Abstract
Water-limiting conditions can severely affect rice yield. Therefore, increasing plant tolerance to water stress is a priority for many rice breeding programs. However, improving rice tolerance to this abiotic stress comes with several complications related to the seeding practices, the adopted water management system and the growth stage where water stress occurs. For this reason, it is challenging to outline single ideotypes showing traits suitable for overcoming drought at different times during the life cycle of rice in diverse cropping ecosystems. The current knowledge of genomics and biochemicals can contribute to drawing rice ideotypes flexible towards diverse water availability conditions. Traits identified in accessions of the wild ancestor of cultivated rice, as well as other wild rice species, in Oryza glaberrima and weedy rice were demonstrated to confer enhanced tolerance to water stress, while screenings of cultivated rice germplasms identified several genes/loci improving water stress resistance. New frontiers are represented by the dissection of the epigenetic control of stress tolerance and the implementation of the contribution of favorable microbiota. Innovative breeding technologies, whose feasibility is related to advancements in genomic analyses, are contributing to enhancing the knowledge-based development of water stress-tolerant rice varieties.
Keywords:
Oryza; water deficit; climate change; stress; genes; molecular changes; germplasm; microbiota 1. Introduction
Food production is continuously affected by climate change. The global average temperature is rising, and extreme climatic events, e.g., heavy rainfalls and severe drought periods often combined with heat waves, that determine the evaporation of soil moisture and salt accumulation, are becoming more recurrent worldwide [1,2,3]. Since most of the genetic advances during the green revolution were directed more toward yield potential than tolerance to abiotic stress factors, many modern crop varieties, for instance, cereals, are sensitive to extreme environmental conditions [3]. This situation addresses the breeders’ search for traits that increase resilience to abiotic stress, preserving yield while reducing input dependence. Among abiotic stresses, physical, economic, or social water scarcity due to the in-progress global warming is one of the main concerns for agriculture.
Rice (Oryza sativa L.) already provides essential food for more than half of the world’s population [4,5], but the expanding world’s population will require an additional increasein rice production [6]. Thus, the availability of rice cultivars able to overcome biotic and abiotic constraints limiting yield will be an increasingly essential requirement. Different rice production ecosystems can be distinguished by considering the field water management applied in different rice growing areas worldwide (Figure 1). Each is differently exposed to water shortage risks in terms of severity and timing along the crop growth cycle.
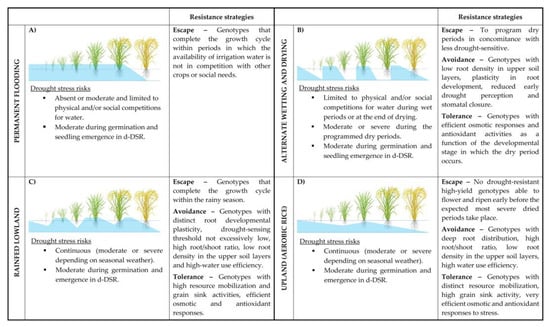
Figure 1.
Water management of rice fields, associated drought-stress risks, and appropriate resistance strategies. Details about the different water management strategies of the paddy field (A–D) and the resistance strategies are reported in the text. Light blue areas indicate the water-saturated (Ψw = 0) soil zones. The areas over the horizontal lines indicate the presence of a 5–20 cm water head above the soil surface. Correspondences between soil water level and plant developmental stages are only indicative. d-DSR: dry Direct Seeding Rice.
In the lowland ecosystem, which covers about 90% of the world’s annual rice production [7,8], high-yield rice cultivars are grown in bunded fields (paddy fields), where the soil is maintained flooded by irrigation (irrigated lowland rice; Figure 1A) or frequent and abundant rainfalls (rain-fed lowland rice; Figure 1C). In the former case, only accentuated climate change or physical and social causes (other seasonal crops and/or civic requirements for water) might generate unpredictable episodes of water scarcity for the culture [9]; thus, currently, drought resistance cannot be considered a trait of primary importance for the irrigated lowland rice cultivars. In rain-fed lowland rice conditions, instead, periods of water shortage, although not entirely predictable in terms of severity and timing could occur, and climate change will increase the frequency and the intensity of adverse conditions, making the availability of better drought-tolerant cultivars a pressing requirement for this rice ecosystem [10,11]. In the upland rice ecosystem, rice is grown in rainfed, naturally well-drained, and not “bunded” fields without surface water accumulation and phreatic water supply. Upland rice is usually cultivated on sloping land with erosion problems, and the soils have poor physical and chemical properties. In these ecosystems, rice varieties are characterized by drought tolerance traits through all growth periods and low yield potential, which makes rice a subsistence crop [12]. In addition to access to supplementary irrigation, wider adoption of the so-called aerobic rice system (Figure 1D) requires specific rice cultivars that combine the drought-tolerance of upland varieties with the high-yielding capacity of lowland varieties. Aerobic rice can be seen as favorable or high-yielding upland rice.
Although the average value of water use efficiency in rice (i.e., the ratio between yield and volume of water transpired by plants) is comparable (about 1.1 g grain yield kg−1 water) with the other major C3 cereal crops [13], in the irrigated lowland systems, rice cultivation requires more than 2.5 kg of water to produce 1 g of rice grain. From this, 40% of the global irrigation freshwater is employed in paddy fields [9,13,14], making this ecosystem scarcely sustainable. Moreover, due to the anaerobic fermentation of soil organic matter during flooding, lowland rice systems have a significant greenhouse footprint, producing approximately 15–20% of the global total anthropogenic emission in the atmosphere of CH4, a potent greenhouse gas [15,16]. To improve the sustainability of lowland rice, the adoption of water-saving practices, such as wet- or dry-Direct Seeding Rice (d-DSR; [17]) rather than either flooded seed sowing or seedling transplantation [18] and Alternate Wetting and Drying (AWD, Figure 1B) [9,19,20] in substitution of permanent flooding (PF), are key strategies.
In d-DSR crop management, rice germplasm is required, which is more tolerant of dry soil conditions in the germination stage. In the case of AWD, consisting of repeated shifts from flooding (15–20 cm of ponded water) to a dry soil condition (soil Ψw about −20 kPa at 20 cm below ground) obtained by draining the field [21], only cultivars not susceptible to events of water scarcity at vegetative stages can limit the yield gaps observed in comparison to PF and make sense for the valuable savings in water, which has been reported to be up to 35% [20,22,23,24].
Since rice is susceptible to drought at the reproductive stage with significant yield penalties, many efforts aimed at obtaining drought-tolerant genotypes at flowering as promising germplasm for upland and lowland rainfed cultivation systems. Similar efforts should be devoted to defining ideotypes and obtaining genotypes able to overcome, without significant penalty, water shortage even at the germination, seedling establishment, and vegetative growth stages in both low- and up-land rainfed rice ecosystems. Nevertheless, the molecular and physiological processes involved in conferring drought tolerance in each indicated stage could be different, and this makes it difficult to outline single ideotypes showing traits suitable for overcoming drought at different moments throughout all rice life cycles in different cultivation ecosystems. Because of the complexity of the involved traits, breeding programs aimed at constituting high-yield and elite rice cultivars with drought resistance still need to be improved. The scarcity of effective donors is one of the main causes and is further complicated if drought tolerance is evaluated not only in terms of quantitative grain yield but also in terms of grain qualitative properties.
This review aims to collect and discuss the current knowledge about genomics and biochemical and microbiological elements exploitable in drawing rice ideotypes overcoming gaps in yield related to different field water management and the significant risks of associated drought events. The review highlights that the growth stage and the adopted water management deeply affect traits involved in rice tolerance to water-limiting conditions, thus complicating the design of single ideotypes showing resistance traits suitable for overcoming water stress during all growth stages and cultivation ecosystems. Notwithstanding, traits, genes, and physiological processes relevant to the water stress response across very diverse water management, agronomical procedures and growth stages were considered and are here described.
2. Resistance Strategies under Diverse Field Water Management
Considering drought tolerance as the ability to produce at maximum yield under water-limited conditions, or at least to reduce losses to a minimum [25], several molecular, physiological, and morphological traits can be identified as relevant for conferring adaptation to rice plants grown under limited water availability. Different strategies conferring drought tolerance can take over depending on the severity, duration, and rate of the drought periods experienced at specific growth stages. They include [26,27]: (i) escape mechanisms by which plants have life cycle duration able to avoid the coincidence of predictable drought periods with the most susceptible growth stages; (ii) avoidance mechanisms, which ensure relatively high tissue water potential (Ψw) when water availability is low; and (iii) tolerance mechanisms to overcome the harmful metabolic conditions established when the tissue Ψw value is low. All these resistance strategies impose metabolic costs that negatively affect crop yield. Thus, it is crucial to define what might be the best solution in relation to the rice production ecosystems case by case.
Among the above-described resistance mechanisms, drought avoidance via efficient water uptake is widely considered the most compatible with high crop yields [28].
In the following sections, rice tolerance to water-limiting conditions will be dissected in terms of genetic factors affecting the deployment of tolerance mechanisms in different growth phases and water management procedures. Gene functions involved in rice responses to water shortage at different growth stages and in different tissues are summarized in Table 1.

Table 1.
Genes affecting traits associated with tolerance for water-limiting and drought stress conditions. The effects are related to the gene expression modifications indicated in the “Gene name” column. KEGG: Kyoto Encyclopedia of Genes and Genomes. GO: Gene Ontology.
3. Rice Adaptation to Dry Direct Seeding
To save water and reduce rice production costs, flooded seed sowing or seedling transplantation technologies are now being replaced by d-DSR [48]. It follows that the availability of rice drought-tolerant germplasm at the germination stage, until now confined to varieties adapted to the aerobic rice system, is becoming fundamental for sustainable rice production under d-DSR [49,50]. Indeed, rice seed germination is hampered by inadequate soil water availability. Despite the strong matrix effect exercised by their tegument and cell wall components, if the value of soil water potential is not sufficient to adequately support the seeds’ imbibition, the reprogramming of molecular and metabolic processes leading to germination of the seeds and seedling emergence [51,52] does not occur or is delayed and not-uniform, while rapid and uniform seed germination is essential for dry/direct seeded rice [18,53].
Seed germination involves various signal transduction pathways where the phytohormones ABA and GA play central antagonistic roles, also interacting with environmental factors [54,55,56]. The better germination ability under drought stress observed in upland compared to lowland rice is related to higher GA3 content and GA3/ABA ratio, promoting the α-amylase activity, thus providing more soluble sugars and energy for seed germination, as well as the action of expansin proteins resulting in cell-wall loosening and the related cell expansion and embryo development [57]. In an RNAseq study, Li and co-workers [58] comparing transcriptomes of germinating seeds of upland and lowland rice under osmotic stress induced by PEG treatments showed that 14 genes related to GA (among which OsGA3ox2 producing bioactive GA3 and four putative gibberellin receptor genes), α-amylase (OsAmy3D and RAmy1A, two putative α-amylase precursor genes, and one putative α-amylase isozyme C) and expansin (among them OsEXPA8, OsEXPA3, and OsEXPA1) were more expressed in upland-adapted than lowland-adapted rice. Concerning the endogenous ABA levels, which negatively control seed germination, a central role is played by the relative activities of the biosynthetic enzyme 9-cis-epoxycarotenoid dioxygenase (NCED) and the catabolic enzyme ABA 8′-hydroxylase (ABA8′ox; [59]). Drought resulted in the accumulation of more ABA in imbibing seeds via enhancing the phytohormone biosynthesis by inducing a dramatic increase in the level of the transcript of the OsNCED3 gene, a member of the NCED gene family in rice; rice lines under-expressing the OsNCED3 gene and over-expressing the ABA8′ox3 gene show a relatively higher tolerance to drought stress than control plants during seed germination [60].
When seeds germinate in conditions of water shortage, the α-amylase activity should be enhanced to provide additional energy for survival [61,62]. Consequently, it can be supposed that rice seeds overexpressing the ABA-responsive AP2-like gene OsARAG1, belonging to the gene family codifying the dehydration-responsive element binding (DREB) proteins and showing increased α-amylase activity, could better survive in stressful conditions [55] such as those experienced in d-DSR.
The osmotic stress/ABA–activated protein kinase 2 (SAPK2) is the primary mediator of ABA signaling along different rice developmental stages with pivotal roles in drought tolerance [61]. Interestingly, the seeds of loss-of-function mutants sapk2, produced with the CRISPR/Cas9 system, showed better germination under hyperosmotic conditions [63]. Thus, the sapk2 mutant less sensitive to ABA would seem potentially useful in d-DSR. However, it must be taken into account that, similarly to OsNCED3 and OsARAG1, a lower activity of the SAPK2 protein favors germination in lower water availability, but with a consequence on drought tolerance traits of the plants along the successive developmental stages. This might not be a problem when adopting d-DSR in the earliest phase of the permanent flooding rice ecosystem.
4. Rice Adaptation under Alternate Wetting and Drying (AWD)
The main aspects related to this water management, in terms of stress risks and resistance strategies, are reported in Figure 1B. Under AWD, the rice plants benefit from a very plastic root system architecture capable of adapting root growth to the variation in soil moisture, assuring the adsorption of immobile nutrients (phosphorus, iron, manganese, and zinc), nutrients transported by diffusion such as nitrate and water ([62], and reference therein).
Best-performing varieties in AWD should keep the S-type lateral roots in a functional state in the dry topsoil, elongate L-type lateral roots in deeper layers for the dry period, initiate the de novo S-type root production on established axile roots in the topsoil upon rewatering, and develop long root hairs to properly support nutrient and water uptake [64]. In addition, the number of nodal roots and root dry weight at 10–20 cm depth 22–30 days after transplanting are key traits to consider in maintaining yield in AWD [65].
A Genome-Wide Association Analysis (GWAS) carried out on 281 accessions of japonica rice characterized the accessions for traits related to phenology, plant, and seed morphology, physiology, and yield for two years in field conditions under PF and limited water (LW). The analysis identified 160 significant marker-trait associations (MTAs): 32 LW-specific, 59 PF-specific, and 69 in common between the two water management systems [66]. Three candidate genes (OsOFP2 on chromosome 1 modulating NOX and BELL transcription factors, which influence plant size, Dlf1 also on chromosome 1 coding for the WRKY11 transcription factor, and OsMADS56 on chromosome 10 coding for a repressor of a putative B3 transcription factor involved in late flowering) were identified to co-localize with LW-specific associations [66]. The authors proposed that some of the identified robust associations could represent suitable targets for genomic selection approaches to improve yield-related traits under LW. Analysis of straw and grain biomass in an aus panel represented by 266 rice accessions grown in PF and AWD [67] was used for GWAS. The study identified 155 QTLs; some genomic regions on chromosomes 1, 10, and 11 can be considered targets for breeding because they are associated with significant increases in grain mass and straw biomass. Only one QTL was shared between the japonica and the aus panels, which concerns days to maturity [67], a trait that could be related to avoidance mechanisms.
Rice cultivation under AWD also implies its adaptation to aerobic growing conditions. Recently, an RNA-seq analysis was performed in root and shoot in aerobic and anaerobic conditions for two cultivars adapted to aerobic and traditional anaerobic conditions, respectively [68]. The investigation highlighted a role for several MADS-box transcription factors, transporters involved in sugar and nutrient uptake, hormones such as ethylene and abscisic acid, and root-related traits. The identified genes coding for TF specific to the aerobic adaptation were genetically localized within water deficit-related QTLs such as qrfw4a, qrfw1b, qbrt1d, and qRDW5-1, and they could be considered as resources to improve rice under aerobic conditions [68].
5. Rice Adaptation to Extreme Water Limitation
Severe water stress typically occurs in upland rainfed and lowland rainfed rice ecosystems (Figure 1D,C). Drought can be defined as a long period with insufficient rainfall, which diminishes soil water via transpiration or evaporation, making it inadequate for crop demands [69]. Plant response to drought depends on plant genotype, developmental stage, growth conditions, and stress severity and duration [70]. In the presence of extremely reduced water availability, many morphological, physiological, biochemical, and molecular changes happen, such as cell turgor loss, a reduction in leaf water potential, leaf curling, stomatal closure, and, therefore, a reduction in CO2, leading to a decrease in photosynthetic activity and consequently to a growth reduction until plant death [3,71]. Drought also determines changes in cell wall polymer structure and composition (e.g., ß-1,4-glucan chains that compose cellulose microfibrils, expansins, pectins, xyloglucan that is the most abundant hemicellulosic polysaccharide) [1].
Drought tolerance is the capacity to maintain growth, flowering, and yields even in sub-optimally hydrated soils [19]. A typical plant response to cope with stress is to close stomata, so the cell turgor and metabolism are kept, even if this affects the photosynthetic rate. From the literature, there is evidence that some accessions of African rice can retain more transpirable water, early closing stomata at the beginning of drought, and mature before, avoiding the severity of the stress at the start of the drought period [72,73]. Fifteen key rice genes and 127 or 233 QTLs have been identified as associated with drought stress tolerance, which were listed by [14,74]. The discovery of the genes was possible thanks to the improvement in omics techniques and the availability of landraces, wild rice accessions, and improved lines that were repositories of resilience genes [74].
6. Rice Adaptation to Water Limitation at the Seedling Stage
Rice plants can encounter this situation mainly under dry seeding, a procedure that can be adopted under diverse water management systems (Figure 1A, B, D). An unweighted network analysis to identify key hub genes or master regulators for multiple abiotic stress tolerance in rice plantlets uncovered that abiotic stress management at the seedling stage involved the conservation of the basic cellular machinery based on genes associated with the biogenesis of ribosome and ribonucleoproteins, DNA repair, replication, and conformational changes, splicing and biogenesis of mRNA and ncRNA and gene silencing, together with helicase activity, zinc finger, dehydrins and F-box LRR type transcriptional factors, oxidase, and heat shock proteins [75]. The validation performed on a reduced set of genes allowed the identification of a gene core associated with three different stresses (drought, salt, and heat) but presented in different genetic backgrounds (indica, japonica, aus, and Basmati germplasm): these genes can be considered candidates in molecular breeding to obtain rice varieties tolerant to multiple stresses at the seedling stage [75].
In another investigation addressed to dissecting tolerance during the first growing stages, rice seedlings of two weeks were grown at 28 °C without watering for either two days until the soil moisture content was 18.7%, or three days until the soil moisture content was 15.6%. An extensive transcriptional analysis using a microarray of rice aerial parts identified about 6000 and 5000 genes that were downregulated and upregulated, respectively [76]. Genes hypothesized to play major roles were coding for ABA-depending TF, such as OsbZIP42 and OsSNAC1, which are involved in the tolerance to water deficit [77,78] together with OsMYB2 [79], OsNAP [80], and proteins such as OsPYL/RCAR5 and OsMAPK5, and other TF belonging to WRKY ([74], and reference therein). In particular, Lim et al. [31] demonstrated that OsWRKY5 is a negative regulator of ABA-induced drought stress tolerance, acting as a transcriptional repressor directly binding to the OsMYB2 promoter region, thus determining downregulation of ABA regulatory and stress genes and suggesting that OsWRKY5 inactivation or modification of its downstream targets could be suitable to improve drought tolerance in rice cultivars. Gu et al. [33] demonstrated that basic helix–loop–helix (bHLH) proteins are a diverse group of TF that regulate different metabolisms and stress responses in rice; among these, OsbHLH130 could activate the expression of OsWIH2, which encodes for an enzyme involved in the fatty acid synthesis. In the presence of OsWIH2 overexpression, rice plants showed more chlorophyll content, an increase in peroxidase activity, and less ROS accumulation, together with an augment of C15, C17, and C26 alkanes under drought conditions (10 days without water) [33].
The role of ribosomal protein (RP) genes in the responses to different water stress treatments of one-month-old rice plantlets was also verified by qRT-PCR gene expression analysis [29]. In this study, it was demonstrated that the RP gene family, including both small (RPS) and large (RPL) subunits, is involved in the response under limited water conditions; in particular, some genes are upregulated both in root and shoots under water-limited conditions and drought, suggesting a common role in inducing tolerance to these two abiotic stresses. The regulation of these genes could be a cellular necessity to keep the integrity and stability of the ribosomal complex, thus allowing the translation of other proteins and conferring an early defense to the plant against abiotic stress. In particular, RPL23A allows the proper functioning of protein secretion, resulting in a key gene in water use efficiency (WUE), increasing growth, tillering, and seed yield, and high chlorophyll fluorescence [29].
7. Plant and Cell Wall Architecture Affecting Tolerance under Limited Water
Genes controlling root architecture impact rice yield under water-liming conditions [81]. A drought-tolerant plant has a good WUE since it has deep and long roots, root length density, and small fine root diameters that reach wider spaces, maintaining productivity and regulating water levels [82]. Kinandang Patong is an example of deeper rooting due to the presence of a DEEPER ROOTING 1 (DRO1) Quantitative Trait Loci (QTL) on chromosome 9, which controls the gravitropic response of root growth angle, increasing it and allowing yield performance equal to unstressed plants under moderate drought conditions. At the same time, the gene did not affect yield under non-stressed conditions [83]. A wide variation in deep rooting was observed, suggesting that deep-rooting QTLs other than DRO1 may exist in rice, thus opening the way for new breeding strategies using genes influencing root system architecture to develop rice cultivars with high adaptability to drought [83].
In addition to root architecture, the above-ground rice plant shape also deeply affects responses to water shortage. The rice OsNAC016 transcription factor was demonstrated to regulate brassinosteroid (BR)-mediated plant architecture positively and modulate ABA-mediated drought tolerance negatively [34]. The loss-function mutant (osnac016) was characterized by erect leaves and shortened internodes, less root growth, reduced cell length in the primary root, and more drought tolerance, with decreased water loss rate and enhanced stomatal closure when exogenous ABA is applied; conversely, lines overexpressing the gene had opposite phenotypes (enlarged leaf inclination), with reduced drought tolerance and ABA sensitivity [34].
Genes encoding enzymes associated with cell wall remodeling were highlighted as involved in rice plants’ response to water-limiting conditions. The glycoside hydrolases (GH) were demonstrated to play an important role under drought stress in Nipponbare: at least two GH genes are activated under drought (LOC_Os01g71670.1; LOC_Os04g56930.1) in rice roots, while another gene (LOC_Os04g51460.1) is repressed by drought and salt stress, suggesting that cell wall remodeling represents a common mechanism in abiotic stress response [84]. In addition, genes coding for cell wall- and plasma membrane-localized REPETITIVE PROLINE-RICH PROTEIN (RePRPs) are involved in root development inhibition under stress or ABA: knockdown of all RePRPs expression in rice reduces the inhibitory effect of ABA on root growth [85]. Recently, a gene (PSL1) encoding for a pectin-degrading enzyme (i.e., a cell wall-localized polygalacturonase) has been characterized. A 260 bp deletion in this gene caused leaf rolling in response to high light intensity and/or low humidity, a different cell wall composition, and, interestingly, higher WUE under drought stress conditions and enhanced drought tolerance [45]. Based on this result, it could be useful to reduce the PSL1 gene expression to enhance plant adaptability to a high light and drought environment. Recently, it was demonstrated that germin-like proteins (GLP), able to conduct oxalate oxidase and super oxide dismutase activities, play a key role in cell wall reinforcement by the cross-linking of cell wall components under stress [86]. A gene expression profile carried out on two indica rice varieties allowed the GLP regulated under drought conditions to be identified [32]. OsGLP8-7 was over-expressed under drought stress in leaves and roots more in KS282 than in Super Basmati, while OsGLP8-4 and OsGLP8-12 in roots of Super Basmati than KS282; OsGLP3-6 was the most expressed in both of the varieties (with a significantly higher expression in KS282 than Super Basmati) and both under drought and salt stress [29]. This study suggested the dual (general and specific) role of OsGLP genes in regulatory mechanisms and the effect of rice variety, stress condition, and tissue type on the role itself [32]. Even lignin biosynthesis mediated by CINNAMOYL-CoA REDUCTASE 10 (OsCCR10) contributes to drought tolerance since the accumulation in root sclerenchyma and fibers reduces water loss [46]. Lignin is composed of p-hydroxyphenyl, guiacyl, and sinapyl units derived from monolignols that form a secondary cell wall, which is involved in controlling water penetration and thus transpiration, and maintaining the cell osmotic balance and in providing further mechanical strength and water impermeability in specific tissues [87]. OsCCR10 catalyzes the conversion of p-coumaroyl-CoA and feruloyl-CoA to coumaraldehyde and coniferaldehyde, respectively; when it is over-expressed, it increases lignin content in the root, drought tolerance at the vegetative stages, the photosynthetic efficiency, the grain yield, and reduces water loss [46].
8. Contribution of Germplasm Biodiversity to Improve Rice Responses to Water Limiting Conditions
The wild ancestor of cultivated rice, O. rufipogon, is considered the donor of drought-tolerant traits to introgress in the domesticated cultivars. Mapping studies were carried out on introgression lines obtained with a back-crossing program using Guichao 2 (O. sativa ssp. indica) as the recipient and Dongxiang accession (O. rufipogon) as the donor. The work identified 12 QTLs related to drought tolerance, and an identified drought-tolerant line (IL23) showed the presence of two QTLs (qSDT2-1 and qSDT12-2) located on chromosomes 2 and 12 [88]. Physiological characterization demonstrated that IL23 had a superior ability to conserve water content, a higher leaf temperature, more soluble sugar content, lower osmotic potential, and lower electrolytic leakage level and MDA content, suggesting that the line can better regulate stomata and osmotic potential, producing compatible osmolyte and soluble sugars [88].
Similarly, traits introgressed from African species such as O. glaberrima into O. sativa increased tolerance to limited water in the latter [89]. Furthermore, other Oryza species like O. meridionalis, O. longistaminata, and O. barthii, which are localized in regions where extreme temperature and moisture conditions are present, could be considered good candidates as donors for water stress tolerance [3]. Some Oryza species (O. australiensis, O. glaberrima, O. longistaminata, and O. meridionalis) are characterized by thick leaves and high mesophyll conductance to CO2 diffusion, which are traits associated with good WUE [90]. Better membrane stability and greater stomatal conductance, compared to O. sativa, were observed in O. rufipogon and O. longistaminata accessions under water deficit [91]. In addition, weedy rice such as Oryza sativa f. spontanea, which is typical of paddy fields, shows tolerance to drought: an analysis based on transcriptomics and proteomics of 133 weedy rice accessions suggested that some proteins (Os09T0478300-01, Os09T0530300-01, and Os01T0800500-01) provide defenses against water deficit [92]. These proteins are seen as a network core and act as a bridge in the crop defense system. In particular, OS09T0478300-01 and related proteins are associated with the Golgi-localized beta-glycan synthase, which polymerizes the hemicellulose backbones, and is involved in plant response to water retention system; OS09T0530300-01 and related proteins are putative cytochrome P450s, involved in crop resistance to oxidation, and members of the scavenging system; OS01T0800500-01 is a purple acid phosphatase (PAP) protein, essential in responding to drought stress conditions [92]. Regarding this last result, a recent combination of GWAS, transcriptome, and proteome analyses of 501 weedy rice accessions allowed the identification of a gene (Os01g0800500) coding for purple acid phosphatase (PAPH1) as a candidate involved in the drought tolerance [93]. Knockout and overexpression of PAPH1 lowered and increased drought stress tolerance compared to the wild types, suggesting a capacity to regulate cell homeostasis, affecting Ca2+ and K+ flux. The authors hypothesized that during the evolution of weedy rice, which originated from cultivated rice by de-domestication [94], chromosome regions associated with drought resistance had been selected thus providing potential genetic resources that can be exploited for the improvement of cultivated rice varieties [93].
Thermal imaging was recently applied to screen rice germplasm for tolerance to water stress [95]; the approach was applied at the seedling stage on 28 rice genotypes (26 advanced lines and one tolerant IR-55419-04, one susceptible Super Basmati variety) in order to integrate morphological, physiological, and biochemical approaches with improved in situ methods, i.e., infrared thermal imaging. The best indica genotype, identified as NIBGE-DT-02, showed significant production of osmoregulators such as proline, antioxidants, high relative water content, better yield, and tolerance to water stress irrespective of the evaluated growth stage, making it a useful genetic material for breeding programs aimed at increasing rice cultivation under water stress condition.
The genetic variability for drought tolerance was also evaluated in breeding lines obtained from an exotic drought-tolerant landrace (Chao Khaw) and a high-yielding aromatic rice cultivar (Kasturi); the research discovered promising breeding lines for different rice environments and drought breeding programs [96]. Twenty-one obtained lines produced more than the parents under drought and well-watered conditions and in a stable way across the years, suggesting a possible role as drought-tolerance donors in breeding programs.
Tolerance to water-limiting conditions is also present in renowned currently cultivated rice varieties; as an example, DT3, called Tainung83 in 2020, is an elite japonica rice line with fine grain quality and drought tolerance, selected from a crossing combination between Taikeng9 and the drought-tolerant cultivar Hang-yu15 [97]. Similarly, the Indonesian pigmented rice cultivars Merah Pari Eja showed enhanced drought tolerance on the basis of a screening performed under drought treatment evaluating the percentage of leaf rolling and leaf damage [98]. Analyzing the gene expression by RT-qPCR, genes like DEHYDRATION RESPONSE ELEMENT B1A (OsDREB1A), NAM, ATAF and CUC 6 (OsNAC6), SODIUM/HYDROGEN EXCHANGER1 (OsNHX1), COPPER/ZINC SUPEROXIDE DISMUTASE2 (OsCuZnSOD2), and CATALASE (OsOSCAT2 and OsCAT3) were upregulated in this variety, suggesting an increase in transcriptional drought-responsive factors associated with scavenging enzymes. In contrast, they were downregulated in the sensitive ones (Inpari 24 and Putih Payo) [98]. In parallel biochemical and physiological analyses based on antioxidant enzyme activity, free radicals, chlorophyll, and anthocyanin contents further demonstrated the drought tolerance of Merah Pari Eja—able, in fact, to produce antioxidants to remove cell-damaging free radicals [98].
Ahmad et al. [99] performed experiments under irrigated and drought conditions (imposed for 30 days at the pre-anthesis stage) on 22 Green Super Rice (GSR) indica lines. The NGSR-15 line demonstrated higher drought tolerance compared to the other lines, showing higher pollen viability, plant biomass, cell membrane stability, and harvest index under drought. From a molecular point of view, the molecular basis was related to drought-responsive known genes, including OsSADRI (Salt-, ABA- and Drought-Induced RING Finger Protein 1), OsDSM1 (Drought-Hypersensitive Mutant1), and OsDT11 (Drought tolerance 11) that are upregulated in this genotype and new responsive genes such as LOC_Os11g36190 (a receptor kinase), LOC_Os12g04500 (a response regulator receiver domain-containing protein), LOC_Os12g26290 (an alpha-DOX2), and LOC_Os02g11960 (an ABC transporter, ATP-binding protein) that were upregulated in this genotype [99].
Subtle dissection of genes differentially expressed in the roots can support the identification of gene markers for tolerant rice accessions; a transcriptome analysis performed on different root zones of drought-sensitive (IR64) and drought-tolerant (Azucena) varieties highlighted that the tolerant variety showed higher expression of genes involved in cell cycle and division and root growth and development and more prominent lateral root system expansion. It was shown that Azucena prevents water stress through increasing growth and root exploration to intercept water, while IR64 tries to cope with the stress by activating antioxidant systems (based, for example, on heat shock proteins) [81]. Known genes belonging to NAC, AP2/ERF, AUX/IAA, EXPANSIN, WRKY, MYB, and other new candidates were identified as related to the root system architecture under drought conditions, potentially usable after further investigations to enhance drought adaptation in rice [81].
Several examples of obtaining drought-tolerant rice lines with enhanced yield by using molecular marker-assisted selection (MAS) are present in the literature. However, the linkage drag associated with the introgression suggests the use of multiple advanced genomics-assisted breeding approaches, e.g., background selection by genome-wide selected molecular markers ([100], and reference therein). A drought-tolerant line (RBL-112) was obtained after five back-crosses on the F1 between the Egyptian variety Giza-178 (recurrent parent) and a Philippines drought-tolerant variety IR60080-46A (donor parent). This line was evaluated under drought and normal irrigation together with the parents and IR64, and its tolerance was ascribed to an improved root system, higher relative water and proline content, lower malondialdehyde content, a more efficient photosystem II, and higher superoxide dismutase activity promoting scavenging of stress-related ROS. In addition, the improved line showed a better capacity to regulate physiological activities and drought-induced gene expression levels to cope the stress while keeping yield potential [101].
Another example of the exploitation of germplasm resources involved the high-yielding japonica line IR91648, derived from the cross Moroberekan x Swarna. This line was used to develop multiple stress-tolerant and climate-resilient rice lines by crossing it with eight different donors of QTL/genes for drought, submergence tolerance, gall midge, blast, brown plant hopper, and bacterial leaf blight resistance for a total of 15 genetic loci [102]. The cross strategy was based on individually crossing the line with four parents, and then the four F1 were crossed with the remaining four donors. In the subsequent seasons, the F1 products were intercrossed to develop a complex F1 population where, by MAS, the presence of QTLs and genes was confirmed. The lines were then brought to homozygosity by selfing for six generations. Different pyramided homozygous lines showed the needed grain quality profiling together with tolerance/resistance to abiotic/biotic stresses. They might be used as parental lines for other breeding programs or varieties to be released after multi-location evaluation in national and provincial coordinated trials [102].
A MABC approach was used to obtain drought and submergence-tolerant indica rice lines in the background of the elite variety UKM5, which harbor the drought yield QTLs (qDTY3.1 and qDTY12.1), using IR64-Sub1 as the donor of the submergence tolerance locus Sub1. The best-performing combination was that with Sub1 and qDTY3.1, which was characterized by higher grain yield under drought in the reproductive stage, demonstrating a positive effect of the Sub1 locus on drought tolerance when it was combined with the drought yield QTLs [103]. A similar approach was adopted to increase water-stress tolerance in Improved White Ponni (IWB), a rice variety popular in South India, which is characterized by high yield, fine grain, and high cooking quality, but it is susceptible to drought, salinity, and submergence [104]. A marker-assisted backcross was adopted to pyramid three QTLs related to drought tolerance (qDTY1.1, qDTY2.1), salinity (Saltol), and submergence (Sub1). Superior lines were obtained with the agronomic and grain quality characters of IWB and greater performance against dehydration, salinity, and submergence stress compared with the recurrent parent [104]. Also, in this work, the new lines harboring pyramided QTLs for tolerance to drought and flood showed effective responses to both stresses. This response agrees with the hypothesis that molecular mechanisms associated with these diverse stressful conditions may be regulated by cross-talking signaling pathways [105].
9. Drought Memory and Epigenetic Control
If plants experience recurrent and repetitive mild drought-stress periods, their responses in the subsequent events can differ from those put in place during the first, and they progressively acquire an improved tolerance to water shortage [106,107,108]. In rice, these differences making up the so-called drought memory may concern the levels of specific transcripts (transcriptional memory), the contents of phytohormones, such as ABA and jasmonic acid, and those of proline [58].
Li and co-workers (2019) [58], exposing rice plants to successive mild drought and re-watering treatments, identified, by a whole-transcriptome strand-specific RNA sequencing approach, 6885 transcripts and 238 long non-coding RNAs involved in the drought memory phenomena and about 5400 of these memory-related transcripts resulted probably regulated by DNA methylation. Recently, new insights into how DNA methylation is involved in this epigenetic memory concerning several genes and, among them, TEs have been provided [109,110]. Interesting pieces of evidence concern the two homologous genes (LOC_Os05g38150 and LOC_Os01g62900) encoding the proline biosynthetic enzyme Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1). Indeed, memory-related DMRs (Differently Methylated Regions) were identified in the promoter region of the former gene and the gene body of the latter [110], coherently with the memory behaviors of the P5CS1 transcript level and the proline content.
This novel information opens the perspective of carrying out future research to develop epigenetic markers to breed drought-resistant rice particularly adapted for irrigated lowland and AWD rice systems in which plants are exposed to repetitive dry periods.
10. Role of Rice Microbiota
Currently, increasing consideration is given to the microbiome, which is represented by microbes in the rhizosphere (soil surrounding roots), endosphere, phyllosphere, and pollen ([111], and reference therein). The host plant and associated microbes considered together are called holobiont [112] and environmental stresses affect not only plants but also soil microbes; consequently, the effect of abiotic stress on the plant also affects microbes in terms of number, types, and functions [113,114]. There is, therefore, a strong, dynamic, and complex interconnection among plants, soil, microbes, and abiotic stress due to several factors related to stress duration, host plant species, the degree of stress levels, and other environmental factors [114]. It was recently highlighted that water availability influences rice microbial community composition and structure by microbiome analysis performed on soil and root DNA in lowland and upland conditions [115]. In rice roots under upland management, arbuscular mycorrhizal fungi (AMF), Streptomyces, and fungal pathogens increase, while Methanobacterial hubs are destroyed; in soil, the impact of flooding/drying is, on the contrary, more limited. Another recent analysis of roots and soil microbiota under two water management systems (PF and AWD) was carried out by high-throughput sequencing of the 16S rRNA in paddy fields with two diverse japonica rice cultivars [116]. While root communities were strongly affected by the water management (with the enrichment of aerobic and potentially plant-growth-promoting bacteria under the AWD treatment), the soil communities were more adapted and more tolerant to changes in the water treatment. In both water management, the rice variety did not affect the soil or root microbial composition.
In the presence of stress, plants can adopt the strategy of ‘cry-for-help’, calling beneficial microbial endophytes to alleviate the stress-induced damage [113]. In rice, the positive effects of endophytes are related to the release of phytohormones and siderophores, nitrogen fixation, and metal detoxification [117]. For example, the production of indole acetic acid, auxins and gibberellins, and other organic acids enhances rice growth under salt stress by reducing endogenous ABA levels and increasing glutathione and sugar content [118]. Conversely, under drought conditions, other endophytes can contribute to increasing ABA, reducing stomatal conductance and density, reducing leaf water potential, and enhancing WUE [119]. Different studies (reviewed by [110]) highlighted that under abiotic stress conditions, fungal and bacterial endophytes act as modulators of antioxidant enzyme (superoxide dismutase, peroxidase, and catalase) activities and metabolic pathways, mitigating oxidative stress and enhancing crop performance.
Endophytes and other plant growth-promoting (PGP) bacteria promote plant growth under water stresses through several other mechanisms that include the production of phytohormones that improve morphological root traits useful to overcome drought conditions, or the production of 1-aminocyclopropane-1-carboxylic acid deaminase (ACCD) that degrade the ethylene precursor 1-Aminocyclopropane-1-carboxylic acid (ACC), or again the production of exopolysaccharides (EPS) capable of maintaining higher soil moisture content. Indeed EPS produced by PGPR form rhizosheath around the roots and thus protect the plant roots from desiccation for a longer period [110]. Recent studies demonstrated that under drought conditions it is evident that enrichment in Actinobacteria (such as Streptomyces) colonization within the root and rhizosphere is beneficial due both to the high tolerance of the spores to dehydration and their active adaptation to this condition. Consequently, this colonization can increase root development during drought [120]. Fitzpatrick et al. [121], studying the microbiome of 30 angiosperms, also observed a significant correlation between the relative abundance of the genus Streptomyces in plant roots and host tolerance to water stress.
Groen et al. [122] underlined that interactions with AMF could improve rice growth under water-limiting conditions. Different papers demonstrated the absence of AMF in rice roots for four months [123] and two months [124] after flooding and the increase in AMF in dryland systems [124]. It was then demonstrated that the water regime is the driving force that affects AM colonization and the root architecture and anatomy [125] and that roots hosting or not hosting AM fungal colonization differ from the anatomical and transcriptional points of view [126]. Indeed, the proportion of large lateral roots (LLR) was higher under aerobic conditions, while fine lateral roots (FLR) prevailed under anaerobic (flooding) conditions; these two morphologies display opposite responses with respect to AM colonization, with LLR supporting AM colonization while FLR do not. These investigations [125,126] also indicated that AMF can colonize rice roots even in flooded soil and that flooding itself was not responsible for the lack of symbiosis; indeed, continuous flooding was leading to a progressive decrease in the tissues available for colonization, including both diminishing LLR formation and widening the area occupied by the aerenchyma, thus decreasing the degree of AMF colonization progressively, since over time from flooding, deep morphological changes occur in the root apparatus.
In conclusion, since in several plant species, including rice, it has been observed that the microbiota can increase WUE (e.g., reviewed in [107,111,127], it is also expected that an increased aptitude of rice plants to associate with PGPB could allow increased WUE in this crop plant. It was clearly demonstrated that O. rufipogon possesses more allelic variation than domesticated rice, and therefore it represents a donor of genetic variability for cultivated rice [128,129]. Analysis of the root microbiome of various crop plants and their wild relatives highlighted differences in the composition of the root/rhizosphere microbiome as well as genetic loci involved in establishing these differences in tomato and barley [130,131,132]. These differences are particularly relevant because the microbiota associated with crop wild relatives evolved under marginal soil conditions and can therefore represent an untapped resource for low-input agriculture, including adaptation to water-limiting conditions [133,134].
11. Conclusions
In this review, we highlighted how complex the process of improving rice tolerance under water-limiting conditions is. Several levels of difficulties need to be taken into consideration, including the growth stage during which water stress occurs (germination, seedling establishment, vegetative or flowering growth stages) and the adopted water management, thus making it very difficult to outline single ideotypes showing resistance traits suitable for overcoming water stress throughout all rice growth stages and in different cultivation ecosystems. However, we have described traits and genes relevant to the response to water stress under diverse water management, agronomical procedures (e.g., dry seeding), and growth stages. The plethora of genes/traits involved in tolerance to water stress was highlighted, and for several of them, functional validations confirmed major roles in affecting the tolerant phenotype. These genes/traits (also derived from wild germplasm) should be considered primary targets for biotechnological/allele mining approaches addressed to introducing alleles effective in improving tolerance to water stress. In addition to plant traits, it is currently well-defined that also the microbiota can protect rice plants from water stress, and it is emerging that plant responses to the stress can shape the root microbiota. Considering that, in turn, plant traits can affect the root microbiota composition, it could therefore be expected that in the near future breeding for the root microbiota will also contribute to improving rice response to water stress. Information supplied in this review in terms of genes, loci, and physiological processes provides perspectives on new horizons in genomic breeding to breeding companies and research institutions toward delivering rice varieties with enhanced tolerance to water-limiting conditions.
12. Future Perspectives
New agronomic methods and breeding technologies, as well as the development of genetic diversity and the use of genomic resources, are needed to discover and integrate different gene functions useful to cope with water stress [135]. A CerealESTdb, based on rice, maize, wheat, and sorghum Expressed Sequence Tags (EST) whose expression is modulated under various abiotic stresses, including water stress, provides an updated picture of genes involved in the response to this stress and supports in accelerating efforts to develop abiotic stress-resistant cultivars without compromising the nutritional quality through introgression and gene pyramiding [136]. In addition, advances in next-generation sequencing (NGS) technologies and the reduction in genome sequencing costs have allowed the decoding of many crop genomes and the development of molecular markers to select and improve traits [137]. Different rice genotypes (cultivated and wild) and varieties have been sequenced, providing better insights into genetic diversity, structural variations, genes or novel genetic factors, and also candidate regions selected during domestication, allowing genomics-assisted breeding (GAB) to tackle abiotic stress to tailor resilient climate crops ([135], and references therein). Numerous genomic resources (e.g., genome sequence assemblies, germplasm sequencing data, and gene expression atlases), specialized genetic populations (e.g., multi-parent advanced generation intercross (MAGIC) and nested association mapping (NAM) populations), high-throughput and cost-effective genotyping/phenotyping platforms, and quicker trait mapping approaches are allowing the discovery and utilization of molecular markers/diagnostic markers associated with essential traits to improve crops [135,137]. The relevance and perspectives of these innovations for crop improvement have recently been elucidated as the importance of 5 Gs: Genome assembly, Germplasm characterization, Gene(s)/marker(s) associated with breeding traits, Genomic Breeding, and Gene editing [138]. Finally, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) can be considered as an additional tool for enhancing rice tolerance to water stress through its various applications that include gene knock-out, repair (allelic improvement) or replacement (allelic substitution) of important genes, to modify in a punctual way the gene sequence, to reprogram gene expression operating on the promoter: all these applications can be used to develop climate-smart rice varieties [139].
Author Contributions
Conceptualization, E.Z., M.P., F.F.N., G.A.S. and G.V.; Writing–Original Draft Preparation, E.Z., M.P. and G.A.S.; Writing–Review & Editing, F.F.N., G.A.S. and G.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| d-DSR | dry Direct Seeding Rice |
| AWD | Alternate Wetting and Drying |
| Ψw | water potential |
References
- Ezquer, I.; Salameh, I.; Colombo, L.; Kalaitzis, P. Plant Cell Walls Tackling Climate Change: Insights into Plant Cell Wall Remodeling, Its Regulation, and Biotechnological Strategies to Improve Crop Adaptations and Photosynthesis in Response to Global Warming. Plants 2020, 9, 212. [Google Scholar] [CrossRef]
- Francini, A.; Sebastiani, L. Abiotic stress effects on performance of horticultural crops. Horticulturae 2019, 5, 67. [Google Scholar] [CrossRef]
- Toulotte, J.M.; Pantazopoulou, C.K.; Sanclemente, M.A.; Voesenek, L.A.C.J.; Sasidharan, R. Water stress resilient cereal crops: Lessons from wild relatives. J. Integr. Plant Biol. 2022, 64, 412–430. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 17031. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Data FAOSTAT Provides Free Access to Food and Agriculture Data for Over 245 Countries and Territories and Covers all FAO Regional Groupings from 1961 to the Most Recent Year Available. 2021. Available online: http://faostat.fao.org (accessed on 1 December 2021).
- Ezin, V.; Ahanchede, W.W.; Ayenan, M.A.T.; Ahanchede, A. Physiological and agronomical evaluation of elite rice varieties for adaptation to heat stress. BMC Plant Biol. 2022, 22, 236. [Google Scholar] [CrossRef]
- GRiSP (Global Rice Science Partnership). Rice Almanac, 4th ed.; International Rice Research Institute: Los Baños, Philippines, 2013. [Google Scholar]
- Dixit, S.; Singh, A.; Kumar, A. Rice breeding for high grain yield under drought: A strategic solution to a complex problem. Int. J. Agron. 2014, 491, 711–716. [Google Scholar] [CrossRef]
- Bouman, B.A.M.; Humphreys, E.; Tuong, T.P.; Barker, R. Rice and water. Adv. Agr. 2007, 92, 187–237. [Google Scholar]
- Fukai, S.; Mitchell, J. Factors determining water use efficiency in aerobic rice. Crop Environ. 2022, 1, 24–40. [Google Scholar] [CrossRef]
- Pandey, V.; Shukla, A. Acclimation and tolerance strategies of rice under drought stress. Rice Sci. 2015, 22, 147–161. [Google Scholar] [CrossRef]
- Haefele, S.M.; Kato, Y.; Singh, S. Climate ready rice: Augmenting drought tolerance with best management practices. Field Crop Res. 2016, 190, 60–69. [Google Scholar] [CrossRef]
- Ringler, C.; Zhu, T. Water resources and food security. Agron. J. 2015, 107, 1533–1538. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef]
- Linquist, B.A.; van Groenigen, K.J.; Adviento-Borbe, M.A.; Pittelkow, C.; van Kessel, C. An agronomic assessment of greenhouse gas emissions from major cereal crops. Glob. Change Biol. 2012, 18, 194–209. [Google Scholar] [CrossRef]
- Sun, H.; Zhou, S.; Fu, Z.; Chen, G.; Zou, G.; Song, X. A two-year field measurement of methane and nitrous oxide fluxes from rice paddies under contrasting climate conditions. Sci. Rep. 2016, 6, 28255. [Google Scholar] [CrossRef]
- Pathak, H.; Tewari, A.N.; Sankhyan, S.; Dubey, D.S.; Mina, U.; Singh, V.; Jain, V.; Bhatia, A. Direct-seeded rice: Potential, performance and problems—A review. Curr. Adv. Agric. Sci. 2011, 3, 77–88. [Google Scholar]
- Liu, H.Y.; Hussain, S.; Zheng, M.M.; Sun, L.M.; Shad, F.; Huang, J.L.; Cui, K.H.; Nie, L.X. Progress and constraints of dry direct-seeded rice in China. J. Food Agr. Environ 2014, 12, 465–472. [Google Scholar]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Price, A.H.; Norton, G.J.; Salt, D.E.; Ebenhoeh, O.; Meharg, A.A.; Meharg, C.; Islam, R.M.; Sarma, R.N.; Dasgupta, T.; Isamil, A.M. Alternate wetting and drying irrigation for rice in Bangladesh: Is it sustainable and has plant breeding something to offer? Food Energy Secur. 2013, 2, 120–129. [Google Scholar] [CrossRef]
- Siopongco, J.D.L.C.; Wassmann, R.; Sander, B.O. Alternate Wetting and Drying in Philippine Rice Production: Feasibility Study for a Clean Development Mechanism (No. 2215-2019-1632); International Rice Research Institute: Laguna, Philippines, 2013; p. 14, IRRI Tech. Bulletin No. 17. [Google Scholar]
- Lampayan, R.M.; Rejesus, R.M.; Singleton, G.R.; Bouman, B.A.M. Adoption and economics of alternate wetting and drying water management for irrigated lowland rice. Field Crop Res. 2015, 170, 95–108. [Google Scholar] [CrossRef]
- Carrijo, D.R.; Lundy, M.E.; Linquist, B.A. Rice yields and water use under alternate wetting and drying irrigation: A meta-analysis. Field Crop Res. 2017, 203, 173–180. [Google Scholar] [CrossRef]
- Monaco, S.; Volante, A.; Orasen, G.; Cochrane, N.; Oliver, V.; Price, A.H.; Teh, Y.A.; Martínez-Eixarch, M.; Thomas, C.; Courtois, B.; et al. Effects of the application of a moderate alternate wetting and drying technique on the performance of different European varieties in Northern Italy rice system. Field Crop Res. 2021, 270, 108220. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, S.; Ram, T.; Yadaw, R.B.; Mishra, K.K.; Mandal, N.P. Breeding high-yielding drought tolerance rice: Genetic variations and conventional and molecular approaches. J. Exp. Bot. 2014, 65, 6265–6278. [Google Scholar] [CrossRef]
- Levitt, J. Responses of Plants to Environmental Stresses; Academic Press: Cambridge, MA, USA, 1980. [Google Scholar]
- Panda, D.; Mishra, S.S.; Behera, P.K. Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Sci. 2021, 28, 119–132. [Google Scholar] [CrossRef]
- Bodner, G.; Nakhforoosh, A.; Kaul, H.P. Management of crop water under drought: A review. Agron. Sustain. Dev. 2015, 35, 401–442. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Expression Profiling of Ribosomal Protein Gene Family in Dehydration Stress Responses and Characterization of Transgenic Rice Plants Overexpressing RPL23A for Water-Use Efficiency and Tolerance to Drought and Salt Stresses. Front. Chem. 2017, 5, 97. [Google Scholar] [CrossRef]
- Gong, X.; Huang, Y.; Liang, Y.; Yuan, Y.; Liu, Y.; Han, T.; Li, S.; Gao, H.; Lv, B.; Huang, X.; et al. OsHYPK-mediated protein N-terminal acetylation coordinates plant development and abiotic stress responses in rice. Mol. Plant 2022, 15, 740–754. [Google Scholar] [CrossRef]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef] [PubMed]
- Anum, J.; O’Shea, C.; Zeeshan Hyder, M.; Farrukh, S.; Skriver, K.; Malik, S.I.; Yasmin, T. Germin like protein genes exhibit modular expression during salt and drought stress in elite rice cultivars. Mol. Biol. Rep. 2022, 49, 293–302. [Google Scholar] [CrossRef]
- Gu, X.; Gao, S.; Li, J.; Song, P.; Zhang, Q.; Guo, J.; Wang, X.; Han, X.; Wang, X.; Zhu, Y.; et al. The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice. Plant Physiol. Biochem. 2021, 169, 269–279. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, Y.; Xie, Z.; Yu, B.; Sun, Y.; Huang, J. OsNAC016 regulates plant architecture and drought tolerance by interacting with the kinases GSK2 and SAPK8. Plant Physiol. 2022, 189, 1296–1313. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiao, S.; Li, W.; Feng, W.; Li, J.; Wu, Z.; Gao, X.; Liu, F.; Shao, M. Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J. Exp. Bot. 2011, 62, 4229–4238. [Google Scholar] [CrossRef]
- Wei, S.; Hu, W.; Deng, X.; Zhang, Y.; Liu, X.; Zhao, X.; Luo, Q.; Jin, Z.; Li, Y.; Zhou, S.; et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014, 14, 133. [Google Scholar] [CrossRef]
- Jin, Y.; Pan, W.; Zheng, X.; Cheng, X.; Liu, M.; Ma, H.; Ge, X. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol. Biol. 2018, 98, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Basu, S.; Singla-Pareek, S.L.; Pareek, A. Unraveling the contribution of OsSOS2 in conferring salinity and drought tolerance in a high-yielding rice. Physiol. Plant 2022, 174, e13638. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Jena, P.; Mondal, S.; Dash, G.K.; Ray, S.; Baig, M.J.; Swain, P. Relative contribution of different members of OsDREB gene family to osmotic stress tolerance in indica and japonica ecotypes of rice. Plant Biol. 2022, 24, 356–366. [Google Scholar] [CrossRef]
- Zhu, M.; He, Y.; Zhu, M.; Ahmad, A.; Xu, S.; He, Z.; Jiang, S.; Huang, J.; Li, Z.; Liu, S.; et al. ipa1 improves rice drought tolerance at seedling stage mainly through activating abscisic acid pathway. Plant Cell Rep. 2022, 41, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Nagar, P.; Sharma, N.; Jain, M.; Sharma, G.; Prasad, M.; Mustafiz, A. OsPSKR15, a phytosulfokine receptor from rice enhances abscisic acid response and drought stress tolerance. Physiol. Plant 2022, 174, e13569. [Google Scholar] [CrossRef]
- Manavalan, L.P.; Chen, X.; Clarke, J.; Salmeron, J.; Nguyen, H.T. RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 2012, 63, 163–175. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Y.; Zhou, Z.; Zhang, Y.; Yang, Y.; Zan, X.; Li, X.; Wan, J.; Gao, X.; Chen, R.; et al. OsSCL30 overexpression reduces the tolerance of rice seedlings to low temperature, drought and salt. Sci. Rep. 2022, 12, 8385. [Google Scholar] [CrossRef]
- Zhang, G.; Hou, X.; Wang, L.; Xu, J.; Chen, J.; Fu, X.; Shen, N.; Nian, J.; Jiang, Z.; Hu, J.; et al. PHOTO-SENSITIVE LEAF ROLLING 1 encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice. New Phytol. 2021, 229, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.W.; Choi, S.; Jin, X.; Jung, S.E.; Choi, J.W.; Seo, J.S.; Kim, J.K. Transcriptional activation of rice CINNAMOYL-CoA REDUCTASE 10 by OsNAC5, contributes to drought tolerance by modulating lignin accumulation in roots. Plant Biotechnol. J. 2022, 20, 736–747. [Google Scholar] [CrossRef]
- He, H.; Wang, Q.; Wang, L.; Yang, K.; Yang, R.; You, C.; Ke, J.; Wu, L. Photosynthetic physiological response of water-saving and drought-resistant rice to severe drought under wetting-drying alternation irrigation. Physiol. Plant 2021, 173, 2191–2206. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Ladha, J.K. Direct Seeding of Rice: Recent Developments and Future Research Needs. Adv. Agron. 2011, 111, 297–413. [Google Scholar] [CrossRef]
- Sandhu, N.; Torres, R.O.; Sta Cruz, M.T.; Maturan, C.; Jain, R.; Kumar, A.; Henry, A. Traits and QTLs for development of dry direct-seeded rainfed rice. J. Exp. Bot. 2015, 66, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Uzair, M.; Patil, S.B.; Zhang, H.; Kumar, A.; Mkumbwa, H.; Zafar, S.A.; Chun, Y.; Fang, J.; Zhao, J.; Khan, M.R.; et al. Screening Direct Seeding-Related Traits by Using an Improved Mesocotyl Elongation Assay and Association between Seedling and Maturity Traits in Rice. Agronomy 2022, 12, 975. [Google Scholar] [CrossRef]
- Hussain, M.; Farooq, M.; Lee, D.J. Evaluating the role of seed priming in improving drought tolerance of pigmented and non-pigmented rice. J. Agr. Crop Sci. 2017, 203, 269–276. [Google Scholar] [CrossRef]
- Islam, M.M.; Kayesh, E.; Zaman, E.; Urmi, T.A.; Haque, M.M. Evaluation of rice (Oryza sativa L.) genotypes for drought tolerance at germination and early seedling stage. Agriculturists 2018, 16, 44–54. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K. Early seedling vigour, an imperative trait for direct-seeded rice: An overview on physio-morphological parameters and molecular markers. Planta 2015, 241, 1027–1050. [Google Scholar] [CrossRef]
- Angelovici, R.; Galili, G.; Fernie, A.R.; Fait, A. Seed desiccation: A bridge between maturation and germination. Trends Plant Sci. 2010, 15, 211–218. [Google Scholar] [CrossRef]
- Zhao, L.; Hu, Y.; Chong, K.; Wang, T. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann. Bot. 2010, 105, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Li, M.Q.; Han, Y.C.; Sun, H.Z.; Du, Y.X.; Zhao, Q.Z. The crucial role of gibberellic acid on germination of drought-resistant upland rice. Biol. Plantarum. 2019, 63, 529–535. [Google Scholar] [CrossRef]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and Transcriptome Analyses Reveal Short-Term Responses and Formation of Memory Under Drought Stress in Rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Cai, S.; Jiang, G.; Ye, N.; Chu, Z.; Xu, X.; Zhang, J.; Zhu, G. A key ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. PLoS ONE 2015, 10, e0116646. [Google Scholar] [CrossRef]
- Plaxton, W.C. Metabolic flexibility helps plants to survive stress. In Plant Physiology, 4th ed.; Taiz, L., Zeiger, E., Eds.; Sinauer Associates: Heidelberg, Germany, 2006. [Google Scholar]
- Plaxton, W.C. Plant response to stress: Biochemical adaptations to phosphate deficiency. In Encyclopedia of Plant and Crop Science; Goodman, R.M., Ed.; Marcel Dekker: New York, NY, USA, 2004; pp. 976–980. [Google Scholar]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef]
- Heredia, M.C.; Kant, J.; Prodhan, M.A.; Dixit, S.; Wissuwa, M. Breeding rice for a changing climate by improving adaptations to water saving technologies. Theor. Appl. Genet. 2022, 135, 17–33. [Google Scholar] [CrossRef]
- Sandhu, N.; Subedi, S.R.; Yadaw, R.B.; Chaudhary, B.; Prasai, H.; Iftekharuddaula, K.; Thanak, T.; Thun, V.; Battan, K.R.; Ram, M.; et al. Root traits enhancing rice grain yield under alternate wetting and drying condition. Front Plant Sci 2017, 8, 1879. [Google Scholar] [CrossRef]
- Volante, A.; Desiderio, F.; Tondelli, A.; Perrini, R.; Orasen, G.; Biselli, C.; Riccardi, P.; Vattari, A.; Cavalluzzo, D.; Urso, S.; et al. Genome-Wide Analysis of japonica Rice Performance under Limited Water and Permanent Flooding Conditions. Front. Plant Sci. 2017, 8, 1862. [Google Scholar] [CrossRef]
- Norton, G.J.; Travis, A.J.; Douglas, A.; Fairley, S.; Alves, E.P.; Ruang-Areerate, P.; Naredo, M.E.B.; McNally, K.L.; Hossainm, M.; Islam, M.R.; et al. Genome Wide Association Mapping of Grain and Straw Biomass Traits in the Rice Bengal and Assam Aus Panel (BAAP) Grown Under Alternate Wetting and Drying and Permanently Flooded Irrigation. Front. Plant Sci. 2018, 9, 1223. [Google Scholar] [CrossRef]
- Phule, A.S.; Barbadikar, K.M.; Maganti, S.M.; Seguttuvel, P.; Subrahmanyam, D.; Babu, M.B.B.P.; Kumar, P.A. RNA-seq reveals the involvement of key genes for aerobic adaptation in rice. Sci. Rep. 2019, 9, 5235. [Google Scholar] [CrossRef]
- Kebede, A.; Kang, M.S.; Bekele, E. Advances in mechanisms of drought tolerance in crops, with emphasis on barley. Adv. Agron. 2019, 156, 265–314. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Avsar, B.; Lucas, S.J.; Budak, H. Plant abiotic stress signaling. Plant Signal. Behav. 2012, 7, 1450–1455. [Google Scholar] [CrossRef]
- Itam, M.; Mega, R.; Tadano, S.; Abdelrahman, M.; Matsunaga, S.; Yamasaki, Y.; Akashi, K.; Tsujimoto, H. Metabolic and physiological responses to progressive drought stress in bread wheat. Sci. Rep. 2020, 10, 17189. [Google Scholar] [CrossRef]
- Bimpong, I.K.; Serraj, R.; Chin, J.H.; Mendoza, E.M.T.; Hern, J.; Mendioro, M.S. Determination of genetic variability for physiological traits related to drought tolerance in African rice (Oryza glaberrima). J. Plant Breed Crop Sci. 2011, 3, 60–67. [Google Scholar]
- Shaibu, A.A.; Uguru, M.I.; Sow, M.; Maji, A.T.; Ndjiondjop, M.N.; Venuprasad, R. Screening African rice (Oryza glaberrima) for tolerance to abiotic stresses: II. Lowland drought. Crop Sci. 2018, 58, 133–142. [Google Scholar] [CrossRef]
- Zargar, S.M.; Mir, R.A.; Ebinezer, L.B.; Masi, A.; Hami, A.; Manzoor, M.; Salgotra, R.K.; Sofi, N.R.; Mushtaq, R.; Rohila, J.S.; et al. Physiological and Multi-Omics Approaches for Explaining Drought Stress Tolerance and Supporting Sustainable Production of Rice. Front. Plant Sci. 2022, 12, 803603. [Google Scholar] [CrossRef]
- Ramkumar, M.K.; Mulani, E.; Jadon, V.; Sureshkumar, V.; Krishnan, S.G.; Senthil Kumar, S.; Raveendran, M.; Singh, A.K.; Solanke, A.U.; Singh, N.K.; et al. Identification of major candidate genes for multiple abiotic stress tolerance at seedling stage by network analysis and their validation by expression profiling in rice (Oryza sativa L.). 3 Biotech 2022, 12, 127. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.; Lee, Y.H.; Song, S.I. OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 2019, 249, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Abdirad, S.; Ghaffari, M.R.; Majd, A.; Irian, S.; Soleymaniniya, A.; Daryani, P.; Koobaz, P.; Shobbar, Z.S.; Farsad, L.K.; Yazdanpanah, P.; et al. Genome-Wide Expression Analysis of Root Tips in Contrasting Rice Genotypes Revealed Novel Candidate Genes for Water Stress Adaptation. Front. Plant Sci. 2022, 13, 792079. [Google Scholar] [CrossRef]
- Comas, L.H.; Becker, S.R.; Cruz, V.M.; Byrne, P.F.; Dierig, D.A. Root traits contributing to plant productivity under drought. Front. Plant Sci. 2013, 4, 442. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Hausman, J.-F.; Guerriero, G.; Esposito, S. Poaceae vs. Abiotic Stress: Focus on Drought and Salt Stress, Recent Insights and Perspectives. Front. Plant Sci. 2017, 8, 1214. [Google Scholar] [CrossRef]
- Tseng, I.C.; Hong, C.Y.; Yu, S.M.; Ho, T.H.D. Abscisic Acid- and Stress-Induced Highly Proline-Rich Glycoproteins Regulate Root Growth in Rice. Plant Physiol. 2013, 163, 118–134. [Google Scholar] [CrossRef]
- Yasmin, T.; Mumtaz, A.; Mahmood, T.; Hyder, M.Z.; Naqvi, S.M.S. A germin-like protein gene of rice increased superoxide dismutase activity in transformed tobacco. Biol Plant 2015, 59, 456–462. [Google Scholar] [CrossRef]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, S.; Fu, Y.; Su, Z.; Wang, X.; Sun, C. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol. Biol. 2006, 62, 247–259. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.N.; Henry, R. Advances in molecular genetics and genomics of African Rice (Oryza glaberrima Steud). Plants 2019, 8, 376. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, R.; Koteyeva, N.; Voznesenskaya, E.; Evans, M.A.; Cousins, A.B.; Edwards, G.E. Coordination of leaf photosynthesis, transpiration, and structural traits in rice and wild relatives (genus Oryza). Plant Physiol. 2013, 162, 1632–1651. [Google Scholar] [CrossRef]
- Neelam, K.; Sahi, G.K.; Kumar, K.; Singh, K. Identification of drought stress tolerance in wild species germplasm of rice based on leaf and root morphology. Plant Genet. Resour. 2018, 16, 289–295. [Google Scholar] [CrossRef]
- Han, B.; Ma, X.; Cui, D.; Geng, L.; Cao, G.; Zhang, H.; Han, L. Parallel reaction monitoring revealed tolerance to drought proteins in weedy rice (Oryza sativa f. spontanea). Sci. Rep. 2020, 10, 12935. [Google Scholar] [CrossRef]
- Han, B.; Cui, D.; Ma, X.; Cao, G.; Zhang, H.; Koh, H.J.; Han, L. Evidence for evolution and selection of drought-resistant genes based on high-throughput resequencing in weedy rice. J. Exp. Bot. 2022, 73, 1949–1962. [Google Scholar] [CrossRef]
- Qiu, J.; Jia, L.; Wu, D.; Weng, X.; Chen, L.; Sun, J.; Chen, M.; Mao, L.; Jiang, B.; Ye, C.; et al. Diverse genetic mechanisms underlie worldwide convergent rice feralization. Genome Biol. 2020, 21, 70. [Google Scholar] [CrossRef]
- Mahreen, N.; Yasmin, S.; Asif, M.; Yousaf, S.; Yahya, M.; Ejaz, K.; Shahid Hussain, H.; Sajjid, Z.I.; Arif, M. Integrated Analysis of Osmotic Stress and Infrared Thermal Imaging for the Selection of Resilient Rice Under Water Scarcity. Front. Plant Sci. 2022, 13, 834520. [Google Scholar] [CrossRef]
- Venkateshwarlu, C.; Kole, P.C.; Paul, P.J.; Singh, A.K.; Singh, V.K.; Kumar, A. Capturing Genetic Variability and Identification of Promising Drought-Tolerant Lines in Exotic Landrace Derived Population Under Reproductive Drought Stress in Rice. Front. Plant Sci. 2022, 13, 814774. [Google Scholar] [CrossRef]
- Lin, K.H.; Kuo, C.W.; Hsu, Y.C.; Lin, Y.R.; Wu, Y.P. Increasing drought tolerance of rice cultivar TK90 using marker- assisted selection. Crop Environ. Bioinform. 2014, 11, 143–164. [Google Scholar]
- Sebastian, A.; Nugroho, I.C.; Putra, H.S.D.; Susanto, F.A.; Wijayanti, P.; Yamaguchi, N.; Nuringtyas, T.R.; Purwestri, Y.A. Identification and characterization of drought-tolerant local pigmented rice from Indonesia. Physiol. Mol. Biol. Plants 2022, 28, 1061–1075. [Google Scholar] [CrossRef]
- Ahmad, H.; Zafar, S.A.; Naeem, M.K.; Shokat, S.; Inam, S.; Rehman, M.A.U.; Naveed, S.A.; Xu, J.; Li, Z.; Ali, G.M.; et al. Impact of Pre-Anthesis Drought Stress on Physiology, Yield-Related Traits, and Drought-Responsive Genes in Green Super Rice. Front. Genet. 2022, 13, 832542. [Google Scholar] [CrossRef] [PubMed]
- Giri, J.; Parida, S.K.; Raghuvanshi, S.; Tyagi, A.K. Emerging Molecular Strategies for Improving Rice Drought Tolerance. Curr. Genom. 2021, 22, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Freeg, H.A.; Attia, K.A.; Casson, S.; Fiaz, S.; Ramadan, E.A.; Banna, A.E.; Zoulias, N.; Aboshosha, A.; Alamery, S. Physio-biochemical responses and expressional profiling analysis of drought tolerant genes in new promising rice genotype. PLoS ONE 2022, 17, e0266087. [Google Scholar] [CrossRef]
- Yadav, S.; Sandhu, N.; Dixit, S.; Singh, V.K.; Catolos, M.; Mazumder, R.R.; Rahman, M.A.; Kumar, A. Genomics-assisted breeding for successful development of multiple-stress-tolerant, climate-smart rice for southern and southeastern Asia. Plant Genome 2021, 14, e20074. [Google Scholar] [CrossRef]
- Mohd Ikmal, A.; Noraziyah, A.A.S.; Wickneswari, R. Incorporating Drought and Submergence Tolerance QTL in Rice (Oryza sativa L.)-The Effects under Reproductive Stage Drought and Vegetative Stage Submergence Stresses. Plants 2021, 10, 225. [Google Scholar] [CrossRef]
- Muthu, V.; Abbai, R.; Nallathambi, J.; Rahman, H.; Ramasamy, S.; Kambale, R.; Thulasinathan, T.; Ayyenar, B.; Muthurajan, R. Pyramiding QTLs controlling tolerance against drought, salinity, and submergence in rice through marker assisted breeding. PLoS ONE 2020, 15, e0227421. [Google Scholar] [CrossRef]
- Bin Rahman, A.N.M.R.; Zhang, J. Flood and drought tolerance in rice: Opposite but may coexist. Food Energy Secur. 2016, 5, 76–88. [Google Scholar] [CrossRef]
- Avramova, Z. Transcriptional ‘memory’ of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef]
- Abdelaal, K.; AlKahtani, M.; Attia, K.; Hafez, Y.; Király, L.; Künstler, A. The Role of Plant Growth-Promoting Bacteria in Alleviating the Adverse Effects of Drought on Plants. Biology 2021, 10, 520. [Google Scholar] [CrossRef]
- Jacques, C.; Salon, C.; Barnard, R.L.; Vernoud, V.; Prudent, M. Drought stress memory at the plant cycle level: A review. Plants 2021, 10, 1873. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.; Farrona, S. Plant epigenetic stress memory induced by drought: A physiological and molecular perspective. Methods Mol. Biol. 2020, 2093, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Kou, S.; Gu, Q.; Duan, L.; Liu, G.; Yuan, P.; Li, H.; Wu, Z.; Liu, W.; Huang, P.; Liu, L.J. Genome-Wide Bisulphite Sequencing Uncovered the Contribution of DNA Methylation to Rice Short-Term Drought Memory Formation. Plant Growth Regul. 2022, 41, 2903–2917. [Google Scholar] [CrossRef]
- Ganie, S.A.; Bhat, J.A.; Devoto, A. The influence of endophytes on rice fitness under environmental stresses. Plant Mol. Biol. 2022, 109, 447–467. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-mediated stress resistance in plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Omae, N.; Tsuda, K. Plant-Microbiota Interactions in Abiotic Stress Environments. Mol. Plant Microbe Interact. 2022, 35, 511–526. [Google Scholar] [CrossRef]
- Chialva, M.; Ghignone, S.; Cozzi, P.; Lazzari, B.; Bonfante, P.; Abbruscato, P.; Lumini, E. Water management and phenology influence the root-associated rice field microbiota. FEMS Microbiol. Ecol. 2020, 96, fiaa146. [Google Scholar] [CrossRef]
- Hester, E.R.; Vaksmaa, A.; Valè, G.; Monaco, S.; Jetten, M.S.M.; Lüke, C. Effect of water management on microbial diversity and composition in an Italian rice field system. FEMS Microbiol. Ecol. 2022, 98, 1–11. [Google Scholar] [CrossRef]
- Kumar, V.; Jain, L.; Jain, S.K.; Chaturvedi, S.; Kaushal, P. Bacterial endophytes of rice (Oryza sativa L.) and their potential for plant growth promotion and antagonistic activities. S. Afr. J. Bot. 2020, 134, 50–63. [Google Scholar] [CrossRef]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Adhikari, A.; Jan, R.; Ali, S.; Imran, M.; Kim, K.M.; Lee, I.J. Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. 2020, 22, 850–862. [Google Scholar] [CrossRef]
- Rho, H.; van Epps, V.; Wegley, N.; Doty, S.L.; Kim, S.H. Salicaceae endophytes modulate stomatal behavior and increase water use efficiency in rice. Front. Plant Sci. 2018, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef]
- Groen, S.C.; Joly-Lopez, Z.; Platts, A.E.; Natividad, M.; Fresquez, Z.; Mauck, W.M.; Quintana, M.R.; Cabral, C.L.U.; Torres, R.O.; Satija, R.; et al. Evolutionary systems biology reveals patterns of rice adaptation to drought-prone agroecosystems. Plant Cell 2022, 34, 759–783. [Google Scholar] [CrossRef]
- Vallino, M.; Greppi, D.; Novero, M.; Bonfante, P.; Lupotto, E. Rice root colonisation by mycorrhizal and endophytic fungi in aerobic soil. Ann. Appl. Biol. 2009, 154, 195–204. [Google Scholar] [CrossRef]
- Lumini, E.; Vallino, M.; Alguacil, M.M.; Romani, M.; Bianciotto, V. Different farming and water regimes in Italian rice fields affect arbuscular mycorrhizal fungal soil communities. Ecol. Appl. 2011, 21, 1696–1707. [Google Scholar] [CrossRef]
- Vallino, M.; Fiorilli, V.; Bonfante, P. Rice flooding negatively impacts root branching and arbuscular mycorrhizal colonization, but not fungal viability. Plant Cell Environ. 2014, 37, 557–572. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vallino, M.; Biselli, C.; Faccio, A.; Bagnaresi, P.; Bonfante, P. Host and non-host roots in rice: Cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front. Plant Sci. 2015, 6, 636. [Google Scholar] [CrossRef]
- Vurukonda, S.S.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jung, J.; Singh, N.; Greenberg, A.; Doyle, J.J.; Tyagi, W.; Chung, J.W.; Kimball, J.; Hamilton, R.S.; McCouch, S.R. Population Dynamics Among six Major Groups of the Oryza rufipogon Species Complex, Wild Relative of Cultivated Asian Rice. Rice 2016, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; de Hollander, M.; Raaijmakers, J.M. The wild side of plant microbiomes. Microbiome 2018, 6, 143. [Google Scholar] [CrossRef]
- Oyserman, B.O.; Flores, S.S.; Griffioen, T.; Pan, X.; van der Wijk, E.; Pronk, L.; Lokhorst, W.; Nurfikari, A.; Paulson, J.N.; Movassagh, M.; et al. Disentangling the genetic basis of rhizosphere microbiome assembly in tomato. Nat. Commun. 2022, 13, 3228. [Google Scholar] [CrossRef]
- Escudero-Martinez, C.; Coulter, M.; Alegria Terrazas, R.; Foito, A.; Kapadia, R.; Pietrangelo, L.; Maver, M.; Sharma, R.; Aprile, A.; Morris, J.; et al. Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 2022, 13, 3443. [Google Scholar] [CrossRef] [PubMed]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef]
- Escudero-Martinez, C.; Bulgarelli, D. Tracing the evolutionary routes of plant-microbiota interactions. Curr. Opin. Microbiol. 2019, 49, 34–40. [Google Scholar] [CrossRef]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef]
- Kumar, S.; Bhati, J.; Saha, A.; Lal, S.B.; Pandey, P.K.; Mishra, D.C.; Farooqi, M.S.; Kumar, A.; Chaturvedi, K.K.; Rai, A. CerealESTDb: A Comprehensive Resource for Abiotic Stress-Responsive Annotated ESTs With Predicted Genes, Gene Ontology, and Metabolic Pathways in Major Cereal Crops. Front. Genet. 2022, 13, 842868. [Google Scholar] [CrossRef]
- Bohra, A.; Jha, U.C.; Godwin, I.D.; Varshney, R.K. Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Sinha, P.; Singh, V.K.; Kumar, A.; Zhang, Q.; Bennetzen, J.L. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020, 56, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Biswal, A.K.; Mangrauthia, S.K.; Reddy, M.R.; Yugandhar, P. CRISPR mediated genome engineering to develop climate smart rice: Challenges and opportunities. Semin. Cell Dev. Biol. 2019, 96, 100–106. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).