Mwakio, E.W.; Wakoli, D.M.; Amwoma, J.G.; et al. Therapeutic response to artemisinin combination therapies among individuals with Plasmodium falciparum single infection vs mixed Plasmodium species infections: A retrospective posthoc analysis in Kisumu County, western Kenya. Int. J. Infect. Dis. 2023, 132, 17–25. [Google Scholar] [CrossRef]
- Xu, S.-J.; Shen, H.-M.; Cui, Y.-B.; Chen, S.-B.; Xu, B.; Chen, J.-H. Genetic diversity and natural selection of rif gene (PF3D7_1254800) in the Plasmodium falciparum global populations. Mol. Biochem. Parasitol. 2023, 254, 111558. [Google Scholar] [CrossRef]
- Dong, L.; Li, W.; Xu, Q.; Gu, J.; Kang, Z.; Chen, J.; Xu, X.; Zhang, X.; Zhang, X.; Jiang, H.; et al. A rapid multiplex assay of human malaria parasites by digital PCR. Clin. Chim. Acta 2023, 539, 70–78. [Google Scholar] [CrossRef]
- Taghdiri, A.; Almani, P.G.N.; Sharifi, I.; Mohammadi, M.A.; Salari, S. Detection of malaria with light microscopy and Nested polymerase chain reaction (Nested PCR) methods in peripheral blood expansions and investigation of the genetic diversity of Plasmodium species by 18S rRNA gene in Southeast of Iran. Microb. Pathog. 2019, 137, 103782. [Google Scholar] [CrossRef]
- Chagas, C.R.F.; Harl, J.; Valkiūnas, G. Co-infections of Plasmodium relictum lineages pSGS1 and pGRW04 are readily distinguishable by broadly used PCR-based protocols, with remarks on global distribution of these malaria parasites. Acta Trop. 2021, 217, 105860. [Google Scholar] [CrossRef]
- Frickmann, H.; Wegner, C.; Ruben, S.; Behrens, C.; Kollenda, H.; Hinz, R.; Rojak, S.; Schwarz, N.G.; Hagen, R.M.; Tannich, E. Evaluation of the multiplex real-time PCR assays RealStar malaria S&T PCR kit 1.0 and FTD malaria differentiation for the differentiation of Plasmodium species in clinical samples. Travel Med. Infect. Dis. 2019, 31, 101442. [Google Scholar] [CrossRef]
- Mirahmadi, H.; Rahmati-Balaghaleh, M.; Afzalaghaee, M.; Zarean, M.; Shamsian, S.A.; Mehravaran, A.; Raissi, V.; Etemadi, S. Detection of malaria using blood smear by light microscopy, RDT and nested-PCR for suspected patients in south-eastern Iran. Gene Rep. 2021, 25, 101339. [Google Scholar] [CrossRef]
- Xiong, X.; Hou, A.; Yi, S.; Guo, Y.; Zhao, Z.; Wu, Z.; Cheng, H.; Li, K.; Li, Z.; Ren, Y.; et al. Analysis of oral microorganism diversity in healthy individuals before and after chewing areca nuts using PCR-denatured gradient gel electrophoresis. Anim. Nutr. 2018, 4, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Hesham, A.E.-L.; Gupta, V.K.; Singh, B.P. Use of PCR-denaturing gradient gel electrophoresis for the discrimination of Candida species isolated from natural habitats. Microb. Pathog. 2018, 120, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, C.; Nag, S.; Goyal, M.; Saha, D.; Siddiqui, A.A.; Mazumder, S.; Debsharma, S.; Pramanik, S.; Bandyopadhyay, U. Nuclease activity of Plasmodium falciparum Alba family protein PfAlba3. Cell Rep. 2023, 42, 112292. [Google Scholar] [CrossRef]
- Wu, Y.; Leyk, S.; Torabi, H.; Höhn, K.; Honecker, B.; Tauler, M.D.P.M.; Cadar, D.; Jacobs, T.; Bruchhaus, I.; Metwally, N.G. Plasmodium falciparum infection reshapes the human microRNA profiles of red blood cells and their extracellular vesicles. IScience 2023, 26, 107119. [Google Scholar] [CrossRef]
- Zhao, X.; Li, M.; Liu, Y. Microfluidic-Based Approaches for Foodborne Pathogen Detection. Microorganisms 2019, 7, 381. [Google Scholar] [CrossRef]
- Wang, X.; Hong, X.-Z.; Li, Y.-W.; Li, Y.; Wang, J.; Chen, P.; Liu, B.-F. Microfluidics-based strategies for molecular diagnostics of infectious diseases. Mil. Med. Res. 2022, 9, 11. [Google Scholar] [CrossRef]
- Hewlin, R.L.; Edwards, M.; Schultz, C. Design and Development of a Traveling Wave Ferro-Microfluidic Device and System Rig for Potential Magnetophoretic Cell Separation and Sorting in a Water-Based Ferrofluid. Micromachines 2023, 14, 889. [Google Scholar] [CrossRef]
- Hewlin, R.; Edwards, M.; Smith, M. A 2D Transient Computational Multi-Physics Model for Analyzing Magnetic and Non-Magnetic Particle (Red Blood Cells and E. Coli bacteria) Dynamics in a Travelling Wave Ferro-Magnetic Microfluidic Device for Potential Cell Separation and Sorting. J. Eng. Sci. Med. Diagn. Ther. 2023, 14, 1–47. [Google Scholar] [CrossRef]
- Bakuova, N.; Toktarkan, S.; Dyussembinov, D.; Azhibek, D.; Rakhymzhanov, A.; Kostas, K.; Kulsharova, G. Design, Simulation, and Evaluation of Polymer-Based Microfluidic Devices via Computational Fluid Dynamics and Cell Culture “On-Chip”. Biosensors 2023, 13, 754. [Google Scholar] [CrossRef]
- Xiang, Z.; Li, D.; Wang, S.; Shen, T.; He, W.; Li, M.; Zeng, W.; Chen, X.; Wu, Y.; Cui, L.; et al. A simple alkali lysis method for Plasmodium falciparum DNA extraction from filter paper blood samples. Mol. Biochem. Parasitol. 2023, 254, 111557. [Google Scholar] [CrossRef] [PubMed]
- Choubey, D.; Deshmukh, B.; Rao, A.G.; Kanyal, A.; Hati, A.K.; Roy, S.; Karmodiya, K. Genomic analysis of Indian isolates of Plasmodium falciparum: Implications for drug resistance and virulence factors. Int. J. Parasitol. Drugs Drug Resist. 2023, 22, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimzadeh, A.; Fouladi, B.; Fazaeli, A. High rate of detection of mixed infections of Plasmodium vivax and Plasmodium falciparum in South-East of Iran, using nested PCR. Parasitol. Int. 2007, 56, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Wångdahl, A.; Bogale, R.T.; Eliasson, I.; Broumou, I.; Faroogh, F.; Lind, F.; Vashchuk, G.; Hildell, A.; Franson, S.; Hallberg, E.; et al. Malaria parasite prevalence in Sub-Saharan African migrants screened in Sweden: A cross-sectional study. Lancet Reg. Health-Eur. 2023, 27, 100581. [Google Scholar] [CrossRef] [PubMed]
- Fall, A.K.D.J.; Dechavanne, C.; Sabbagh, A.; Garcia, A.; Courtin, D.; Migot-Nabias, F. Combined polymorphisms involving the IgG heavy chain and Fc gamma receptors among Fulani and non-Fulani in Benin: Implications for the natural protection of young Fulani against Plasmodium falciparum malaria infections. Infect. Genet. Evol. 2023, 112, 105461. [Google Scholar] [CrossRef]
- Hermsen, C.C.; Telgt, D.S.C.; Linders, E.H.P.; Van De Locht, L.A.T.F.; Eling, W.M.C.; Mensink, E.J.B.M.; Sauerwein, R.W. Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol. Biochem. Parasitol. 2001, 118, 247–251. [Google Scholar] [CrossRef]
- Fernandas, N.E.P.; Silveira, H.; Franco, A.S.; Arez, A.P.; Forte, J.M.; Rosário, V.E.D. Detection of malaria parasites in paraffin-embedded spleen and placental tissues by nested PCR. Trans. R. Soc. Trop. Med. Hyg. 2001, 95, 293–294. [Google Scholar] [CrossRef]
- Maier, C.; Calafut, T. Polypropylene: The Definitive User’s Guide and Databook; Elsevier Science: Amsterdam, The Netherlands, 2008; Available online: https://books.google.dk/books?id=AWaSJd9Non8C (accessed on 22 July 2023).
- Bair, H.E.; Falcone, D.R.; Hellman, M.Y.; Johnson, G.E.; Kelleher, P.G. Hydrolysis of polycarbonate to yield BPA. J. Appl. Polym. Sci. 1981, 26, 1777–1786. [Google Scholar] [CrossRef]
- Gale, B.K.; Eddings, M.A.; Sundberg, S.O.; Hatch, A.; Kim, J.; Ho, T.; Karazi, S.M. Low-Cost MEMS Technologies. In Reference Module in Materials Science and Materials Engineering; Elsevier Science: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- Yang, X.D.; Melnik, R.V.N. Effect of internal viscosity on Brownian dynamics of DNA molecules in shear flow. Comput. Biol. Chem. 2007, 31, 110–114. [Google Scholar] [CrossRef][Green Version]
- Agrawal, A.P.; Ali, S.; Rathore, S. Finite element stress analysis for shape optimization of spur gear using ANSYS. Mater. Today Proc. 2022, 64, 1147–1152. [Google Scholar] [CrossRef]
- Karthick, M.; Ramakrishna, C.S.; Pugazhenthi, R.; Gudadhe, N.; Baskar, S.; Renu; Kumar, R. Contact stress analysis of xylon coated spur gear using ANSYS workbench. Mater. Today Proc. 2023, 4, 232–237. [Google Scholar] [CrossRef]
- Bhasha, A.C.; Balamurugan, K. Fracture analysis of fuselage wing joint developed by aerodynamic structural materials. Mater. Today Proc. 2021, 38, 2555–2562. [Google Scholar] [CrossRef]
- Malika, M.; Bhad, R.; Sonawane, S.S. ANSYS simulation study of a low volume fraction CuO–ZnO/water hybrid nanofluid in a shell and tube heat exchanger. J. Indian Chem. Soc. 2021, 98, 100200. [Google Scholar] [CrossRef]
- Bhola, M.; Singh, S. Analysis of heat transfer of turbulent channel using different geometries using ANSYS. Mater. Today Proc. 2022, 64, 1223–1228. [Google Scholar] [CrossRef]
- Costales, C.; Broadhurst, M.J.; Budvytiene, I.; Banaei, N. Clinical accuracy of malaria loop-mediated isothermal amplification assay as a stand-alone screening tool at a non-endemic Northern California regional health system. Diagn. Microbiol. Infect. Dis. 2022, 103, 115680. [Google Scholar] [CrossRef]
- Pian, H.; Yang, M.; Sun, X.; Zheng, Z. Sandwich hybridization-based loop-mediated isothermal amplification (SHB-LAMP) for high-throughput detection of malaria RNA from asymptomatic infections. Sens. Actuators B Chem. 2022, 365, 131973. [Google Scholar] [CrossRef]
- Zen, L.P.Y.; Lai, M.Y.; Rozlan, S.I.B.; Hamid, M.H.A.; Jelip, J.; Mudin, R.N.; Lau, Y.L. End-point detection of loop-mediated isothermal amplification (LAMP) on malaria by direct observation with colorimetric dyes. Exp. Parasitol. 2022, 239, 108310. [Google Scholar] [CrossRef]
- Picot, S.; Cucherat, M.; Bienvenu, A.L. Systematic review and meta-analysis of diagnostic accuracy of loop-mediated isothermal amplification (LAMP) methods compared with microscopy, polymerase chain reaction and rapid diagnostic tests for malaria diagnosis. Int. J. Infect. Dis. 2020, 98, 408–419. [Google Scholar] [CrossRef]
- Dong, X.; Liu, L.; Tu, Y.; Zhang, J.; Miao, G.; Zhang, L.; Ge, S.; Xia, N.; Yu, D.; Qiu, X. Rapid PCR powered by microfluidics: A quick review under the background of COVID-19 pandemic. Trac. Trends Anal. Chem. 2021, 143, 116377. [Google Scholar] [CrossRef]
- Tsai, Y.S.; Wang, C.H.; Tsai, H.P.; Shan, Y.S.; Lee, G.B. Electromagnetically-driven integrated microfluidic platform using reverse transcription loop-mediated isothermal amplification for detection of severe acute respiratory syndrome coronavirus 2. Anal. Chim. Acta 2022, 1219, 340036. [Google Scholar] [CrossRef]
- Struijk, R.; van den Ouden, A.; Louwerse, J.; Čurová, K.; Burggrave, R.; McNally, B.; de Groot, T.; Mulder, B.; de Vos, G. Ultrafast RNA extraction-free SARS-CoV-2 detection by direct RT-PCR using a rapid thermal cycling approach. Diagn. Microbiol. Infect. Dis. 2023, 107, 115975. [Google Scholar] [CrossRef] [PubMed]
- Brons, J.K.; Vink, S.N.; de Vos, M.G.J.; Reuter, S.; Dobrindt, U.; van Elsas, J.D. Fast identification of Escherichia coli in urinary tract infections using a virulence gene based PCR approach in a novel thermal cycler. J. Microbiol. Methods 2020, 169, 105799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, C.; Shi, Y.; Wu, J.; Wu, J.; Chen, H. Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application. Talanta 2020, 214, 120818. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.K.; Qin, H.; Molodysky, E.; Morris, B.J. Simple microwave and thermal cycler boiling methods for preparation of cervicovaginal lavage cell samples prior to PCR for human papillomavirus detection. J. Virol. Methods 1993, 44, 77–81. [Google Scholar] [CrossRef]
- Uribe, M.R.; Adams, S.; Barracchini, K.; Hackett, J.; Marincola, F.; Stroncek, D. An alternative PCR-based method for monitoring thermal cycler performance. Hum. Immunol. 2003, 64, S177. [Google Scholar] [CrossRef]
- Nabuti, J.; Elbab, A.R.F.; Abdel-Mawgood, A.; Yoshihisa, M.; Shalaby, H.M.H. Highly efficient photonic PCR system based on plasmonic heating of gold nanofilms. Biosens. Bioelectron. X 2023, 14, 100346. [Google Scholar] [CrossRef]
- Higashi, T.; Minegishi, H.; Echigo, A.; Nagaoka, Y.; Fukuda, T.; Usami, R.; Maekawa, T.; Hanajiri, T. Nanomaterial-assisted PCR based on thermal generation from magnetic nanoparticles under high-frequency AC magnetic fields. Chem. Phys. Lett. 2015, 635, 234–240. [Google Scholar] [CrossRef]
- Gama, B.E.; Silva-Pires, F.D.E.S.; Lopes, M.N.K.R.; Cardoso, M.A.B.; Britto, C.; Torres, K.L.; de Mendonça Lima, L.; de Souza, J.M.; Daniel-Ribeiro, C.T.; de Fátima Ferreira-da-Cruz, M. Real-time PCR versus conventional PCR for malaria parasite detection in low-grade parasitemia. Exp. Parasitol. 2007, 116, 427–432. [Google Scholar] [CrossRef]
- Godin, M.J.; Sebastian, A.; Albert, I.; Lindner, S.E. Long-read genome assembly and gene model annotations for the rodent malaria parasite Plasmodium yoelii 17XNL. J. Biol. Chem. 2023, 299, 104871. [Google Scholar] [CrossRef]
- Cao, Y.; Ye, C.; Zhang, C.; Zhang, G.; Hu, H.; Zhang, Z.; Fang, H.; Zheng, J.; Liu, H. Simultaneous detection of multiple foodborne bacteria by loop-mediated isothermal amplification on a microfluidic chip through colorimetric and fluorescent assay. Food Control 2022, 134, 108694. [Google Scholar] [CrossRef]
- Cao, X.; Ge, S.; Zhou, X.; Mao, Y.; Sun, Y.; Lu, W.; Ran, M. A dual-signal amplification strategy based on pump-free SERS microfluidic chip for rapid and ultrasensitive detection of non-small cell lung cancer-related circulating tumour DNA in mice serum. Biosens. Bioelectron. 2022, 205, 114110. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Lan, X.; Zhu, L.; Zhang, Y.; Chen, K.; Zhang, W.; Xu, W. Portable dual-aptamer microfluidic chip biosensor for Bacillus cereus based on aptamer tailoring and dumbbell-shaped probes. J. Hazard. Mater. 2023, 445, 130545. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Jin, L.; Jia, C.; Wang, X.; Zhao, J.; Feng, S.; Guo, X. A droplet digital PCR chip with passive bubble removal for absolute nucleic acid quantification. Sens. Actuators B Chem. 2023, 392, 134109. [Google Scholar] [CrossRef]
- Su, W.; Qiu, J.; Mei, Y.; Zhang, X.E.; He, Y.; Li, F. A microfluidic cell chip for virus isolation via rapid screening for permissive cells. Virol. Sin. 2022, 37, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Tong, Z.; Wu, Z.; Jian, X.; Zhou, L.; Qiu, S.; Shen, C.; Yin, H.; Mao, H. Multiple exosome RNA analysis methods for lung cancer diagnosis through integrated on-chip microfluidic system. Chin. Chem. Lett. 2022, 33, 3188–3192. [Google Scholar] [CrossRef]
- Kim, E.; Lee, G.Y.; Yang, S.M.; Kim, H.Y. Rapid and accurate on-site identification of Lactobacillus delbrueckii subspecies in dairy products using direct polymerase chain reaction with microfluidic chip. LWT 2023, 179, 114635. [Google Scholar] [CrossRef]
- Wu, C.; Liu, L.; Ye, Z.; Gong, J.; Hao, P.; Ping, J.; Ying, Y. TriD-LAMP: A pump-free microfluidic chip for duplex droplet digital loop-mediated isothermal amplification analysis. Anal. Chim. Acta 2022, 1233, 340513. [Google Scholar] [CrossRef]
- Parvizi, F.; Parvareh, A.; Heydari, R. Fabrication of a hydrophobic surface as a new supported liquid membrane for microfluidic based liquid phase microextraction device using modified boehmite nanoparticles (AlOO-NSPO). Microchem. J. 2023, 189, 108514. [Google Scholar] [CrossRef]
- Zarghampour, F.; Yamini, Y.; Dil, E.A.; Shokrollahi, A.; Javadian, G. A new microfluidic-chip device followed by sensitive image analysis of smart phone for simultaneous determination of dyes with different acidic–basic properties. Talanta 2023, 254, 124168. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, R.; Huang, S.; Huang, Y.; Zhang, J.; Su, X.; Zhang, S.; Ge, S.; Zhang, J.; Xia, N. All-in-one microfluidic chip for 30-min quantitative point-of-care-testing of nucleic acids. Sens. Actuators B Chem. 2023, 390, 133939. [Google Scholar] [CrossRef]
- Yang, K.; Pan, J.; Deng, G.; Hua, C.; Zhu, C.; Liu, Y.; Zhu, L. Mkit: A mobile nucleic acid assay based on a chitosan-modified minimalistic microfluidic chip (CM3-chip) and smartphone. Anal. Chim. Acta 2023, 1253, 341030. [Google Scholar] [CrossRef] [PubMed]
- Kaba, A.M.; Jeon, H.; Park, A.; Yi, K.; Baek, S.; Park, A.; Kim, D. Cavitation-microstreaming-based lysis and DNA extraction using a laser-machined polycarbonate microfluidic chip. Sens. Actuators B Chem. 2021, 346, 130511. [Google Scholar] [CrossRef]
- Su, S.; Jing, G.; Zhang, M.; Liu, B.; Zhu, X.; Wang, B.; Fu, M.; Zhu, L.; Cheng, J.; Guo, Y. One-step bonding and hydrophobic surface modification method for rapid fabrication of polycarbonate-based droplet microfluidic chips. Sens. Actuators B Chem. 2019, 282, 60–68. [Google Scholar] [CrossRef]
- Wang, Y.; He, Q.; Dong, Y.; Chen, H. In-channel modification of biosensor electrodes integrated on a polycarbonate microfluidic chip for micro flow-injection amperometric determination of glucose. Sens. Actuators B Chem. 2010, 145, 553–560. [Google Scholar] [CrossRef]
- Bi, W.; Cai, S.; Lei, T.; Wang, L. Implementation of blood-brain barrier on microfluidic chip: Recent advance and future prospects. Ageing Res. Rev. 2023, 87, 101921. [Google Scholar] [CrossRef]
- Açıkgöz, H.N.; Karaman, A.; Şahin, M.A.; Çaylan, Ö.R.; Büke, G.C.; Yıldırım, E.; Eroğlu, İ.C.; Erson-Bensan, A.E.; Çetin, B.; Özer, M.B. Assessment of silicon, glass, FR4, PDMS and PMMA as a chip material for acoustic particle/cell manipulation in microfluidics. Ultrasonics 2023, 129, 106911. [Google Scholar] [CrossRef]
- Guler, M.T. Fabricating plasma bonded microfluidic chips by CO2 laser machining of PDMS by the application of viscoelastic particle focusing and droplet generation. J. Manuf. Process. 2022, 73, 260–268. [Google Scholar] [CrossRef]
- Jiang, B.; Guo, H.; Chen, D.; Zhou, M. Microscale investigation on the wettability and bonding mechanism of oxygen plasma-treated PDMS microfluidic chip. Appl. Surf. Sci. 2022, 574, 151704. [Google Scholar] [CrossRef]
- Tang, Q.; Li, X.; Lai, C.; Li, L.; Wu, H.; Wang, Y.; Shi, X. Fabrication of a hydroxyapatite-PDMS microfluidic chip for bone-related cell culture and drug screening. Bioact. Mater. 2021, 6, 169–178. [Google Scholar] [CrossRef]
- Liu, X.; Li, S. Fabrication of a Three-Layer PDMS Pneumatic Microfluidic Chip for Micro Liquid Sample Operation. SLAS Technol. 2020, 25, 151–161. [Google Scholar] [CrossRef]
- Song, Q.; Sun, J.; Mu, Y.; Xu, Y.; Zhu, Q.; Jin, Q. A new method for polydimethylsiloxane (PDMS) microfluidic chips to maintain vacuum-driven power using Parylene C. Sens. Actuators B Chem. 2018, 256, 1122–1130. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, J.H.; Oh, S.W. Combination of filtration and immunomagnetic separation based on real-time PCR to detect foodborne pathogens in fresh-cut apple. J. Microbiol. Methods 2022, 201, 106577. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Choi, S.; Cho, I.S.; Yoon, J.Y.; Jeong, R.D. Development and application of a reverse transcription droplet digital PCR assay for detection and quantification of Plantago asiatica mosaic virus. Crop Prot. 2023, 169, 106255. [Google Scholar] [CrossRef]
- Kheiroddin, P.; Gaertner, V.D.; Schöberl, P.; Fischer, E.; Niggel, J.; Pagel, P.; Lampl, B.M.J.; Ambrosch, A.; Kabesch, M. Gargle pool PCR testing in a hospital during medium and high SARS-CoV-2 incidence. J. Hosp. Infect. 2022, 127, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Guan, N.; Li, Y.; Yang, H.; Hu, P.; Lu, S.; Ren, H.; Liu, Z.; Park, K.S.; Zhou, Y. Dual-functionalized gold nanoparticles probe based bio-barcode immuno-PCR for the detection of glyphosate. Food Chem. 2021, 338, 128133. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.A.; Morrison, H.G.; Huse, S.M.; Neal, P.R.; Sogin, M.L.; Welch, D.B.M. Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Env. Microbiol. 2009, 11, 1292. [Google Scholar] [CrossRef]
- Hofmann, N.; Mwingira, F.; Shekalaghe, S.; Robinson, L.J.; Mueller, I.; Felger, I. Ultra-Sensitive Detection of Plasmodium falciparum by Amplification of Multi-Copy Subtelomeric Targets. PLoS Med. 2015, 12, e1001788. [Google Scholar] [CrossRef]


| Material Property | Unit | [28] | [29] | [30] |
|---|---|---|---|---|
| Melting Point | 432.15 | 428.15 | 408.15 | |
| Thermal Conductivity | 0.8 | 0.24 | 0.15 | |
| Specific Heat | 1.8 | 1.2 | 1.46 | |
| Density | 920 | 1200 | 970 |
| Total Length Calculation | ||||
|---|---|---|---|---|
| Design | Radius (mm) | Circumference (mm) | Length (mm) | Total Length (mm) |
| - | - | 30 | 30 | |
| 0.25 | 1.570796327 | 14 | 30 | |
| 0.25 | 1.570796327 | 27 | 30 | |
| Region | Residence Time (s) | Length (µm) | Passes | Volume | Volumetric Flowrate | Speed (µm/s) |
|---|---|---|---|---|---|---|
| Annealing (58 °C) | 15 | 15,000 | 1.5 | 2.94 × 109 | 1.96 × 108 | 1500 |
| Extension (72 °C) | 20 | 10,000 | 3 | 3.92 × 109 | 1.96 × 108 | 1500 |
| Denaturation (95 °C) | 10 | 15,000 | 1 | 1.96 × 109 | 1.96 × 108 | 1500 |
| Spaces | 5.3 | 8000 | 1.05 × 109 | 1.96 × 108 | 1500 | |
| Total | 153 | 75,500 | 3 | 9.87 × 109 |
| Design | Relative Deviation | |
|---|---|---|
| No Loop | 0.65 | 1.12 |
| One Loop | 0.80 | 1.39 |
| Two Loops | 0.88 | 1.52 |
| 58 °C, 72 °C, 95 °C | 60 °C, 74 °C, 97 °C | 63 °C, 77 °C, 100 °C | ||||
|---|---|---|---|---|---|---|
| Material | Relative Deviation | Relative Deviation | Relative Deviation | |||
| 0.94 | 3.21 | 2.84 | 5.82 | 6.90 | 25.00 | |
| 1.92 | 8.61 | 3.15 | 10.00 | 6.92 | 26.70 | |
| 2.78 | 14.00 | 3.47 | 14.00 | 6.93 | 29.00 | |
| Parameter | Design 1 | Design 2 |
|---|---|---|
| Diameter | 300 | 500 |
| Length | 200 | 300 |
| Relative Deviation | 0.94 | 2.31 |
| 3.21 | 5.96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Austria, M.; Garcia, J.P.; Caparanga, A.; Tayo, L.; Doma, B., Jr. Designing Microfluidic PCR Chip Device Using CFD Software for the Detection of Malaria. Computation 2023, 11, 190. https://doi.org/10.3390/computation11100190
Austria M, Garcia JP, Caparanga A, Tayo L, Doma B Jr. Designing Microfluidic PCR Chip Device Using CFD Software for the Detection of Malaria. Computation. 2023; 11(10):190. https://doi.org/10.3390/computation11100190
Chicago/Turabian StyleAustria, Meynard, Jon Patrick Garcia, Alvin Caparanga, Lemmuel Tayo, and Bonifacio Doma, Jr. 2023. "Designing Microfluidic PCR Chip Device Using CFD Software for the Detection of Malaria" Computation 11, no. 10: 190. https://doi.org/10.3390/computation11100190
APA StyleAustria, M., Garcia, J. P., Caparanga, A., Tayo, L., & Doma, B., Jr. (2023). Designing Microfluidic PCR Chip Device Using CFD Software for the Detection of Malaria. Computation, 11(10), 190. https://doi.org/10.3390/computation11100190






