Addition of Exogenous Organic Ameliorants Mediates Soil Bacteriome and Microbial Community Carbon Source Utilization Pattern in Coastal Saline–Alkaline Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site and Experimental Layout
2.2. Soil Physicochemical and Biolog Microplate Assays
2.3. DNA Extraction and Real-Time Quantitative PCR
2.4. Miseq Sequencing and Bioinformatics Analysis
2.5. Statistical Analyses
3. Results
3.1. Soil Physicochemical Characteristics
3.2. Soil Microbial Metabolic Utilization of Carbon Sources
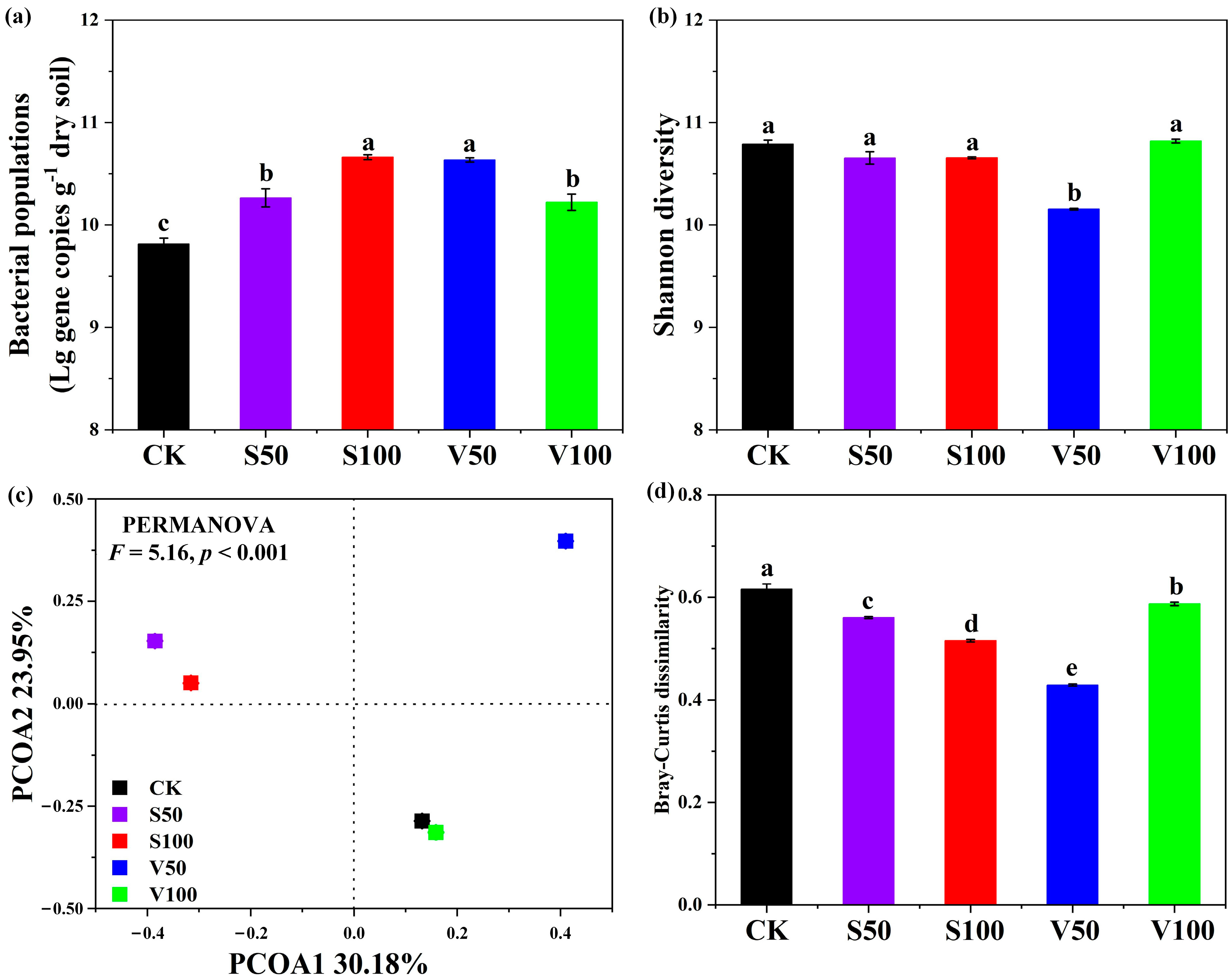
3.3. Compositional and Structural Diversities of Bacterial Community
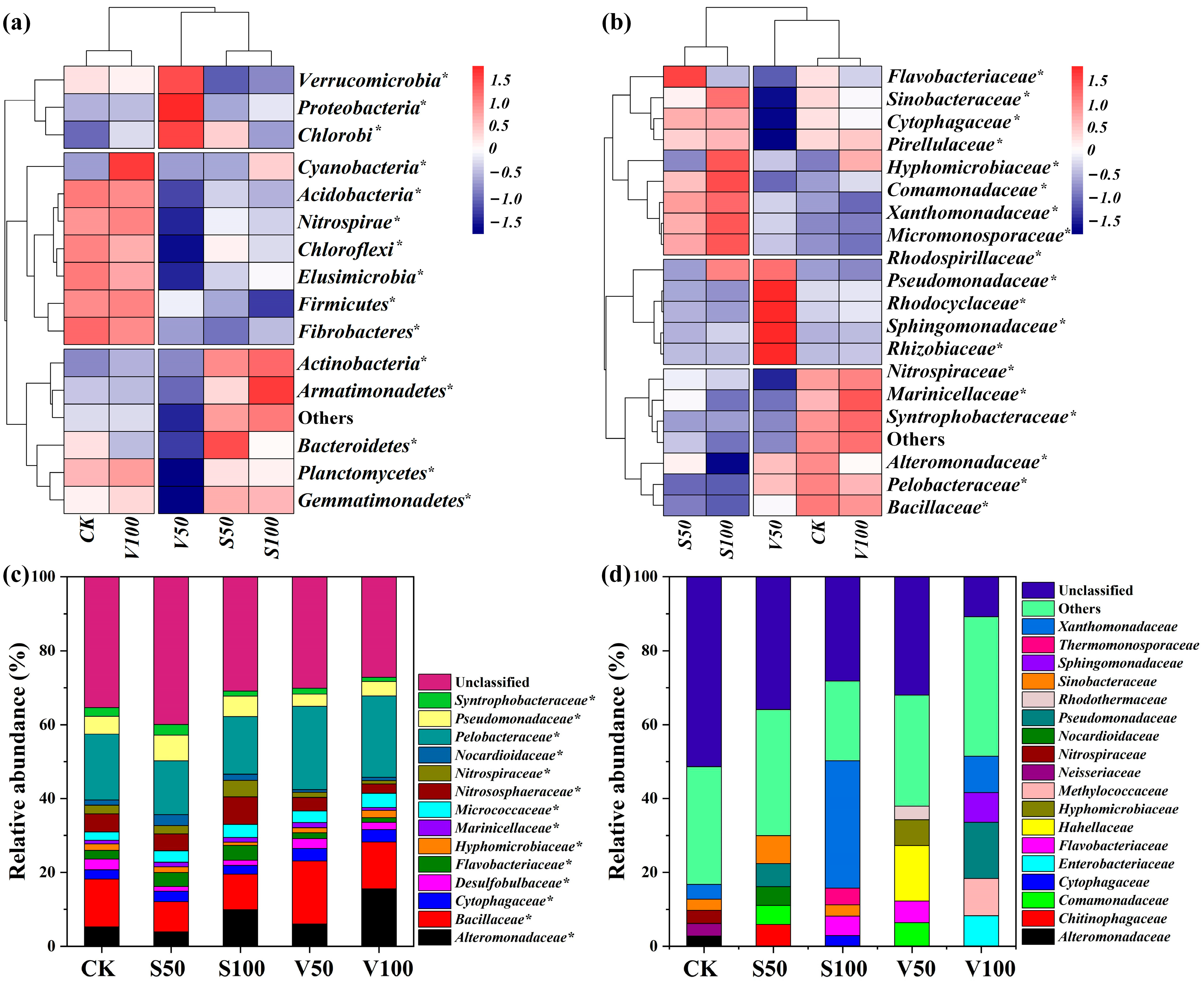
3.4. Soil Bacterial Community Composition
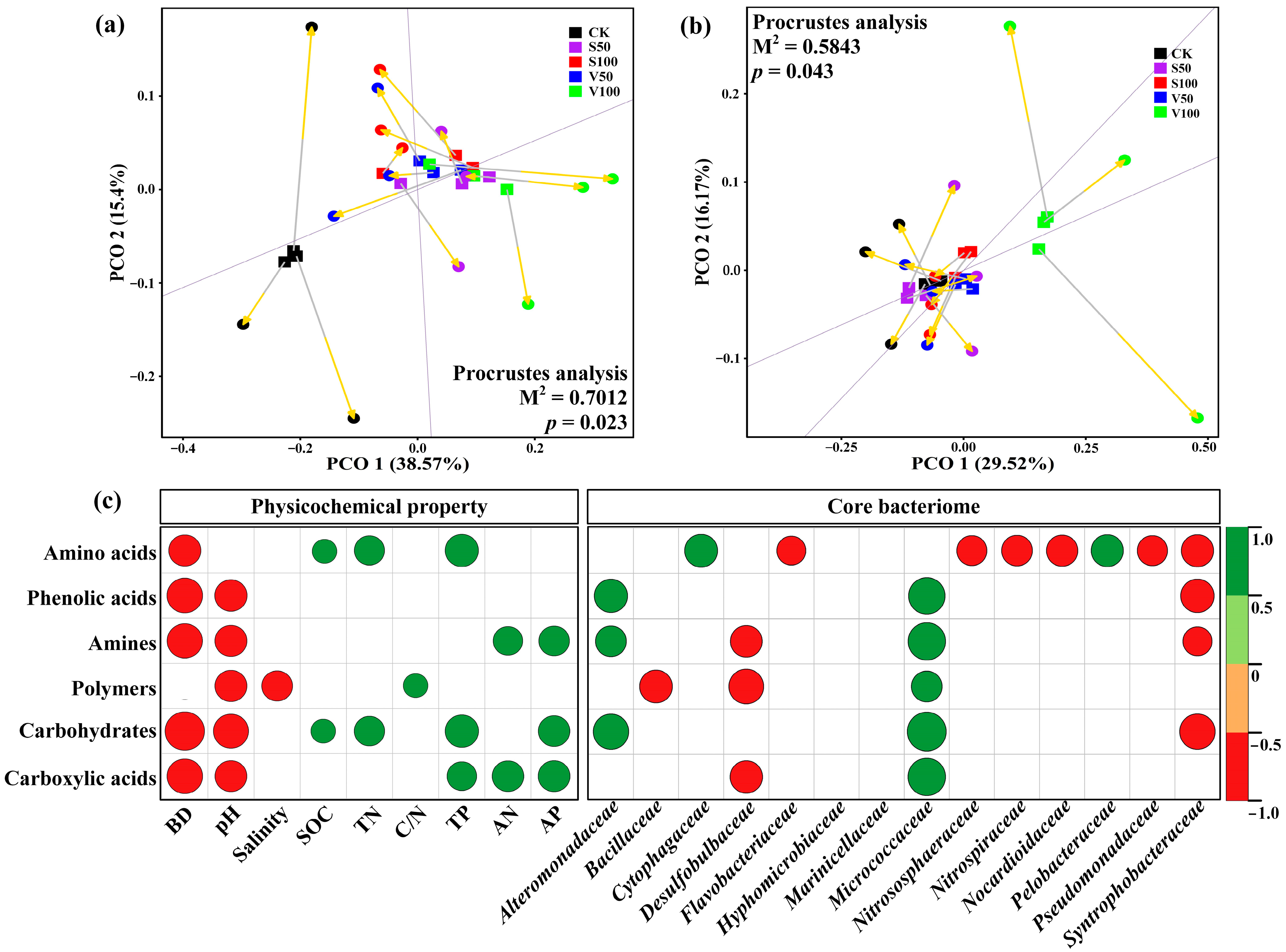
3.5. Correlating the Microbial Function with Soil Physicochemical Property and Microbial Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xie, X.F.; Pu, L.J.; Zhu, M.; Meadows, M.; Sun, L.C.; Wu, T.; Bu, X.G.; Xu, Y. Differential effects of various reclamation treatments on soil characteristics: An experimental study of newly reclaimed tidal mudflats on the east China coast. Sci. Total Environ. 2021, 768, 144996. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wall, G. Mudflat development in Jiangsu Province, China: Practices and experiences. Ocean Coast. Manag. 2010, 53, 691–699. [Google Scholar] [CrossRef]
- Cao, W.Z.; Wong, M.H. Current status of coastal zone issues and management in China: A review. Environ. Int. 2007, 33, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Dahlawi, S.; Naeem, A.; Rengel, Z.; Naidu, R. Biochar application for the remediation of salt-affected soils: Challenges and opportunities. Sci. Total Environ. 2018, 625, 320–335. [Google Scholar]
- Jiang, S.Q.; Yu, Y.N.; Gao, R.W.; Wang, H.; Zhang, J.; Li, R.; Long, X.H.; Shen, Q.R.; Chen, W.; Cai, F. High-throughput absolute quantification sequencing reveals the effect of different fertilizer applications on bacterial community in a tomato cultivated coastal saline soil. Sci. Total Environ. 2019, 687, 601–609. [Google Scholar] [CrossRef]
- Ritz, K.; Young, I.M. Interactions between soil structure and fungi. Mycologist 2004, 18, 52–59. [Google Scholar] [CrossRef]
- Goldford, J.E.; Lu, N.; Bajić, D.; Estrela, S.; Tikhonov, M.; Sanchez-Gorostiaga, A.; Segrè, D.; Mehta, P.; Sanchez, A. Emergent simplicity in microbial community assembly. Science 2018, 361, 469–474. [Google Scholar] [CrossRef]
- Palmer, M.A.; Zedler, J.B.; Falk, D.A. Ecological Theory and Restoration Ecology; Foundations of Restoration Ecology; Island Press: Washington, DC, USA, 2016; pp. 3–26. [Google Scholar] [CrossRef]
- Bai, Y.C.; Tao, T.Y.; Gu, C.H.; Wang, L.; Feng, K.; Shan, Y.H. Mudflat soil amendment by sewage sludge: Soil physicochemical properties, perennial ryegrass growth, and metal uptake. Soil Sci. Plant Nutr. 2013, 59, 942–952. [Google Scholar] [CrossRef]
- Wu, L.P.; Wang, Y.D.; Zhang, S.R.; Wei, W.L.; Kuzyakov, Y.; Ding, X.D. Fertilization effects on microbial community composition and aggregate formation in saline-alkaline soil. Plant Soil 2021, 463, 523–535. [Google Scholar] [CrossRef]
- Yao, R.J.; Yang, J.S.; Wang, X.P.; Xie, W.P.; Zheng, F.L.; Li, H.Q.; Tang, C.; Zhu, H. Response of soil characteristics and bacterial communities to nitrogen fertilization gradients in a coastal salt-affected agroecosystem. Land Degrad. Dev. 2021, 32, 338–353. [Google Scholar] [CrossRef]
- Sastre-Conde, I.; Lobo, M.C.; Beltrán-Hernández, R.I.; Poggi-Varaldo, H.M. Remediation of saline soils by a two-step process: Washing and amendment with sludge. Geoderma 2015, 247, 140–150. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, T.; Liu, K.; Wang, L.; Wang, K.; Zhou, Y. Effects of different amendments for the reclamation of coastal saline soil on soil nutrient dynamics and electrical conductivity responses. Agr. Water Manag. 2015, 159, 115–122. [Google Scholar] [CrossRef]
- Aksakal, E.L.; Sari, S.; Angin, I. Effects of vermicompost application on soil aggregation and certain physical properties. Land Degrad. Dev. 2016, 27, 983–995. [Google Scholar] [CrossRef]
- Moorhead, D.L.; Sinsabaugh, R.L. A theoretical model of litter decay and microbial interaction. Ecol. Monogr. 2006, 76, 151–174. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Lauber, C.L.; Weintraub, M.N.; Ahmed, B.; Allison, S.D.; Crenshaw, C.; Contosta, A.R.; Cusack, D.; Frey, S.; Gallo, M.E. Stoichiometry of soil enzyme activity at global scale. Eol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Omirou, M.; Rousidou, C.; Bekris, F.; Papadopoulou, K.K.; Menkissoglou-Spiroudi, U.; Ehaliotis, C.; Karpouzas, D.G. The impact of biofumigation and chemical fumigation methods on the structure and function of the soil microbial community. Microb. Ecol. 2011, 61, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.C.; Zuo, W.G.; Yan, Y.Y.; Gu, C.H.; Guan, Y.X.; Mei, L.J.; Xue, W.J.; Shan, Y.H.; Feng, K. Sewage sludge amendment combined with green manuring to a coastal mudflat salt-soil in eastern China: Effects on soil physicochemical properties and maize yield. Int. J. Agron. 2017, 2017, 8526598. [Google Scholar] [CrossRef]
- Li, Y.L.; Wang, Y.M.; Shen, C.; Xu, L.; Yi, S.Q.; Zhao, Y.L.; Zuo, W.G.; Gu, C.H.; Shan, Y.H.; Bai, Y.C. structural and predicted functional diversities of bacterial microbiome in response to sewage sludge amendment in coastal mudflat soil. Biology 2021, 10, 1302. [Google Scholar] [CrossRef]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 2017, 57, 2351–2359. [Google Scholar] [CrossRef]
- Choi, K.H.; Dobbs, F.C. Comparison of two kinds of Biolog microplates (GN and ECO) in their ability to distinguish among aquatic microbial communities. J. Microbiol. Meth. 1999, 36, 203–213. [Google Scholar] [CrossRef]
- Fisk, M.C.; Ruether, K.F.; Yavitt, J.B. Microbial activity and functional composition among northern peatland ecosystems. Soil Biol. Biochem. 2003, 35, 591–602. [Google Scholar] [CrossRef]
- Feigl, V.; Ujaczki, É.; Vaszita, E.; Molnár, M. Influence of red mud on soil microbial communities: Application and comprehensive evaluation of the Biolog EcoPlate approach as a tool in soil microbiological studies. Sci. Total Environ. 2017, 595, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.; Zhang, R.F. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Wang, Y.M.; Gu, C.H.; Shen, C.; Xu, L.; Zhao, Y.L.; Yi, S.Q.; Zuo, W.G.; Shan, Y.H.; Zhang, Z.Q.; et al. Differential effects of organic ameliorants on the reassembly of bacterial communities in newly amended coastal mudflat salt-affected soil. Agronomy 2022, 12, 2525. [Google Scholar] [CrossRef]
- Lai, J.; Zou, Y.; Zhang, J.; Peres-Neto, P.R. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Jorenush, M.H.; Sepaskhah, A.R. Modelling capillary rise and soil salinity for shallow saline water table under irrigated and non-irrigated conditions. Agr. Water Manage. 2003, 61, 125–141. [Google Scholar] [CrossRef]
- Wu, W.; Huang, H.L.; Biber, P.; Bethel, M. Litter decomposition of Spartina alterniflora and Juncus roemerianus: Implications of climate change in salt marshes. J. Coastal Res. 2017, 33, 372–384. [Google Scholar] [CrossRef]
- Kacprzak, M.; Neczaj, E.; Fijakowski, K.; Grobelak, A.; Grosser, A.; Worwag, M.; Rorat, A.; Brattebo, H.; Almås, Å.; Singh, B.R. Sewage sludge disposal strategies for sustainable development. Environ. Res. 2017, 156, 39–46. [Google Scholar] [CrossRef]
- Bai, Y.C.; Zuo, W.G.; Shao, H.B.; Mei, L.J.; Tang, B.P.; Gu, C.H.; Wang, X.K.; Guan, Y.X. Eastern China coastal mudflats: Salt-soil amendment with sewage sludge. Land Degrad. Dev. 2018, 29, 3803–3811. [Google Scholar] [CrossRef]
- Tian, X.M.; Fan, H.; Wang, J.Q.; Ippolito, J.; Li, Y.B.; Feng, S.S.; An, A.J.; Zhang, F.H.; Wang, K.Y. Effect of polymer materials on soil structure and organic carbon under drip irrigation. Geoderma 2019, 340, 94–103. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2003, 68, 12–26. [Google Scholar] [CrossRef]
- Gomez, E.; Ferreras, L.; Toresani, S. Soil bacterial functional diversity as influenced by organic amendment application. Bioresour. Technol. 2006, 97, 1484–1489. [Google Scholar] [CrossRef] [PubMed]
- Govaerts, B.; Mezzalama, M.; Unno, Y.; Sayre, K.D.; Luna-Guido, M.; Vanherck, K.; Dendooven, L.; Deckers, J. Influence of tillage, residue management, and crop rotation on soil microbial biomass and catabolic diversity. Appl. Soil Ecol. 2006, 37, 18–30. [Google Scholar] [CrossRef]
- Pascual, I.; Avilés, M.; Aguirreolea, J.; Sánchez-Díaz, M. Effect of sanitized and non-sanitized sewage sludge on soil microbial community and the physiology of pepper plants. Plant Soil. 2008, 310, 41–53. [Google Scholar] [CrossRef]
- Liu, X.; Guo, K.L.; Huang, L.; Ji, Z.Y.; Jiang, H.M.; Li, H.; Zhang, J.F. Responses of absolute and specific enzyme activity to consecutive application of composted sewage sludge in a Fluventic Ustochrept. PLoS ONE 2017, 12, e0177796. [Google Scholar] [CrossRef] [PubMed]
- Calleja-Cervantes, M.E.; Aparicio-Tejo, P.M.; Villadas, P.J.; Irigoyen, I.; Irañeta, J.; Fernández-González, A.J.; Fernández-López, M.; Menéndez, S. Rational application of treated sewage sludge with urea increases GHG mitigation opportunities in Mediterranean soils. Agric. Ecosyst. Environ. 2017, 238, 114–127. [Google Scholar] [CrossRef]
- Latare, A.M.; Singh, S.K.; Kumar, O. Impact of sewage sludge application on soil fertility, microbial population and enzyme activities in soil under rice-wheat system. J. Indian Soc. Soil Sci. 2018, 66, 300–309. [Google Scholar] [CrossRef]
- Bhat, S.A.; Singh, J.; Vig, A.P. Earthworms as organic waste managers and biofertilizer producers. Waste Biomass Valori. 2018, 9, 1073–1086. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, L.; Zhao, J.; Niu, Y.; Xiao, H.; Wang, Z.; Yu, S.; Shi, Z. Forty-year-old orchards promote carbon storage by changing aggregate-associated enzyme activities and microbial communities. Catena 2022, 213, 106195. [Google Scholar] [CrossRef]
- Li, F.Q.; Xue, C.; Qiu, P.F.; Liu, Y.X.; Shi, J.X.; Shen, B.; Yang, X.M.; Shen, Q.R. Soil aggregate size mediates the responses of microbial communities to crop rotation. Eur. J. Soil Biol. 2018, 88, 48–56. [Google Scholar] [CrossRef]
- Six, J.; Paustian, K.; Elliott, E.T.; Combrink, C. Soil structure and organic matter I. Distribution of aggregate-size classes and aggregate-associated carbon. Soil Sci. Soc. Am. J. 2018, 64, 681–689. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Tian, C.Y. Soil pH is equally important as salinity in shaping bacterial communities in saline soils under halophytic vegetation. Sci. Rep. 2018, 8, 4550. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Luo, N.Y.; Ji, C.L.; Li, J.; Zhang, L.; Xiao, L.; She, X.L.; Liu, Z.; Li, Y.L.; Liu, C.S.; et al. Liquid organic fertilizer amendment alters rhizosphere microbial community structure and co-occurrence patterns and improves sunflower yield under salinity-alkalinity stress. Microb. Ecol. 2021, 84, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.S.; Wang, S.J.; Tian, L.; Shi, S.H.; Xu, S.Q.; Yang, F.; Li, X.J.; Wang, Z.C.; Tian, C.J. Aggregate-related changes in soil microbial communities under different ameliorant applications in saline-sodic soils. Geoderma 2018, 329, 108–117. [Google Scholar] [CrossRef]


| Items | Treatments $ | Organics | |||||
|---|---|---|---|---|---|---|---|
| CK | S50 | S100 | V50 | V100 | Sewage Sludge | Vermicompost | |
| BD (g cm−3) | 1.34 ± 0.00 a | 1.31 ± 0.00 b | 1.28 ± 0.00 cd | 1.29 ± 0.00 bc | 1.26 ± 0.01 d | / | / |
| pH | 9.33 ± 0.09 a | 9.11 ± 0.04 b | 8.78 ± 0.02 c | 9.04 ± 0.06 b | 8.96 ± 0.08 bc | 6.32 | 6.33 |
| Salinity (‰) | 3.15 ± 0.16 ab | 2.61 ± 0.16 c | 2.79 ± 0.06 bc | 3.41 ± 0.02 a | 3.30 ± 0.03 a | 32.9 | 8.43 |
| SOC (g kg−1) | 3.60 ± 0.32 b | 11.27 ± 1.13 a | 11.92 ± 1.83 a | 11.73 ± 1.32 a | 13.59 ± 3.44 a | 216.2 | 464.9 |
| TN (g kg−1) | 0.33 ± 0.01 a | 0.67 ± 0.06 a | 0.70 ± 0.13 a | 0.68 ± 0.03 a | 0.90 ± 0.25 a | 51.2 | 19.27 |
| C/N | 10.78 ± 3.19 a | 16.90 ± 4.60 a | 17.12 ± 6.35 a | 17.27 ± 5.16 a | 15.06 ± 5.56 a | 4.22 | 24.33 |
| TP (mg kg−1) | 0.4 ± 0.02 b | 0.61 ± 0.04 ab | 0.68 ± 0.14 a | 0.72 ± 0.06 a | 0.82 ± 0.10 a | 5.51 | 24.12 |
| AN (mg kg−1) | 37.98 ± 0.30 b | 107.10 ± 14.8 a | 82.82 ± 11.36 a | 83.89 ± 7.43 a | 112.39 ± 23.36 a | 3440 | 15.87 |
| AP (mg kg−1) | 21.43 ± 1.77 b | 84.50 ± 11.44 a | 94.75 ± 21.61 a | 84.96 ± 7.61 a | 108.18 ± 15.38 a | 813 | 2467 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, B.; Qin, T.; Qiu, M.; Yu, J.; Zhang, L.; Li, Y. Addition of Exogenous Organic Ameliorants Mediates Soil Bacteriome and Microbial Community Carbon Source Utilization Pattern in Coastal Saline–Alkaline Soil. Agriculture 2024, 14, 44. https://doi.org/10.3390/agriculture14010044
Gu B, Qin T, Qiu M, Yu J, Zhang L, Li Y. Addition of Exogenous Organic Ameliorants Mediates Soil Bacteriome and Microbial Community Carbon Source Utilization Pattern in Coastal Saline–Alkaline Soil. Agriculture. 2024; 14(1):44. https://doi.org/10.3390/agriculture14010044
Chicago/Turabian StyleGu, Binxian, Tianyang Qin, Meihua Qiu, Jie Yu, Li Zhang, and Yunlong Li. 2024. "Addition of Exogenous Organic Ameliorants Mediates Soil Bacteriome and Microbial Community Carbon Source Utilization Pattern in Coastal Saline–Alkaline Soil" Agriculture 14, no. 1: 44. https://doi.org/10.3390/agriculture14010044






