Comparative Metabolomics Analysis of Weedy Rice (Oryza spp.) across Peninsular Malaysia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Materials
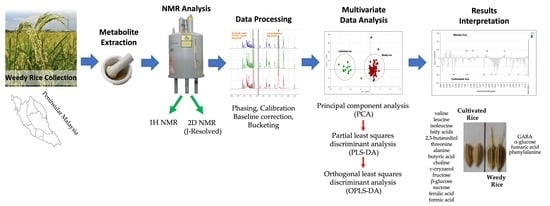
2.3. Metabolite Extraction
2.4. NMR Analysis
2.5. Multivariate Data Analysis
3. Results and Discussion
3.1. Comparative Metabolomics Analysis of Weedy Rice and Cultivated Rice
3.2. Metabolomic Analysis of Malaysian Weedy Rice Associated with Grain Morphology
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saha, S.; Patra, B.C.; Munda, S.; Mohapatra, T. Weedy rice: Problems and its management. Indian J. Weed Sci. 2014, 46, 14–22. [Google Scholar]
- Nadir, S.; Xiong, H.B.; Zhu, Q.; Zhang, X.L.; Xu, H.Y.; Li, J.; Dongchen, W.; Henry, D.; Guo, X.Q.; Khan, S.; et al. Weedy rice in sustainable rice production. A review. Agron. Sustain. Dev. 2017, 37, 46. [Google Scholar] [CrossRef]
- Wahab, A.; Suhaimi, O. Weedy rice characteristics, adverse effects and methods of its eradication. Teknologi Padi 1991, 7, 21–31. (In Malay) [Google Scholar]
- Abdullah, M.Z.; Vaughan, A.; Watanabe, H.; Okuno, K. The origin of weedy rice in Peninsular Malaysia. MARDI Res. J. 1996, 24, 169–174. [Google Scholar]
- Anuar, N.S.; Mazlan, N.; Ariff, E.E.; Juraimi, A.S.; Yusop, M.R. A comparative study of vegetative and reproductive growth of local weedy and Clearfield® rice varieties in Malaysia. J. Int. Soc. Southeast Asian Agric. Sci. 2014, 20, 41–51. [Google Scholar]
- Cao, Q.; Lu, B.R.; Xia, H.; Rong, J.; Sala, F.; Spada, A.; Grassi, F. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in North-eastern China Revealed by simple sequence repeat (SSR) markers. Annals Bot. 2006, 98, 1241–1252. [Google Scholar] [CrossRef]
- Baki, B.B.; Shakirin, M.M. Spatio-temporal distribution pattern of new biotypes of weedy rice (Oryza sativa L.) in Selangor north-west project, Malaysia. Korean J. Weed Sci. 2010, 30, 68–83. [Google Scholar] [CrossRef]
- Zhang, L.; Dai, W.; Wu, C.; Song, X.; Qiang, S. Genetic diversity and origin of Japonica- and Indica-like rice biotypes of weedy rice in the Guangdong and Liaoning provinces of China. Genet. Resour. Crop Evol. 2012, 59, 399–410. [Google Scholar] [CrossRef]
- Song, E.H.; Kim, H.J.; Jeong, J.; Chung, H.J.; Kim, H.Y.; Bang, E.; Hong, Y.S. A 1H HR-MAS NMR-based metabolomic study for metabolic characterization of rice grain from various Oryza sativa L. cultivars. J. Agric. Food Chem. 2016, 64, 3009–3016. [Google Scholar] [CrossRef]
- Chauhan, B.S. Strategies to manage weedy rice in Asia. Crop Prot. 2013, 48, 51–56. [Google Scholar] [CrossRef]
- Olajumoke, B.; Juraimi, A.S.; Uddin, M.K.; Husni, M.H.A.; Alam, M.A. Competitive ability of cultivated rice against weedy rice biotypes—A review. Chil. J. Agric. Res. 2016, 76, 243–252. [Google Scholar] [CrossRef]
- Sudianto, E.; Neik, T.X.; Tam, S.M.; Chuah, T.S.; Idris, A.A.; Olsen, K.M.; Song, B.K. Morphology of Malaysian weedy rice (Oryza sativa): Diversity, origin and implications for weed management. Weed Sci. 2016, 64, 501–512. [Google Scholar] [CrossRef]
- Mohamed, Z.; Terani, R.; Shamsudin, M.N.; Latif, I.A. Paddy farmers’ sustainability practices in granary areas in Malaysia. Resources 2016, 5, 17. [Google Scholar] [CrossRef]
- Dilipkumar, M.; Burgos, N.R.; Chuah, T.S.; Ismail, S. Cross-resistance to imazapic and imazapyr in a weedy rice (Oryza sativa) biotype found in Malaysia. Planta Daninha 2018, 36, e018182239. [Google Scholar] [CrossRef]
- Pramai, P.; Hamid, N.A.A.; Mediani, A.; Maulidiani, M.; Abas, F.; Jiamyangyuen, S. Metabolite profiling, antioxidant, and α-glucosidase inhibitory activities of germinated rice: Nuclear-magnetic-resonance-based metabolomics study. J. Food Drug Anal. 2018, 26, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Uawisetwathana, U.; Graham, S.F.; Kamolsukyunyong, W.; Sukhaket, W.; Klanchui, A.; Toojinda, T.; Vanavichit, A.; Karoonuthaisiri, N.; Elliot, C.T. Quantitative 1H NMR metabolome profiling of Thai jasmin rice (Oryza sativa) reveals primary metabolic response during brown planthopper infestation. Metabolomics 2015, 11, 1640–1655. [Google Scholar] [CrossRef]
- Wijaya, D.N.; Susanto, F.A.; Purwestri, Y.A.; Ismoyowati, D.; Nuringtyas, T.R. NMR metaboliote comparison of local pigmented rice in Yogyakarta. Indones. J. Biotechnol. 2017, 22, 68–75. [Google Scholar] [CrossRef]
- Zarei, I.; Luna, E.; Leach, J.E.; McClung, A.; Vilchez, S.; Koita, O.; Ryan, E.P. Comparative rice bran metabolomics across diverse cultivars and functional rice gene-bran metabolite relationships. Metabolites 2018, 8, 63. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based plant metabolomics: Where do we stand, where do we go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef]
- Du, S.; Cui, M.; Cai, Y.; Xue, A.; Hao, Y.; Huang, X.; Liu, L.; Luo, L. Metabolomic analysis of chilling response in rice (Oryza sativa L.) seedlings by extractive electrospray ionization mass spectrometry. Environ. Exp. Bot. 2020, 180, 104231. [Google Scholar] [CrossRef]
- Nam, M.H.; Bang, E.; Kwon, T.Y.; Kim, Y.; Kim, E.H.; Cho, K.; Yoon, I.S. Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 2015, 16, 21959–21974. [Google Scholar] [CrossRef]
- Schaarschmidt, S.; Lawas, L.M.F.; Glaubitz, U.; Li, X.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Hincha, D.K.; Zuther, E. Season Affects Yield and Metabolic Profiles of Rice (Oryza sativa) under High Night Temperature Stress in the Field. Int. J. Mol. Sci. 2020, 21, 3187. [Google Scholar] [CrossRef]
- Mahmod, I.F.; Saiman, M.Z.; Mohamed, Z.; Ishak, M.N.; Mispan, M.S. Morphological variation, distribution and relationship of weedy rice (Oryza sativa L.) in Peninsular Malaysia. Weed Biol. Manag. 2021, 21, 86–99. [Google Scholar] [CrossRef]
- Saiman, M.Z.; Mustafa, N.R.; Schulte, A.E.; Verpoorte, R.; Choi, Y.H. Induction, characterization, and NMR-based metabolic profiling of adventitious root cultures from leaf explants of Gynura procumbens. Plant Cell Tissue Organ Cult. 2012, 109, 465–475. [Google Scholar] [CrossRef]
- Saiman, M.Z.; Mustafa, N.R.; Choi, Y.H.; Verpoorte, R.; Schulte, A.E. Metabolic alterations and distribution of five-carbon precursors in jasmonic acid-elicited Catharanthus roseus cell suspension cultures. Plant Cell Tissue Organ Cult. 2015, 122, 351–362. [Google Scholar] [CrossRef]
- Heuberger, A.L.; Lewis, M.R.; Chen, M.H.; Brick, M.A.; Leach, J.E.; Ryan, E.P. Metabolomic and functional genomic analyses reveal varietal differences in bioactive compounds of cooked rice. PLoS ONE 2010, 5, e12915. [Google Scholar] [CrossRef]
- Hu, C.; Shi, J.; Quan, S.; Cui, B.; Kleessen, S.; Nikoloski, Z.; Tohge, T.; Alexander, D.; Guo, L.; Lin, H.; et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 2014, 4, 5067. [Google Scholar] [CrossRef]
- Deng, G.F.; Xu, X.R.; Zhang, Y.; Li, D.; Gan, R.Y.; Li, H.B. Phenolic compounds and bioactivities of pigmented rice. Crit. Rev. Food Sci. Nutr. 2013, 53, 296–306. [Google Scholar] [CrossRef]
- Mbanjo, E.G.N.; Kretzschmar, T.; Jones, H.; Ereful, N.; Blanchard, C.; Boyd, L.A.; Sreenivasulu, N. The genetic basis and nutritional benefits of pigmented rice grain. Front. Genet. 2020, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.R.; Jung, E.S.; Lee, S.; Lim, S.H.; Ha, S.H.; Lee, C.H. Combined mass spectrometry-based metabolite profiling of different pigmented rice (Oryza sativa L.) seeds and correlation with antioxidant activities. Molecules 2014, 19, 15673–15686. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, A. Weedy rice, biological features and control. In Weed Management for Developing Countries; Labrada, R., Ed.; FAO: Rome, Italy, 2003; pp. 89–107. [Google Scholar]
- Kanapeckas, K.L.; Vigueira, C.C.; Ortiz, A.; Gettler, K.A.; Burgos, N.R.; Fischer, A.J.; Lawton-Rauh, A.L. Escape to ferality: The endoferal origin of weedy rice from crop rice through de-domestication. PLoS ONE 2016, 11, e0162676. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Pruthi, V. Potential applications of ferulic acid from natural sources. Biotechnol. Rep. 2014, 4, 86–93. [Google Scholar] [CrossRef]
- Goufo, P.; Henrique, T. Rice antioxidants: Phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2014, 2, 75–104. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Karbalaii, M.T.; Jaafar, H.Z.; Rahmat, A. Phytochemical constituents, antioxidant activity, and antiproliferative properties of black, red, and brown rice bran. Chem. Cent. J. 2018, 12, 17. [Google Scholar] [CrossRef]
- Yamuangmorn, S.; Prom-u-Thai, C. The potential of high-anthocyanin purple rice as a functional ingredient in human health. Antioxidants 2021, 10, 833. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Z.; Yun-Jie, G.U. Caryopsis development and anthocyanidin accumulation of colored rice. Chin. J. Rice Sci. 2011, 25, 392–398. [Google Scholar]
- Andriani, R.; Subroto, T.; Ishmayana, S.; Kurnia, D. Enhancement Methods of Antioxidant Capacity in Rice Bran: A Review. Foods 2022, 11, 2994. [Google Scholar] [CrossRef]
- Modgil, R.; Rani, U. Effect of Processing on the Nutritional Quality of Red Rice Cultivars. J. Life Sci. 2016, 8, 12–18. [Google Scholar] [CrossRef]
- Priya, T.R.; Nelson, A.R.L.E.; Ravichandran, K.; Antony, U. Nutritional and functional properties of coloured rice varieties of South India: A review. J. Ethn. Foods 2019, 6, 11. [Google Scholar] [CrossRef]
- Biswas, S.K.; Kim, D.E.; Keum, Y.S.; Saini, R.K. Metabolite profiling and antioxidant activities of white, red, and black rice (Oryza sativa L.) grains. J. Food Meas. Charact. 2018, 12, 2484–2492. [Google Scholar] [CrossRef]
- Frank, T.; Reichardt, B.; Shu, Q.; Engel, K.H. Metabolite profiling of coloured rice (Oryza sativa L. ) grains. J. Cereal Sci. 2012, 55, 112–119. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Cros, G.; Yokota, T.; Crozier, A. Phytochemical profiles of black, red, brown, and white rice from the Camargue region of France. J. Agric. Food Chem. 2013, 61, 7976. [Google Scholar] [CrossRef]
- Lerma-García, M.J.; Herrero-Martínez, J.M.; Simó-Alfonso, E.F.; Mendonça, C.R.B.; Ramis-Ramos, G. Composition industrial processing and applications of rice bran gamma-oryzanol. Food Chem. 2009, 115, 389–404. [Google Scholar] [CrossRef]
- Moongngarm, A.; Saetung, N. Comparison of chemical compositions and bioactive compounds of germinated rough rice and brown rice. Food Chem. 2010, 122, 782–788. [Google Scholar] [CrossRef]
- Gealy, D.R.; Bryant, R.J. Seed physicochemical characteristics of field-grown US weedy red rice (Oryza sativa) biotypes: Contrasts with commercial cultivars. J. Cereal Sci. 2009, 49, 239–245. [Google Scholar] [CrossRef]
- Jia, Y.; Gealy, D. Weedy Red Rice Has Novel Sources of Resistance to Biotic Stress. Crop J. 2018, 6, 443–450. [Google Scholar] [CrossRef]
- Mohd Hanafiah, N.; Cheng, A.; Lim, P.-E.; Sethuraman, G.; Mohd Zain, N.A.; Baisakh, N.; Mispan, M.S. Novel PCR-Based Multiplex Assays for Detecting Major Quality and Biotic Stress in Commercial and Weedy Rice. Life 2022, 12, 1542. [Google Scholar] [CrossRef]









| Metabolites | 1H NMR Signals of Chemical Shift (ppm) and Coupling Constant (Hz) | Weedy Rice | Cultivated Rice |
|---|---|---|---|
| Fatty acids | δ 0.88 (d, J = 7.0), δ 1.30 (m) | 🗸 | 🗸 |
| Butyric acid | δ 0.89 (d, J = 7.0); δ 1.58 (m) | 🗸 | 🗸 |
| Isoleucine | δ 0.95 (t, J = 7.5), δ 1.03 (d, J = 7.0), | 🗸 | 🗸 |
| Leucine | δ 0.96 (d, J = 6.5), δ 0.98 (d, J = 6.5) | 🗸 | 🗸 |
| Valine | δ 1.01 (d, J = 7.0), δ 1.06 (d, J = 7.0) | 🗸 | 🗸 |
| 2,3-Butanediol | δ 1.12 (d, J = 7.0) | 🗸 | 🗸 |
| Threonine | δ 1.33 (d, J = 6.6) | 🗸 | 🗸 |
| Alanine | δ 1.47 (d, J = 6.6) | 🗸 | 🗸 |
| γ-aminobutyric acid (GABA) | δ 1.89 (m), δ 2.32 (d, J = 7.5), δ 2.98 (d, J = 7.5) | 🗸 | 🗸 |
| Choline | δ 3.23 (s) | 🗸 | 🗸 |
| γ-oryzanol | δ 3.92 (s), δ 5.39 (d, J = 5.9) | 🗸 | 🗸 |
| Fructose | δ 4.13 (d, J = 8.2), δ 3.78–3.82 (m) | 🗸 | 🗸 |
| β-glucose | δ 4.53 (d, J = 7.7) | 🗸 | 🗸 |
| α-glucose | δ 5.15 (d, J = 3.7) | 🗸 | 🗸 |
| Sucrose | δ 5.4 (d, J = 3.8), δ 4.13 (d, J = 8.5) | 🗸 | 🗸 |
| Fumaric acid | δ 6.52 (s) | 🗸 | - |
| Ferulic acid | δ 7.14 (d, J = 8.0), δ 7.27 (s), δ 3.90 (s) | 🗸 | 🗸 |
| Phenylalanine | δ 7.38 (t, J = 6.2) | 🗸 | - |
| Formic acid | δ 8.48 (s) | 🗸 | 🗸 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmod, I.F.; Jeyasimman, S.; Mispan, M.S.; Supandi, F.; Khatib, A.; Saiman, M.Z. Comparative Metabolomics Analysis of Weedy Rice (Oryza spp.) across Peninsular Malaysia. Agriculture 2023, 13, 1230. https://doi.org/10.3390/agriculture13061230
Mahmod IF, Jeyasimman S, Mispan MS, Supandi F, Khatib A, Saiman MZ. Comparative Metabolomics Analysis of Weedy Rice (Oryza spp.) across Peninsular Malaysia. Agriculture. 2023; 13(6):1230. https://doi.org/10.3390/agriculture13061230
Chicago/Turabian StyleMahmod, Intan Filzah, Saharshini Jeyasimman, Muhamad Shakirin Mispan, Farahaniza Supandi, Alfi Khatib, and Mohd Zuwairi Saiman. 2023. "Comparative Metabolomics Analysis of Weedy Rice (Oryza spp.) across Peninsular Malaysia" Agriculture 13, no. 6: 1230. https://doi.org/10.3390/agriculture13061230