Involvement of Epigenetic Mechanisms in Herbicide Resistance: The Case of Conyza canadensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Herbicide Treatment
2.3. DNA Isolation and Bisulfite Sequencing
2.4. Statistical and In Silico Analysis
3. Results
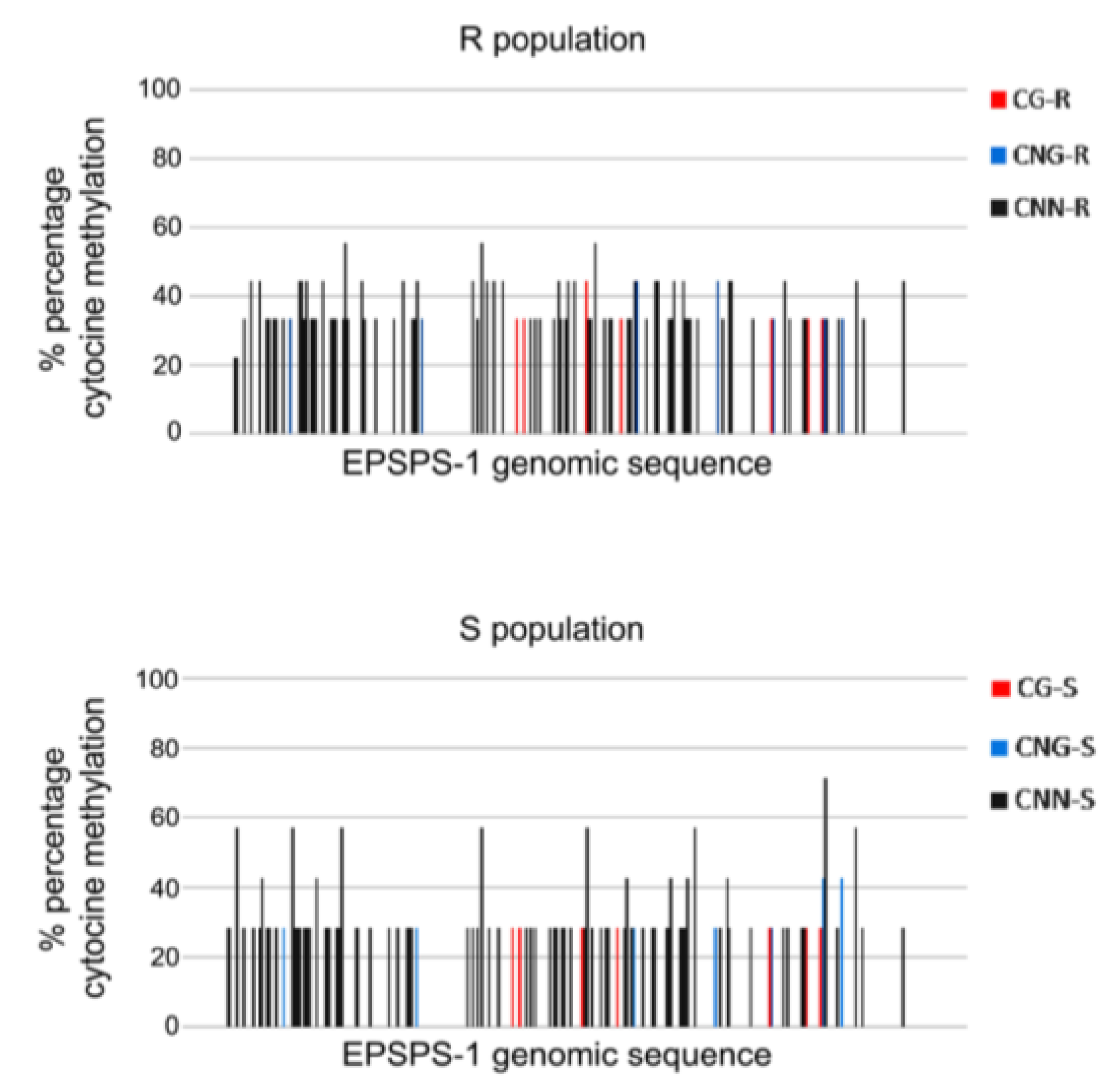
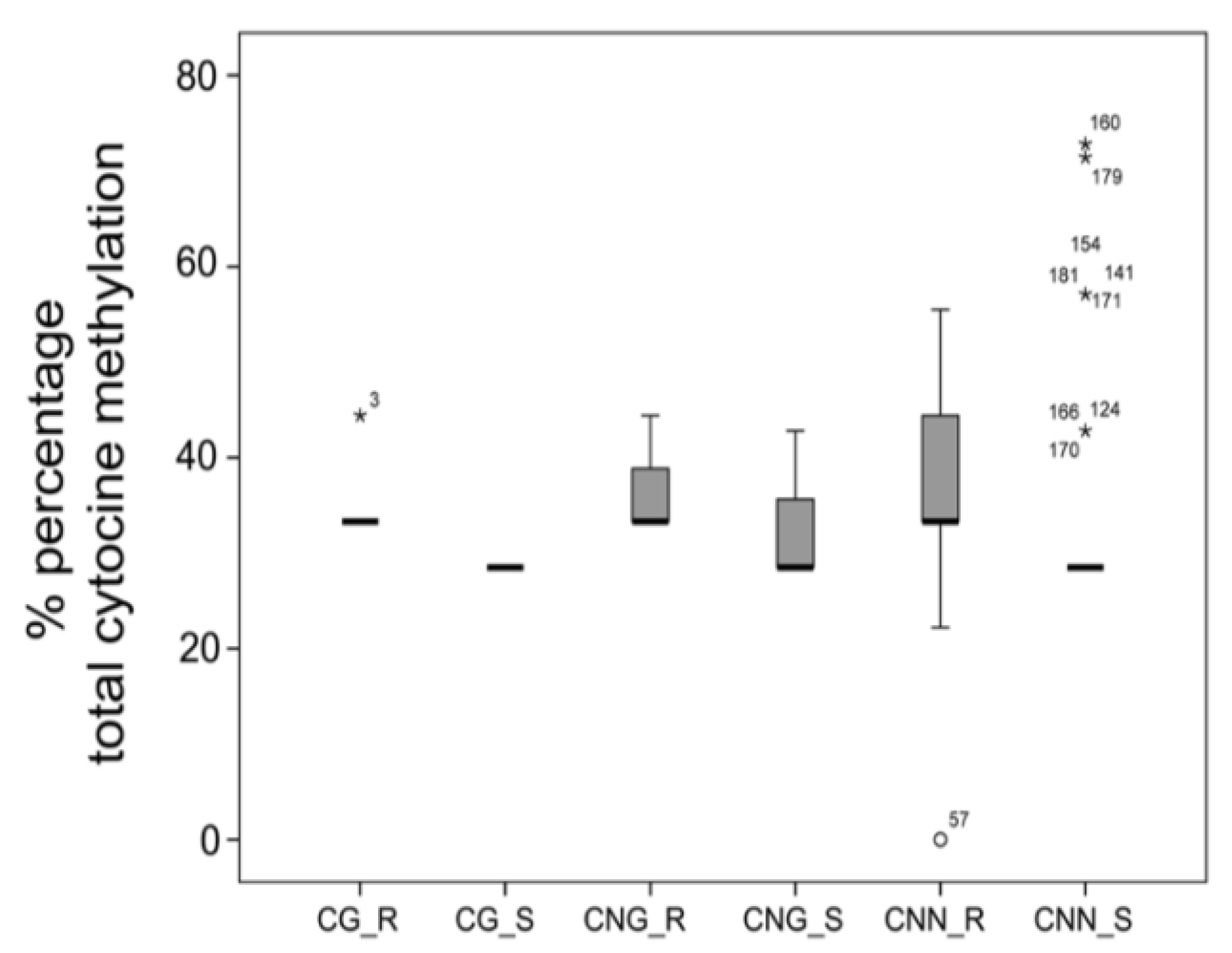
3.1. Differential Epigenetic Landscape in R and S Populations of Conyza canadensis
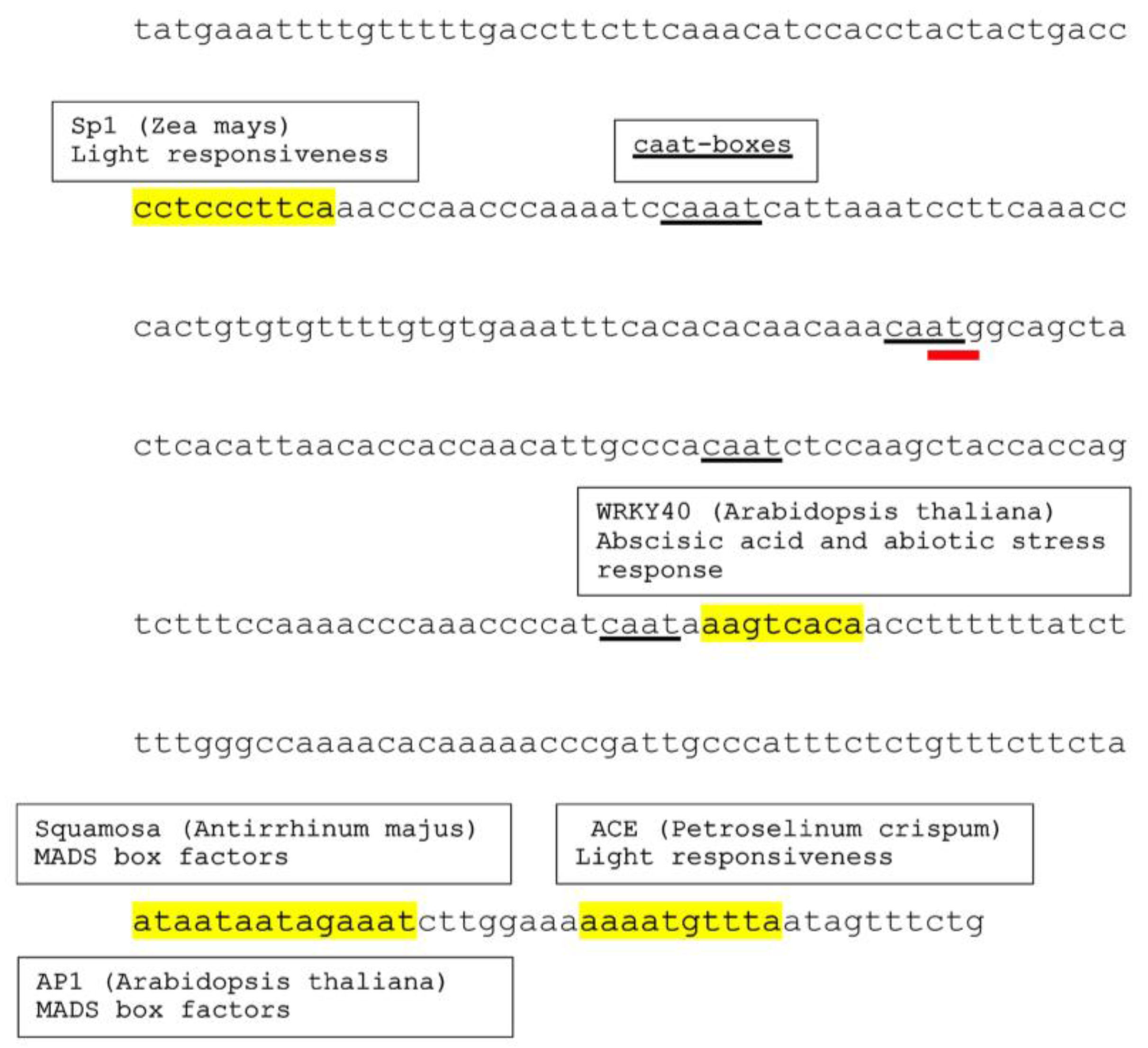
3.2. Various Cis-Acting Elements Are Present on Conyza canadensis Nucleotide Sequence
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heap, I. Global perspective of herbicide-resistant weeds. Pest Manag. Sci. 2014, 70, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Daga, P.R.; Duke, S.O.; Lee, R.M.; Tranel, P.J.; Doerksen, R.J. Biochemical and structural consequences of a glycine deletion in the α-8 helix of protoporphyrinogen oxidase. BBA Proteins Proteom. 2010, 1804, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Avila-Garcia, W.V.; Sanchez-Olguin, E.; Hulting, A.G.; Mallory-Smith, C. Target-site mutation associated with glufosinate resistance in italian ryegrass (Lolium perenne L. ssp. multiflorum). Pest Manag. Sci. 2012, 68, 1248–1254. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Han, H.; Vila-Aiub, M.M.; Powles, S.B. Ahas herbicide resistance endowing mutations: Effect on ahas functionality and plant growth. J. Exp. Bot. 2010, 61, 3925–3934. [Google Scholar] [CrossRef] [PubMed]
- Shaner, D.L.; Lindenmeyer, R.B.; Ostlie, M.H. What have the mechanisms of resistance to glyphosate taught us? Pest Manag. Sci. 2012, 68, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.; Duhieu, B.; Boucansaud, K.; Délye, C. Complex genetic control of non-target-site-based resistance to herbicides inhibiting acetyl-coenzyme a carboxylase and acetolactate-synthase in alopecurus myosuroides huds. Plant Sci. 2010, 178, 501–509. [Google Scholar] [CrossRef]
- Busi, R.; Vila-Aiub, M.; Powles, S. Genetic control of a cytochrome p450 metabolism-based herbicide resistance mechanism in lolium rigidum. Heredity 2011, 106, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.C.; Pratley, J.E.; Bohn, J.A. Resistance to glyphosate in lolium rigidum. Ii. Uptake, translocation, and metabolism. Weed Sci. 1999, 47, 412–415. [Google Scholar]
- Ge, X.; d’Avignon, D.A.; Ackerman, J.J.; Sammons, R.D. Rapid vacuolar sequestration: The horseweed glyphosate resistance mechanism. Pest Manag. Sci. 2010, 66, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Zhang, W.; Wang, D.; Bukun, B.; Chisholm, S.T.; Shaner, D.L.; Nissen, S.J.; Patzoldt, W.L.; Tranel, P.J.; Culpepper, A.S. Gene amplification confers glyphosate resistance in amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2010, 107, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Robertson, R.R. Physiological and Biochemical Characterization of Glyphosate Resistant Ambrosia trifida L.; Purdue University: West Lafayette, IN, USA, 2010. [Google Scholar]
- Lespérance, M.A. Programmed Cell Death and Altered Translocation Cause Glyphosate Resistance in Giant Ragweed (Ambrosia trifida L.); University of Guelph: Guelph, ON, Canada, 2016. [Google Scholar]
- Preston, C.; Wakelin, A.M.; Dolman, F.C.; Bostamam, Y.; Boutsalis, P. A decade of glyphosate-resistant lolium around the world: Mechanisms, genes, fitness, and agronomic management. Weed Sci. 2009, 57, 435–441. [Google Scholar] [CrossRef]
- Bostamam, Y.; Malone, J.M.; Dolman, F.C.; Boutsalis, P.; Preston, C. Rigid ryegrass (Lolium rigidum) populations containing a target site mutation in epsps and reduced glyphosate translocation are more resistant to glyphosate. Weed Sci. 2012, 60, 474–479. [Google Scholar] [CrossRef]
- Kaundun, S.S.; Dale, R.P.; Zelaya, I.A.; Dinelli, G.; Marotti, I.; McIndoe, E.; Cairns, A. A novel p106l mutation in epsps and an unknown mechanism (s) act additively to confer resistance to glyphosate in a south african Lolium rigidum population. J. Agric. Food. Chem. 2011, 59, 3227–3233. [Google Scholar] [CrossRef] [PubMed]
- Tani, E.; Chachalis, D.; Travlos, I.S. A glyphosate resistance mechanism in Conyza canadensis involves synchronization of epsps and abc-transporter genes. Plant Mol. Biol. Rep. 2015, 33, 1721–1730. [Google Scholar] [CrossRef]
- Gressel, J. Evolving understanding of the evolution of herbicide resistance. Pest Manag. Sci. 2009, 65, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Sammons, R.D.; Gaines, T.A. Glyphosate resistance: State of knowledge. Pest Manag. Sci. 2014, 70, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Abercrombie, L.L.; Yuan, J.S.; Riggins, C.W.; Sammons, R.D.; Tranel, P.J.; Stewart, C.N. Characterization of the horseweed (Conyza canadensis) transcriptome using gs-flx 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Manag. Sci. 2010, 66, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Kane, N.C.; Kozik, A.; Hodgins, K.A.; Dlugosch, K.M.; Barker, M.S.; Matvienko, M.; Yu, Q.; Turner, K.G.; Pearl, S.A. Genomics of Compositae weeds: Est libraries, microarrays, and evidence of introgression. Am. J. Bot. 2012, 99, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Délye, C.; Duhoux, A.; Pernin, F.; Riggins, C.W.; Tranel, P.J. Molecular mechanisms of herbicide resistance. Weed Sci. 2015, 63, 91–115. [Google Scholar] [CrossRef]
- Boyko, A.; Kovalchuk, I. Epigenetic control of plant stress response. Environ. Mol. Mutagen. 2008, 49, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Delye, C. Unravelling the genetic bases of non-target-site-based resistance (ntsr) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013, 69, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.P.; Pandey, G.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013, 32, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Bobadilla, R.; Berr, A. Histone methylation-a cornerstone for plant responses to environmental stresses? In Abiotic and Biotic Stress in Plants-Recent Advances and Future Perspectives; InTech: Vienna, Austria, 2016. [Google Scholar]
- Hauser, M.-T.; Aufsatz, W.; Jonak, C.; Luschnig, C. Transgenerational epigenetic inheritance in plants. BBA Gene Regul. Mech. 2011, 1809, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Chevala, V.N.; Shankar, R.; Jain, M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.G.; Waldin, T.R.; Ray, J.A.; Bright, S.W.; Hussey, P.J. Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature 1998, 393, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.M.; Webster, T.M.; Steckel, L.E. Palmer amaranth (Amaranthus palmeri): A review. Weed Technol. 2013, 27, 12–27. [Google Scholar] [CrossRef]
- Travlos, I.; Chachalis, D. Assessment of glyphosate-resistant horseweed (Conyza canadensis L. Cronq.) and fleabane (Conyza albida willd. Ex spreng) populations from perennial crops in greece. Int. J. Plant Prod. 2013, 7. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. In Circular; California Agricultural Experiment Station: Davis, CA, USA, 1950; Volume 347. [Google Scholar]
- Chachalis, D.; Travlos, I. Glyphosate resistant weeds in southern europe: Current status, control strategies and future challenges. In Handbook of Herbicides: Biological Activity, Classification, and Health and Environmental Implications; Kobayashi, D., Watanabe, E., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2014; pp. 175–191. [Google Scholar]
- Tani, E.; Chachalis, D.; Travlos, I.S.; Bilalis, D. Environmental conditions influence induction of key abc-transporter genes affecting glyphosate resistance mechanism in Conyza canadensis. Int. J. Mol. Sci. 2016, 17, 342. [Google Scholar] [CrossRef] [PubMed]
- Aceituno, F.F.; Moseyko, N.; Rhee, S.Y.; Gutiérrez, R.A. The rules of gene expression in plants: Organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genom. 2008, 9, 438. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, M.C.; Dickerson, D.R.; Schmitt, M.; Groudine, M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004, 11, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-W.; Friso, S. Epigenetics: A new bridge between nutrition and health. Adv. Nutr. Int. Rev. J. 2010, 1, 8–16. [Google Scholar] [CrossRef] [PubMed]
- González, R.M.; Ricardi, M.M.; Iusem, N.D. Atypical epigenetic mark in an atypical location: Cytosine methylation at asymmetric (cnn) sites within the body of a non-repetitive tomato gene. BMC Plant Biol. 2011, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Asensi-Fabado, M.-A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. BBA Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Margaritopoulou, T.; Tani, E.; Chachalis, D.; Travlos, I. Involvement of Epigenetic Mechanisms in Herbicide Resistance: The Case of Conyza canadensis. Agriculture 2018, 8, 17. https://doi.org/10.3390/agriculture8010017
Margaritopoulou T, Tani E, Chachalis D, Travlos I. Involvement of Epigenetic Mechanisms in Herbicide Resistance: The Case of Conyza canadensis. Agriculture. 2018; 8(1):17. https://doi.org/10.3390/agriculture8010017
Chicago/Turabian StyleMargaritopoulou, Theoni, Eleni Tani, Demosthenis Chachalis, and Ilias Travlos. 2018. "Involvement of Epigenetic Mechanisms in Herbicide Resistance: The Case of Conyza canadensis" Agriculture 8, no. 1: 17. https://doi.org/10.3390/agriculture8010017
APA StyleMargaritopoulou, T., Tani, E., Chachalis, D., & Travlos, I. (2018). Involvement of Epigenetic Mechanisms in Herbicide Resistance: The Case of Conyza canadensis. Agriculture, 8(1), 17. https://doi.org/10.3390/agriculture8010017







