Distribution of “Characteristic” Terms in MEDLINE Literatures
Abstract
: Given the occurrence frequency of any term within any set of articles within MEDLINE, we define “characteristic” terms as words and phrases that occur in that literature more frequently than expected by chance (at p < 0.001 or better). In this report, we studied how the cut-off criterion varied as a function of literature size and term frequency in MEDLINE as a whole, and have compared the distribution of characteristic terms within a number of journal-defined, affiliation-defined and random literatures. We also investigated how the characteristic terms were distributed among MEDLINE titles, abstracts, and last sentence of abstracts, including “regularized” terms that appear both in the title and abstract of the same paper for at least one paper in the literature. For a set of 10 disciplinary journals, the characteristic terms comprised 18% of the total terms on average. Characteristic terms are utilized in several of our web-based services (Anne O'Tate and Arrowsmith), and should be useful for a variety of other information-processing tasks designed to improve text mining in MEDLINE.1. Introduction
Terms occurring in a given set of articles (i.e., a literature) more than expected by chance form a literature-specific vocabulary that is similar to the concept of a domain “sublanguage” [1-3]. They differ from the keywords extracted from a particular literature [4,5], insofar as keywords occur frequently relative to other terms in that literature, whereas a literature-specific term may occur only a few times (as long as it is more frequent in that literature than in MEDLINE as a whole).
In the present paper, we have computed empirical occurrence frequencies of terms within a number of journal-defined, affiliation-defined and random literatures. We derived statistical criteria for asserting that a single term occurs more often within any given literature than expected by chance, and denote the set of terms that occur more than expected by chance (at p < 0.001) as the “characteristic” terms for that literature. Finally, we have studied their distribution across MEDLINE titles, abstracts, and last sentences of abstracts, including “regularized” characteristic terms that appear both in the title and abstract of the same paper for at least one paper in the literature. These studies set the stage for utilizing characteristic terms as features in text mining models, and in creating thumbnail annotations of the literatures.
2. Results
2.1. Delineating Characteristic Terms
We examined 10 different disciplinary journals published in English, containing abstracts, which comprised 2,000-10,000 papers each (average 5,132 papers), and characterized the distribution of term frequencies within the journal set vs. within MEDLINE as a whole. This distribution was compared with the distribution of terms in an affiliation-defined literature consisting of all articles published in 2000 having the word “California” in the affiliation field, and with a set of 5,000 articles chosen at random within MEDLINE. In each case, the Poisson approximation was used to define the distribution of term occurrence that would be expected by chance.
Figure 1 shows the raw distribution of term occurrence frequencies in the text fields (i.e., title or abstract) for Journal of Biomedical Materials Research compared to a random literature of similar size. Term occurrence frequency was almost exactly linear for the journal when plotted on a log-log scale, indicating that frequencies followed a regular Zipf distribution. The frequencies for the random set followed a parallel curve and were significantly different from that of the journal.
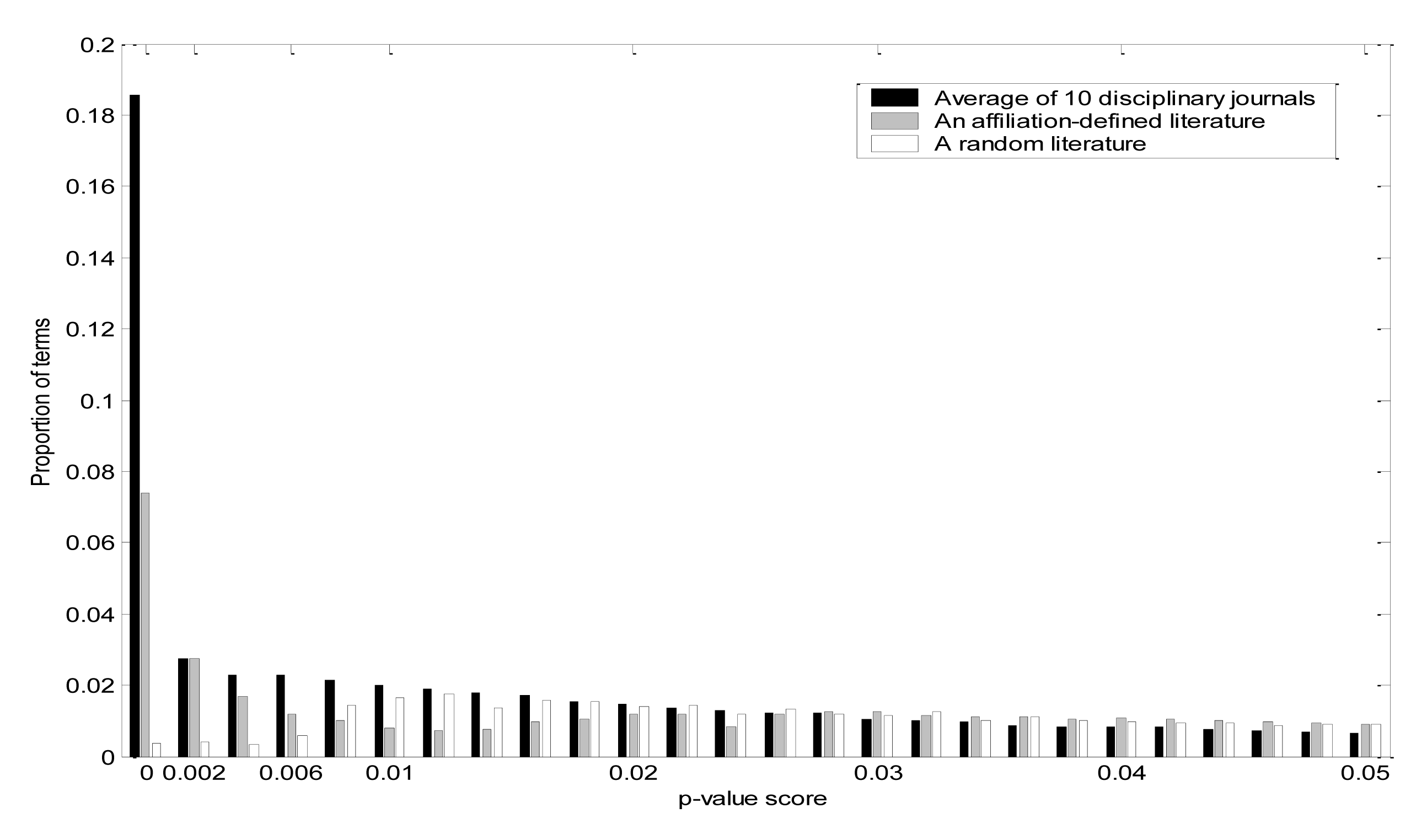
To identify individual terms that were significantly more frequent than expected by chance, we computed p-value scores for each term across 10 disciplinary journals and plotted the average p-value scores in comparison to the California set and to a random set of 5,000 articles (Figure 2). Although terms associated with p-values < 0.05 are nominally significant, that does not take into account the fact that multiple tests are carried out. Figure 2 emphasizes that the difference between journal-defined sets and random sets is most striking at p-values below 0.001. Thus, we have chosen p < 0.001 as our preferred cut-off value, and the set of terms in a literature with p-values below 0.001 will be called the characteristic terms of that literature.
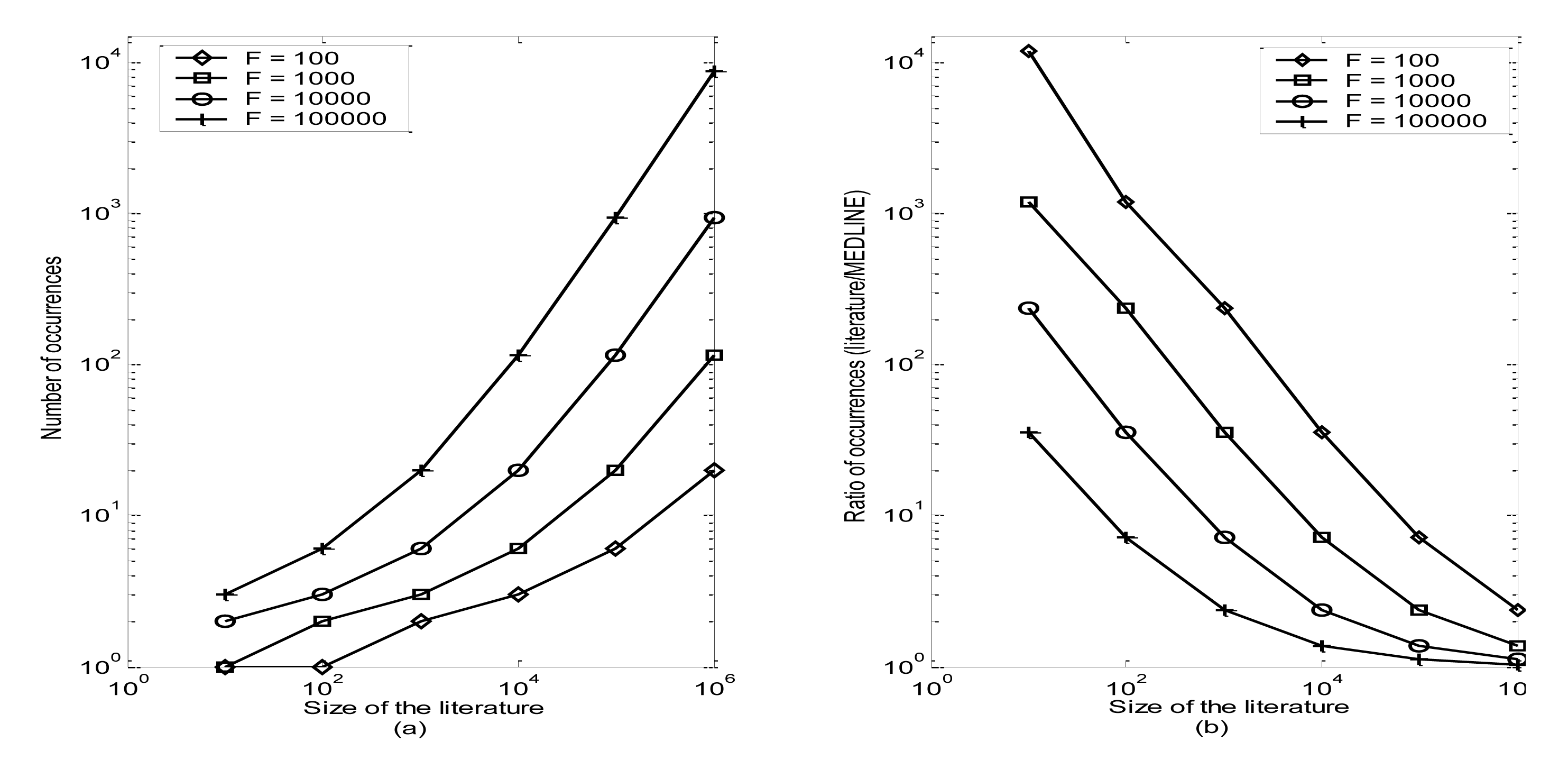
For the set of 10 disciplinary journals, the set of characteristic terms comprise, on average, 18% of the total terms in that literature. The cut-off criteria for deeming a term as “characteristic” vary systematically as functions both of literature size and term frequency within MEDLINE (Figure 3). Among the entire set of characteristic terms for the 10 disciplinary journals, average term occurrence is 23 times within the set of journal articles, which is 88 times more frequent in that literature than in MEDLINE.
To illustrate the types of terms that are characteristic for a specific literature, we show results from International Journal of Food Microbiology. Table 1 shows the 10 characteristic terms with the lowest p-values, 10 having moderate p-values and 10 having p-values near 0.001. Clearly, the top ten terms are closely related to the journal topic (food, listeria, meat, etc.), as are the moderate set (ethanol, shigella, mold, etc.), whereas those at the margin of significance are still relevant but less specific (tbg, gene coding, fever vomiting, sandwich, etc.).
2.2. Distribution of Characteristic Terms within Individual Article Records
Several previous studies have emphasized that specific terms or MeSH concepts may be enriched in particular sections of scientific papers [6,7]. We examined how the set of characteristic terms are distributed among 8 different sections of papers encoded in MEDLINE fields for each of 10 disciplinary journals, the California literature and the random literature: text (comprising title and abstract fields); ti (title); ab (abstract); lastsen (last sentence of the abstract); ti + ab (present both in the title and in the abstract of at least one paper in the literature, though not necessarily the same paper); tiab (in the title and the abstract of the same paper, for at least one paper in the literature); ti + lastsen (in title and last sentence of the abstract, for at least one paper in the literature, though not necessarily the same paper); and tiab + lastsen (in tiab and in last sentence of the abstract for at least one paper in the literature).
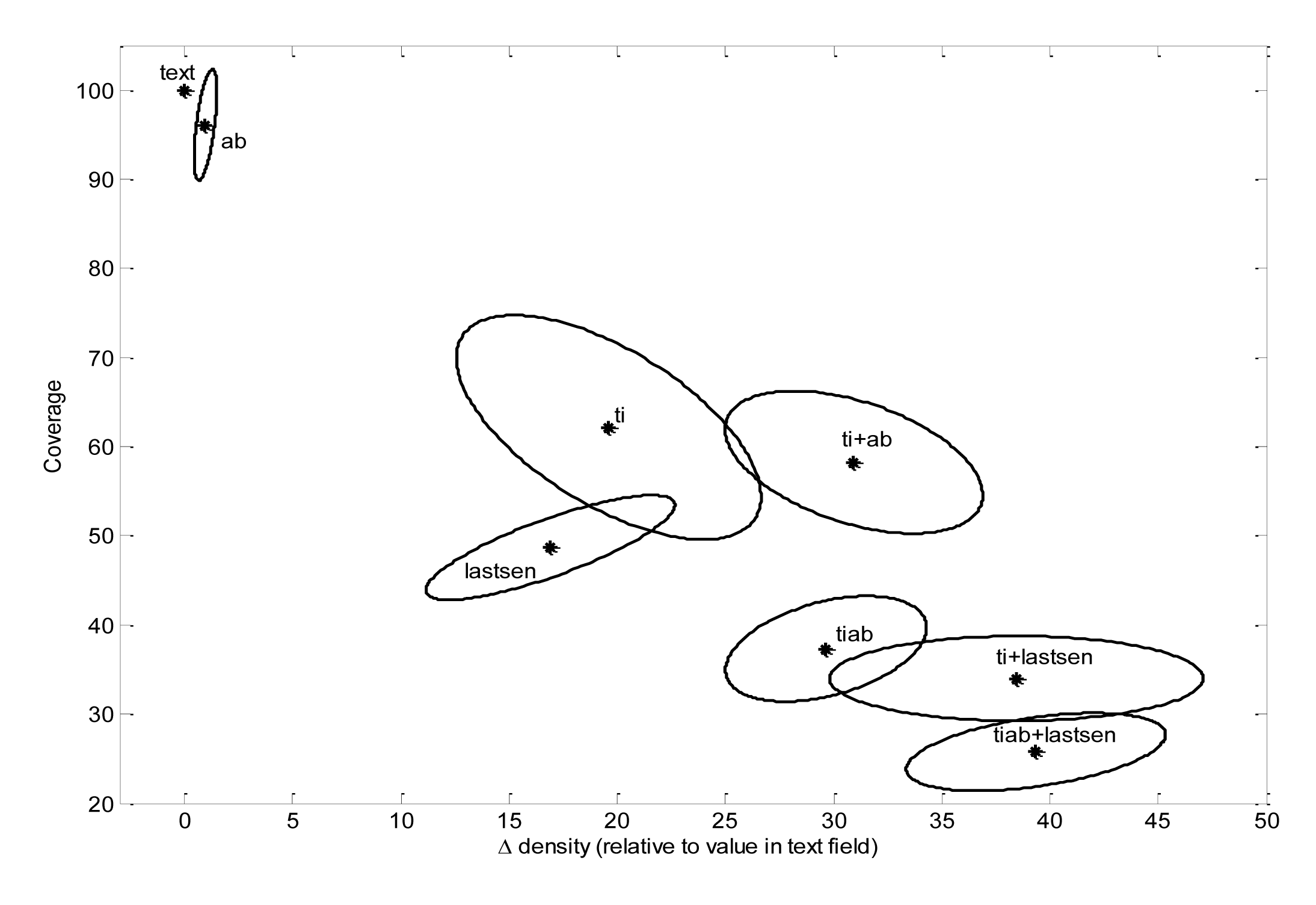
One basic measure is the “density”—this is the percentage of all terms in each section that are comprised of characteristic terms. Those sections that are high in density are relatively rich in characteristic terms. Another measure is the “coverage”—defined as the number of characteristic terms found in each section, as a percentage of the total characteristic terms for that journal. Those sections that are high in coverage have the most characteristic terms overall.
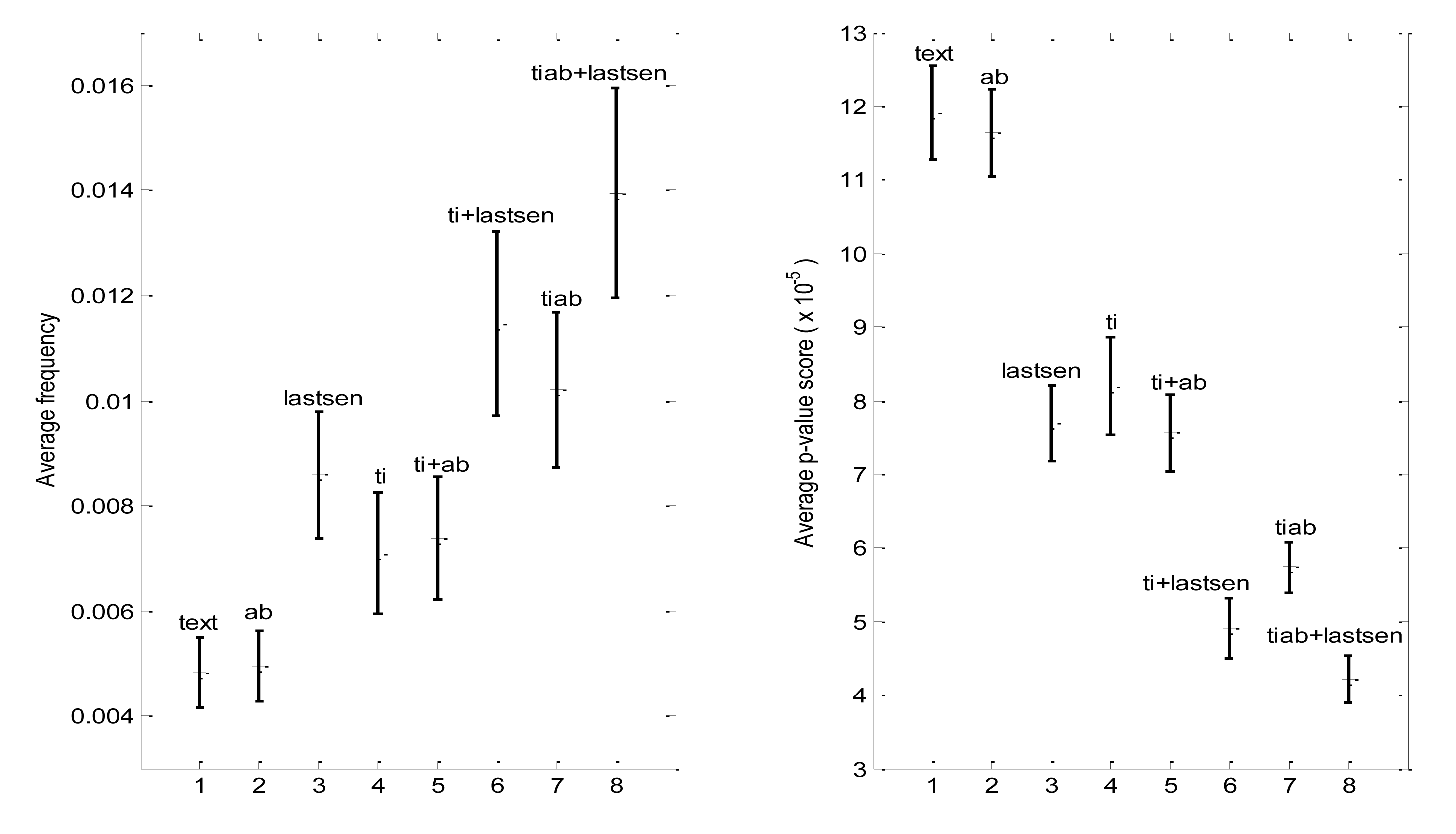
The average density value varied significantly from journal to journal within our set of 10 disciplinary journals (Table 2), presumably due to different journal policies such as limits on abstract length and structured vs. unstructured abstracts. However, after normalizing the density values for each journal, one could readily observe systematic section-related differences in density and coverage that were similar across journals (Figure 4). Title and last sentence of the abstract had significantly more density than the abstract field, whereas terms that appeared in multiple sections had significantly more density than those appearing in a single section (Figure 4). Not only were these fields progressively richer in characteristic terms, but the characteristic terms that they contained had higher average frequency of occurrence than the overall set of characteristic terms, and were more specific insofar as they had lower average p-values (Figure 5). Interestingly, the set of “regularized” characteristic terms (tiab) appearing in the title and abstract of the same paper had significantly higher average frequency and lower p-values than terms which appeared in titles and abstract of different papers (ti + ab) (Figure 5) (each parameter significantly different at p < 0.00001, using paired t-test).
Regularized terms (tiab) that also appeared in the last sentence of at least one paper (tiab + lastsen) had the highest average frequency and lowest average p-value of all (Figure 5), suggesting that this subset of characteristic terms comprises, in some sense, the most important terms associated with the journal. Of the 20 characteristic terms having the lowest p-values overall in one journal, International Journal of Food Microbiology, all were found in the tiab + l astsen set as well. Thus, two independent methods—lowest p-value vs. presence in multiple sections of papers—agree in giving the most “important” characteristic terms.
We also considered whether, given two characteristic terms with equal p-values, the term appearing in the greater number of papers in the literature should be considered the more important. For the characteristic terms in International Journal of Food Microbiology, we calculated a “corrected” p-value score by dividing the raw p-value by the fraction of papers in the journal containing the term; however, this correction did not alter the top 20 characteristic terms and had only a very minor effect on their relative ranking (Table 3). Thus, at least for the task of choosing the few most important characteristic terms, it does not seem to be necessary to take this factor into account as a separate variable.
3. Experimental Methods
The universe of terms was defined in the following manner, consistent with the larger aims of the Arrowsmith Project [8]. Specifically, the titles of all papers in MEDLINE were extracted, stemmed and stoplisted using the short PubMed 364-word stoplist [9]. Words were kept only if they appeared in the abstract of at least three papers in MEDLINE, and up to three word phrases were kept only if they appeared in at least 10 abstracts. Finally, terms were mapped through the NIH MetaMap program keeping only those terms that mapped to at least one UMLS semantic category. (This removes most of the nonsensical phrases but includes many that do not correspond exactly to UMLS concepts.) After filtering, the total number of words = 52,997, two word phrases = 747,484, and three word phrases = 429,566. (Note that if a term occurred at all within an abstract, it was scored as 1 occurrence regardless of how many times the term occurred within the same abstract.) For each occurrence of a term within a MEDLINE record, we noted its location within title, abstract, or last sentence in abstract. Sentence boundaries were identified using the Sentence Splitter [10].
Modeling the expected term occurrence in a literature
Think of all the N papers in MEDLINE as a collection of N balls in an urn, where f1 black balls correspond to papers that contain a certain term, and the remaining N − f1 balls are white (do not contain the term.) In constructing a random literature of f2 papers, we randomly select f2 distinct balls from the urn. The number of black balls selected, X, is a random variable that follows a hypergeometric distribution defined by:
In other words, if a literature and a given term are independent of each other, then the number of papers within that literature that contain the term should follow the hypergeometric distribution.
The Poisson distribution is a good approximation when N is large relative to f1 and f2:
Where λ = f1f2/N is the expected value of X. We have verified that the Poisson distribution is an extremely close approximation for the hypergeometric distribution in the full range of literature sizes and term frequencies considered in this paper.
4. Conclusions
In the present paper, we have calculated and empirically validated statistical criteria for saying that a term occurs in a given literature more often than by chance, and have analyzed the resulting set of “characteristic” terms (having p-values < 0.001) in some detail. Note that the characteristic terms for a literature are not necessarily the most frequent in that literature. Nor, for topically-defined literatures, do they need to have any semantic relation to the query term that generated the literature.
Characteristic terms of a literature have proven useful for different information-processing tasks. In the Anne O'Tate tool [11] that combines PubMed literature retrieval with additional post-retrieval analyses, the set of characteristic terms gives a thumbnail annotation of any retrieved literature. For example, in the case of papers describing diabetes research, the set of characteristic terms (restricted to the semantic category of gene names) gives a thumbnail annotation of the genes that have been studied in this field. In the Author-ity author name disambiguation tool [12], characteristic terms provide a thumbnail annotation of any given author's research output. Other possible uses for characteristic terms occur in post-processing of a PubMed query, to replace or supplement other language resources such as Medical Subject Headings, UMLS concepts or keyword thesauri, e.g., to expand the query automatically to include highly related papers [13,14], to cluster the retrieved papers by theme [15], or to reformulate the query in a manner that permits cross-disciplinary retrieval [16]. For example, to expand an original query automatically, one could replace the original terms used in the search with a new Boolean query made up of a small number of characteristic terms. These would not necessarily be the terms with the lowest p-values, but rather would be the set of the terms that (when combined with appropriate AND and OR operations) cover the original literature most accurately and with least redundancy.
The characteristic terms with the lowest p-values are likely to be most useful for annotation; this is similar to the log-entropy term weighting approach taken by Homayouni et al. [17]. Other annotation methods are possible—for example, Erkan and Radev [18] used a graph theoretic-approach to obtain the “most important” terms within document sets—but this is far more computationally complex than the method proposed here, and would not scale well to large literatures. The terms with lowest p-values are likely to be the most important as well, especially since these terms appeared in multiple sections of the papers.
Finally, characteristic terms have been useful for assisting in literature-based discovery. In the Arrowsmith two-node search tool [19,20], the user seeks to assess a possible relationship between literatures A and C; the computer interface presents a list of terms (the “B-list”) in common between the literatures to serve as a conceptual bridge. However, not all B-terms are likely to be of equal value in discovering significant implicit links. Characteristic terms expressed in each literature are computed as a feature in the quantitative model that allows us to rank the B-terms in order of predicted relevance to linking the two literatures in a meaningful way [19]. Moreover, B-terms that are not characteristic in either literature A or C are unlikely to indicate important concepts in either literature, whereas B-terms that are characteristic in both A and C may represent concepts that are already well known. Thus, we are currently exploring the hypothesis that the B-terms most likely to point to new discoveries in two node searches are those that are characteristic in one literature, but not both.

| Terms (lowest p-value) | Terms (moderate p-value) | Terms (p-value near 0.001) | |
|---|---|---|---|
| 1 | food | ph ethanol | strain x |
| 2 | listeria | recurrent neural network | tbg |
| 3 | strain | shigella yersinia | or h |
| 4 | listeria monocytogene | disinfection or | mytilus galloprovincialis |
| 5 | degree c | growth environmental | gene coding |
| 6 | meat | mold growth | fever vomiting |
| 7 | l monocytogene | staphylococcal strain isolated | sandwich |
| 8 | lactic acid | yeast high | growth effect |
| 9 | lactobacillus | longitudinally | density nm |
| 10 | lactic acid bacteria | pathogen human | reliable method |
| Density (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jrn1 | Jrn2 | Jrn3 | Jrn4 | Jrn5 | Jrn6 | Jrn7 | Jrn8 | Jrn9 | Jrn10 | Average of 10 Jrns | California literature | Random literature | |
| Text | 18.31 | 9.01 | 11.82 | 20.74 | 15.54 | 19.94 | 24.65 | 17.26 | 29.29 | 19.01 | 18.55 | 7.39 | 0.37 |
| Ab | 19.94 | 9.14 | 12.71 | 21.65 | 16.41 | 20.82 | 25.65 | 18.14 | 30.68 | 20.19 | 19.53 | 7.87 | 0.40 |
| Lastsen | 34.92 | 18.61 | 21.79 | 39.69 | 31.68 | 38.21 | 43.99 | 35.26 | 53.27 | 37.27 | 35.46 | 19.71 | 0.48 |
| Ti | 34.08 | 21.5 | 23.14 | 44.76 | 34.11 | 43.52 | 51.61 | 37.02 | 53.89 | 38.02 | 38.16 | 18.53 | 0.60 |
| Ti + Ab | 48.54 | 33.74 | 34.73 | 55.57 | 45.81 | 51.98 | 62.33 | 48.01 | 63.79 | 50.43 | 49.49 | 24.87 | 0.93 |
| Ti + Lastsen | 57.11 | 36.94 | 37.51 | 64.49 | 53.84 | 60.45 | 69.06 | 58.76 | 72.19 | 59.82 | 57.01 | 36.51 | 0.65 |
| Tiab | 47.69 | 37.06 | 36.41 | 54.87 | 44.84 | 50.08 | 59.64 | 43.61 | 62.31 | 45.59 | 48.21 | 25.66 | 0.42 |
| Tiab + Lastsen | 57.89 | 42.57 | 41.97 | 65.11 | 55.15 | 59.92 | 68.83 | 57.62 | 72.19 | 57.85 | 57.91 | 37.34 | 0.37 |
| Coverage (%) | |||||||||||||
| Jrn1 | Jrn2 | Jrn3 | Jrn4 | Jrn5 | Jrn6 | Jrn7 | Jrn8 | Jrn9 | Jrn10 | Average of 10 Jrns | California literature | Random literature | |
| Text | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Ab | 95.33 | 85.51 | 89.89 | 98.37 | 98.38 | 98.47 | 98.87 | 98.63 | 98.64 | 98.79 | 96.08 | 99.01 | 93.83 |
| Lastsen | 48.46 | 41.76 | 41.7 | 49.3 | 50.1 | 55.99 | 49.3 | 50.18 | 52.75 | 47.21 | 48.67 | 59.98 | 27.01 |
| Ti | 66.73 | 77.47 | 75.77 | 57.01 | 53.6 | 68.34 | 55.29 | 52.9 | 61.71 | 52.83 | 62.16 | 66.23 | 53.55 |
| Ti + Ab | 62.07 | 62.98 | 65.67 | 55.39 | 51.99 | 66.81 | 62.33 | 48.01 | 63.79 | 50.43 | 58.94 | 65.24 | 47.39 |
| Ti + Lastsen | 34.6 | 33.48 | 33.53 | 33.32 | 33.93 | 42.28 | 30.66 | 31.52 | 36.89 | 29.75 | 33.99 | 45.57 | 15.16 |
| Tiab | 38.04 | 27.89 | 38.49 | 39.07 | 36.79 | 44.94 | 39.39 | 35.04 | 39.4 | 34.12 | 37.31 | 50.38 | 16.11 |
| Tiab + Lastsen | 25.59 | 19.77 | 24.76 | 26.49 | 28.08 | 31.84 | 25.3 | 24.67 | 28.21 | 22.78 | 25.74 | 38.14 | 7.10 |
| Top 20 ranked by raw p-value score | Top 20 ranked by corrected p-value score | ||||||
|---|---|---|---|---|---|---|---|
| Term | f | F | raw p-value score | Term | f | F | corrected p-value score |
| food | 718 | 102,805 | 1.04 × 10−847 | food | 718 | 102,805 | 3.01 × 10−847 |
| listeria | 384 | 6,879 | 2.11 × 10−797 | listeria | 384 | 6,879 | 1.14 × 10−796 |
| strain | 775 | 246,958 | 6.76 × 10−656 | strain | 775 | 246,958 | 1.81 × 10−655 |
| listeria monocytogene | 310 | 5,648 | 6.90 × 10−642 | listeria monocytogene | 310 | 5,648 | 4.62 × 10−641 |
| degree c | 625 | 139,694 | 3.84 × 10−621 | degree c | 625 | 139,694 | 1.28 × 10−620 |
| meat | 309 | 11,582 | 1.81 × 10−543 | meat | 309 | 11,582 | 1.22 × 10−542 |
| l monocytogene | 231 | 2,514 | 2.05 × 10−530 | l monocytogene | 231 | 2,514 | 1.84 × 10−529 |
| lactic acid | 257 | 7,008 | 1.11 × 10−487 | lactic acid | 257 | 7,008 | 9.03 × 100−487 |
| lactobacillus | 238 | 5,441 | 6.39 × 10−470 | lactobacillus | 238 | 5,441 | 5.57 × 10−469 |
| lactic acid bacteria | 188 | 1,324 | 1.52 × 10−467 | lactic acid bacteria | 188 | 1,324 | 1.67 × 10−466 |
| lactic | 270 | 13,490 | 1.07 × 10−441 | lactic | 270 | 13,490 | 8.22 × 10−441 |
| temperature | 467 | 156,235 | 8.00 × 10−387 | temperature | 467 | 156,235 | 3.56 × 10−386 |
| degree | 661 | 405,979 | 3.17 × 10−386 | degree | 661 | 405,979 | 9.95 × 10−386 |
| salmonella | 294 | 32,199 | 6.87 × 10−382 | salmonella | 294 | 32,199 | 4.85 × 10−381 |
| spp | 245 | 15,673 | 5.88 × 10−375 | growth | 662 | 426,410 | 3.91 × 10−374 |
| growth | 662 | 426,410 | 1.25 × 10−374 | spp | 245 | 15,673 | 4.98 × 10−374 |
| ph | 413 | 157,692 | 3.99 × 10−320 | ph | 413 | 157,692 | 2.00 × 10−319 |
| isolate | 331 | 81,127 | 2.75 × 10−317 | isolate | 331 | 81,127 | 1.73 × 10−316 |
| storage | 265 | 47,338 | 1.63 × 10−289 | storage | 265 | 47,338 | 1.28 × 10−288 |
| foodborne | 113 | 1,198 | 8.98 × 10−262 | foodborne | 113 | 1,198 | 1.65 × 10−260 |
Acknowledgments
This Human Brain Project/Neuroinformatics research (LM007292 and LM08364) is funded jointly by the National Library of Medicine and the National Institute of Mental Health. The Medline database and the MMTx program (a Java implementation of the MetaMap algorithm) were graciously provided by the National Library of Medicine.
References and Notes
- Grishman, R.; Kittredge, R. Analyzing Language in Restricted Domains: Sublanguage Description and Processing; Lawrence Erlbaum Associates: Mahwah, NJ, USA, 1986; pp. 19–38. [Google Scholar]
- Liu, Y.; Brandon, M.; Navathe, S.; Dingledine, R.; Ciliax, B.J. Text Mining Functional Keywords Associated with Genes. Medinfo 2004, 107, 292–296. [Google Scholar]
- Tudor, C.O.; Vijay-Shanker, K.; Schmidt, C.J. Mining the Biomedical Literature for Genic Information. Proceedings of BioNLP Workshop in Conjunction with ACL-2008, Columbus Ohio, 28-29 June 2008.
- Andrade, M.A.; Valencia, A. Automatic Extraction of Keywords from Scientific Text: Application to the Knowledge Domain of Protein Families. BMC Bioinform. 1998, 14, 600–607. [Google Scholar]
- Kostoff, R.N.; Block, J.A.; Stump, J.A.; Pfeil, K.M. Information content in Medline record fields. Int. J. Med. Inform. 2004, 73, 515–527. [Google Scholar]
- Schuemie, M.J.; Weeber, M.; Schijvenaars, B.J.; van Mulligen, E.M.; van der Eijk, C.C.; Jelier, R.; Mons, B.; Kors, J.A. Distribution of Information in Biomedical Abstracts and Full-text Publications. Bioinformation 2004, 20, 2597–2604. [Google Scholar]
- Shah, P.K.; Perez-Iratxeta, C.; Bork, P.; Andrade, M.A. Information Extraction from Full Text Scientific Articles: Where Are the Keywords? BMC Bioinform. 2003, 4, 20. [Google Scholar]
- Smalheiser, N.R.; Torvik, V.I.; Bischoff-Grethe, A.; Burhans, L.B.; Gabriel, M.; Homayouni, R.; Kashef, A.; Martone, M.E.; Perkins, G.A.; Price, D.L.; Talk, A.C.; West, R. Collaborative Development of the Arrowsmith Two Node Search Interface Designed For Laboratory Investigators. J. Biomed. Discov. Collab. 2006, 1, 8. [Google Scholar]
- http://www.ncbi.nlm.nih.gov/books/NBK3827/table/pubmedhelp.T43/.
- http://l2r.cs.uiuc.edu/∼cogcomp/tools.php.
- Smalheiser, N.R.; Zhou, W.; Torvik, V.I. Anne O'Tate: A Tool to Support User-Driven Summarization, Drill-Down And Browsing Of Pubmed Search Results. J. Biomed. Discov. Collab. 2008, 3, 2. [Google Scholar]
- Torvik, V.I.; Smalheiser, N.R. Author Name Disambiguation in MEDLINE. ACM Trans. Knowl. Discov. Data 2009, 3, 11. [Google Scholar]
- Hersh, W.; Price, S.; Donohoe, L. Assessing Thesaurus-based Query Expansion Using the UMLS Metathesaurus. Proc. AMIA Symp. 2000, 73, 344–348. [Google Scholar]
- Wilbur, W.J.; Yang, Y. An Analysis of Statistical Term Strength and Its Use in the Indexing and Retrieval of Molecular Biology Texts. Comput. Biol. Med. 1996, 26, 209–222. [Google Scholar]
- Wilbur, W.J. A Thematic Analysis of the AIDS Literature. Pac. Symp. Biocomput. 2002, 73, 386–397. [Google Scholar]
- Chen, H.; Ng, T.D.; Martinez, J.; Schatz, B.R. A Concept Space Approach to Addressing the Vocabulary Problem in Scientific Information Retrieval: an Experiment on the Worm Community System. J. Am. Soc. Inf. Sci. 1997, 48, 17–31. [Google Scholar]
- Homayouni, R.; Heinrich, K.; Wei, L.; Berry, M.W. Gene Clustering By Latent Semantic Indexing of MEDLINE Abstracts. Bioinformatics 2004, 73, 515–527. [Google Scholar]
- Erkan, G.; Radev, D.R. LexRank: Graph-based Centrality as Salience in Text Summarization. J. Artif. Intell. Res. 2004, 22, 457–479. [Google Scholar]
- Torvik, V.I.; Smalheiser, N.R. A Quantitative Model for Linking Two Disparate Sets of Articles in MEDLINE. Bioinformatics 2007, 23, 1658–1565. [Google Scholar]
- http://arrowsmith.psych.uic.edu.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Smalheiser, N.R.; Zhou, W.; Torvik, V.I. Distribution of “Characteristic” Terms in MEDLINE Literatures. Information 2011, 2, 266-276. https://doi.org/10.3390/info2020266
Smalheiser NR, Zhou W, Torvik VI. Distribution of “Characteristic” Terms in MEDLINE Literatures. Information. 2011; 2(2):266-276. https://doi.org/10.3390/info2020266
Chicago/Turabian StyleSmalheiser, Neil R., Wei Zhou, and Vetle I. Torvik. 2011. "Distribution of “Characteristic” Terms in MEDLINE Literatures" Information 2, no. 2: 266-276. https://doi.org/10.3390/info2020266




