OTUD7A Regulates Inflammation- and Immune-Related Gene Expression in Goose Fatty Liver
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiment
2.2. Preparation of Goose Primary Hepatocytes
2.3. Overexpression of Goose OTUD7A
2.4. Transcriptome Analysis
2.5. RNA Interference Assay
2.6. Gene Expression Analysis
2.7. Statistical Analysis
3. Results
3.1. Genes and Pathways Affected by OTUD7A
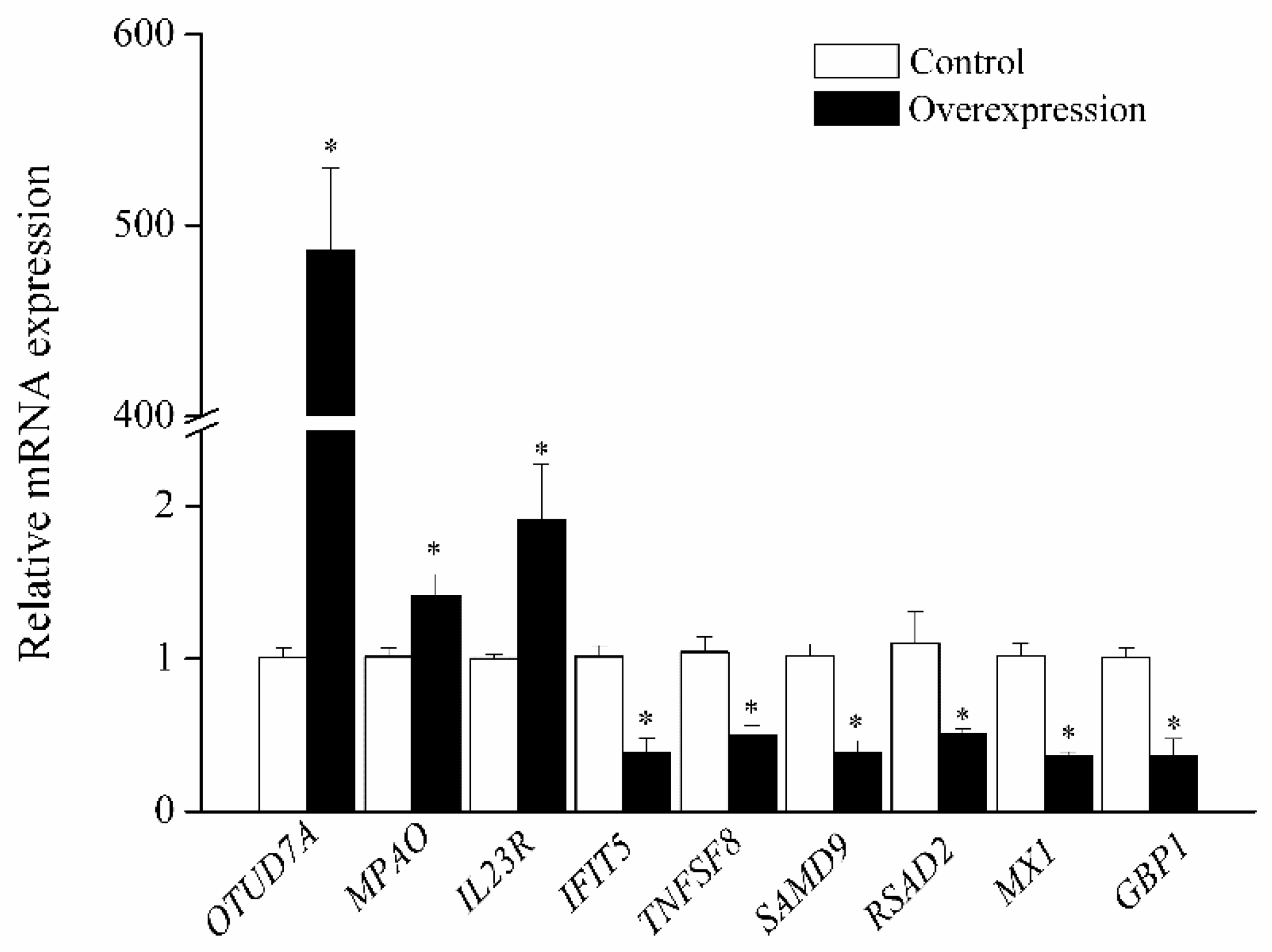
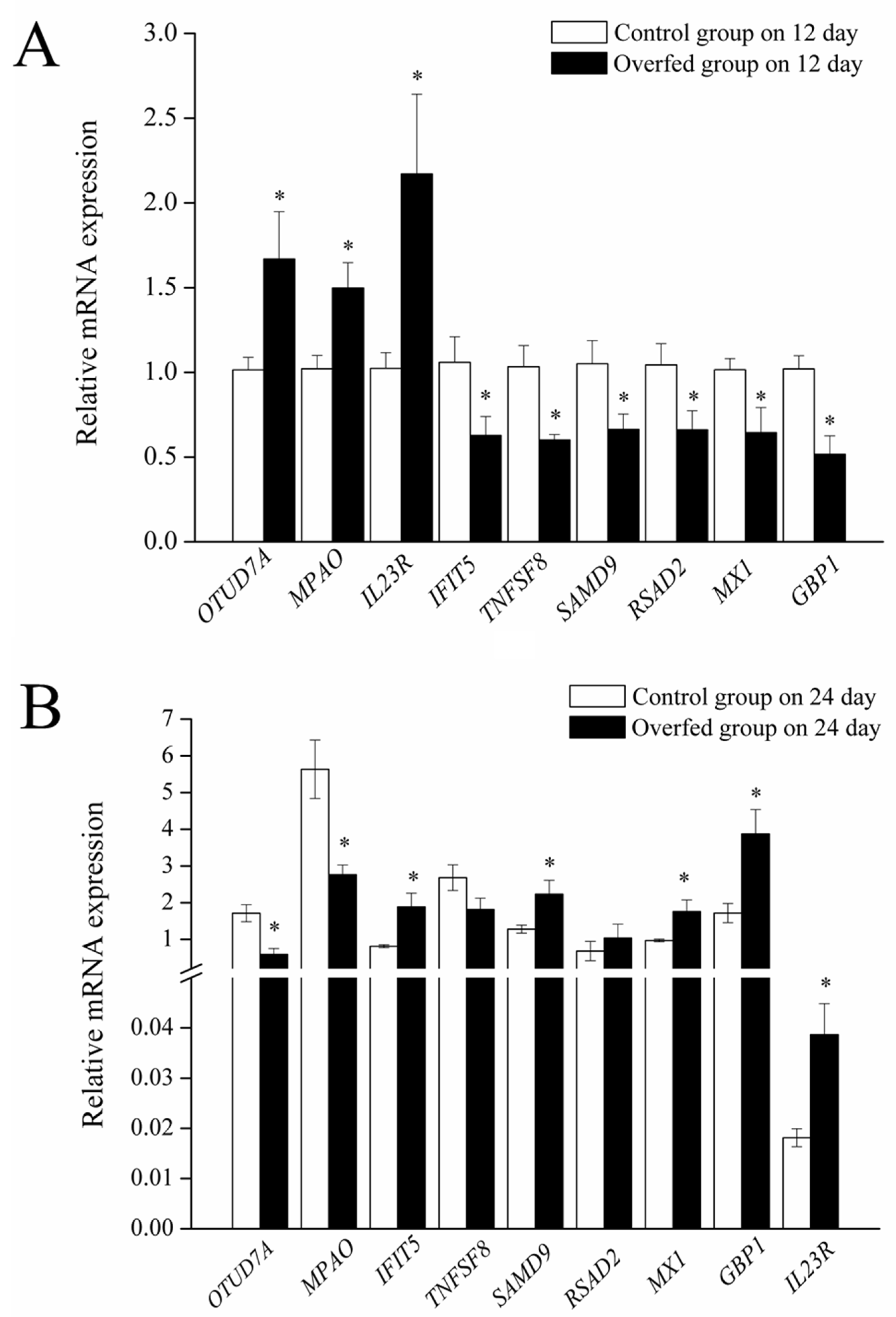
3.2. Expression of OTUD7A and Its Downstream Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hermier, D.; Rousselot-Pailley, D.; Peresson, R.; Sellier, N. Influence of orotic acid and estrogen on hepatic lipid storage and secretion in the goose susceptible to liver steatosis. Biochim. Biophys. Acta 1994, 1211, 97–106. [Google Scholar] [CrossRef]
- Geng, T.Y.; Xia, L.L.; Li, F.Y.; Xia, J.; Zhang, Y.H.; Wang, Q.Q.; Yang, B.; Montgomery, S.; Cui, H.M.; Gong, D.Q. The role of endoplasmic reticulum stress and insulin resistance in the occurrence of goose fatty liver. Biochem. Biophys. Res. Commun. 2015, 465, 83–87. [Google Scholar] [CrossRef]
- Geng, T.Y.; Yang, B.; Li, F.Y.; Xia, L.L.; Wang, Q.Q.; Zhao, X.; Gong, D.Q. Identification of protective components that prevent the exacerbation of goose fatty liver: Characterization, expression and regulation of adiponectin receptors. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 194–195, 32–38. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, X.; Wang, Q.; Sun, X.X.; Xia, L.L.; Wang, Q.Q.; Yang, B.; Zhang, Y.H.; Montgomery, S.; Meng, H.; et al. Prosteatotic and protective components in a unique model of fatty liver: Gut microbiota and suppressed complement system. Sci. Rep. 2016, 6, 31763. [Google Scholar] [CrossRef] [Green Version]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Ding, Y.L.; Zhang, J.L.; Zhang, P.; Wang, J.Q.; Li, Z.H. Alpinetin improved high fat diet-induced non-alcoholic fatty liver disease (NAFLD) through improving oxidative stress, inflammatory response and lipid metabolism. Biomed. Pharmacother. 2018, 97, 1397–1408. [Google Scholar] [CrossRef]
- Hossain, N.; Kanwar, P.; Mohanty, S.R. A comprehensive updated review of pharmaceutical and nonpharmaceutical treatment for NAFLD. Gastroenterol. Res. Pract. 2016, 144, 7109270. [Google Scholar]
- Rahman, A.M.; Mahdavi, A.H.; Rahmani, H.R.; Jahanian, E. Clove bud (Syzygium aromaticum) improved blood and hepatic antioxidant indices in laying hens receiving low n-6 to n-3 ratios. J. Anim. Physiol. Anim. Nutr. 2017, 101, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Marra, F. Nuclear factor-κB inhibition and non-alcoholic steatohepatitis: Inflammation as a target for therapy. Gut 2008, 57, 570–572. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Moschen, A.R. Evolution of inflammation in nonalcoholic fatty liver disease: The multiple parallel hits hypothesis. Hepatology 2010, 52, 1836–1846. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.L.; Ma, J.T.; Wang, W.D.; Zhang, L.J.; Xu, J.; Wang, K.; Li, D.F. Resveratrol supplement inhibited the NF-κB inflammation pathway through activating AMPKα-SIRT1 pathway in mice with fatty liver. Mol. Cell. Biochem. 2016, 422, 75–84. [Google Scholar] [CrossRef]
- Mevissen, T.E.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; Oualid, F.E.I.; et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Pei, L.; Wang, L.; Zhang, F.; Hu, X.; Gui, Y. Snail1-dependent transcriptional repression of Cezanne2 in hepatocellular carcinoma. Oncogene 2014, 33, 2836–2845. [Google Scholar] [CrossRef]
- Zhao, M.M.; Liu, T.J.; Wang, Q.; Zhang, R.; Liu, L.; Gong, D.Q.; Geng, T.Y. Fatty acids modulate the expression of pyruvate kinase and arachidonate-lipoxygenase through PPARγ/CYP2C45 pathway: A link to goose fatty liver. Poult. Sci. 2019, 98, 4346–4358. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Parola, M.; Vajro, P. Nocturnal hypoxia in obese-related obstructive sleep apnea as a putative trigger of oxidative stress in pediatric NAFLD progression. J. Hepatol. 2016, 65, 470–472. [Google Scholar] [CrossRef] [Green Version]
- Wruck, W.; Graffmann, N.; Kawala, M.A.; Adjaye, J. Concise review: Current status and future directions on research related to nonalcoholic fatty liver disease. Stem Cells 2017, 35, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Pirozzi, C.; Lama, A.; Simeoli, R.; Paciello, O.; Pagano, T.B.; Mollica, M.P.; Di Guida, F.; Russo, R.; Magliocca, S.; Canani, R.B.; et al. Hydroxytyrosol prevents metabolic impairment reducing hepatic inflammation and restoring duodenal integrity in a rat model of NAFLD. J. Nutr. Biochem. 2016, 30, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Kondylis, V.; Kumari, S.; Vlantis, K.; Pasparakis, M. The interplay of IKK, NF-κB and RIPK1 signaling in the regulation of cell death, tissue homeostasis and inflammation. Immunol. Rev. 2017, 277, 113–127. [Google Scholar] [CrossRef]
- John, J.L.; Qian, T.; He, L.H.; Kim, J.S.; Elmore, S.P.; Cascio, W.E.; Brenner, D.A. Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxid. Redox. Signal. 2002, 4, 769–781. [Google Scholar]
- Xing, Y.; Xu, C.; Lin, X.; Zhao, M.M.; Gong, D.Q.; Liu, L.; Geng, T.Y. Complement C3 participates in the development of goose fatty liver potentially by regulating the expression of FASN and ETNK1. Anim. Sci. J. 2021, 92, e13527. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Zhao, L.P.; Shen, L.J.; Wang, S.; Zhang, K.; Qi, Y.; Zheng, J.; Zhang, X.J.; Zhu, X.Y.; Bao, R.; et al. USP18 protects against hepatic steatosis and insulin resistance through its deubiquitinating activity. Hepatology 2017, 666, 1866–1884. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Wang, F.; Gao, L.C.; Xu, L.W.; Tong, R.Y.; Lin, N.; Su, Y.Y.; Yan, Y.; Gao, Y.; He, J.; et al. Ubiquitin-specific protease 4 is an endogenous negative regulator of metabolic dysfunctions in nonalcoholic fatty liver disease in mice. Hepatology 2018, 68, 897–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.D.; Shen, Y.; Qiao, S.; Liu, W.W.; Zheng, L.S.; Wang, Y.N.; Cui, N.P.; Wang, Y.F.; Zhao, S.L.; Shi, J.H. Upregulation of OTUD7B (cezanne) promotes tumor progression via AKT/VEGF pathway in lung squamous carcinoma and adenocarcinoma. Front. Oncol. 2019, 9, 862. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.J.; Hou, G.X.; Fang, Z.X.; Liu, J.L.; Lv, X.D. Distinct expression and prognostic value of OTU domain-containing proteins in non-small-cell lung cancer. Oncol. Lett. 2019, 5, 5417–5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccarelli, S.; Panera, N.; Mina, M.; Gnani, D.; Stefanis, C.D.; Crudele, A.; Rychlicki, C.; Petrini, S.; Bruscalupi, G.; Agostinelli, L.; et al. LPS-induced TNF-α factor mediates pro-inflammatory and pro-fibrogenic pattern in non-alcoholic fatty liver disease. Oncotarget 2015, 6, 41434–41452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, W.J.; Lin, J.X. Cytokine receptor signaling pathways. J. Allergy Clin. Immunol. 2000, 105, 877–888. [Google Scholar] [CrossRef]
- Croft, M.; Duan, W.; Choi, H.; Eun, S.Y.; Madireddi, S.; Mehta, A. TNF superfamily in inflammatory disease: Translating basic insights. Trends Immunol. 2012, 33, 144–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, R.; Aizawa, S.; Nagai, M.; Ito, K.; Higashihara, M.; Ishida, T.; Inoue, J.; Watanabe, T. A novel domain in the CD30 cytoplasmic tail mediates NFkappaB activation. Int. Immunol. 1998, 10, 203–210. [Google Scholar] [CrossRef] [Green Version]
- McGillicuddy, F.C.; Chiquoine, E.H.; Hinkle, C.C.; Kim, R.J.; Shah, R.; Roche, H.M.; Smyth, E.M.; Reilly, M.P. Interferon γ attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J. Biol. Chem. 2009, 284, 31936–31944. [Google Scholar] [CrossRef] [Green Version]
- Imada, K.; Leonard, W.J. The Jak-STAT pathway. Mol. Immunol. 2000, 37, 1–11. [Google Scholar] [CrossRef]
- Coskun, M.; Salem, M.; Pedersen, J.; Nielsen, O.H. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacol. Res. 2013, 76, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; Santhakumar, D.; Naggar, R.F.E.; Iqbal, M.; Hussein, H.A.; Munir, M. Chickens expressing IFIT5 ameliorate clinical outcome and pathology of highly pathogenic avian influenza and velogenic newcastle disease viruses. Front. Immunol. 2018, 9, 2025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Liu, X.Y.; Chen, W.; Chen, L. IFIT5 potentiates anti-viral response through enhancing innate immune signaling pathways. Acta Biochim. Biophys. Sin. 2013, 45, 867–874. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.S.; Zheng, Z.H.; Zhang, Z.H.; Meng, J.; Liu, Y.; Ke, X.L.; Hu, Q.X.; Wang, H.Z. IFIT5 positively regulates NF-κB signaling through synergizing the recruitment of IκB kinase (IKK) to TGF-β-activated kinase 1 (TAK1). Cell. Signal. 2015, 27, 2343–2354. [Google Scholar] [CrossRef]
- Chefetz, I.; Ben, A.D.; Browning, S.; Skorecki, K.; Adir, N.; Thomas, M.G.; Kogleck, L.; Topaz, O.; Indelman, M.; Uitto, J.; et al. Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-α responsive protein. J. Investig. Dermatol. 2008, 128, 1423–1429. [Google Scholar] [CrossRef] [Green Version]
- Hershkovitz, D.; Gross, Y.; Nahum, S.; Yehezkel, S.; Sarig, O.; Uitto, J.; Sprecher, E. Functional characterization of SAMD9, a protein deficient in normophosphatemic familial tumoral calcinosis. J. Investig. Dermatol. 2011, 131, 662–669. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Wu, L.-F.; Bing, P.-F.; Xia, W.; Wang, L.; Xie, F.-F.; Lu, X.; Lei, S.-F.; Deng, F.-Y. SAMD9 is a (epi-) genetically regulated anti-inflammatory factor activated in RA patients. Mol. Cell Biochem. 2019, 456, 135–144. [Google Scholar] [CrossRef]
- Nounamo, B.; Li, Y.B.; Byrne, P.O.; Kearney, A.M.; Khan, A.; Liu, J. An interaction domain in human SAMD9 is essential for myxoma virus host-range determinant M062 antagonism of host anti-viral function. Virology 2017, 503, 94–102. [Google Scholar] [CrossRef]
- Parham, C.; Chirica, M.; Timans, J.; Vaisberg, E.; Travis, M.; Cheung, J.; Pflanz, S.; Zhang, R.; Singh, K.P.; Vega, F.; et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rβ1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002, 168, 5699–5708. [Google Scholar] [CrossRef] [Green Version]
- Rivieccio, M.A.; Suh, H.S.; Zhao, Y.M.; Zhao, M.L.; Chin, K.C.; Lee, S.C.; Brosnan, C.F. TLR3 ligation activates an antiviral response in human fetal astrocytes: A role for viperin/cig5. J. Immunol. 2006, 177, 4735–4741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisskirchen, C.; Ludersdorfer, T.H.; Müller, D.A.; Moritz, E.; Pavlovic, J. Interferon-induced antiviral protein MxA interacts with the cellular RNA helicases UAP56 and URH49. J. Biol. Chem. 2011, 286, 34743–34751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Yan, J.; Chen, H.; Li, J.; Tian, Y.; Tang, L.S.; Feng, H. Mx1 of black carp functions importantly in the antiviral innate immune response. Fish Shellfish Immunol. 2016, 58, 584–592. [Google Scholar] [CrossRef]
- Tripal, P.; Bauer, M.; Naschberger, E.; Mörtinger, T.; Hohenadl, C.; Cornali, E.; Thurau, M.; Stürzl, M. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 2007, 27, 44–52. [Google Scholar] [CrossRef]
- Britzen-Laurent, N.; Bauer, M.; Berton, V.; Fischer, N.; Syguda, A.; Reipschläger, S.; Naschberger, E.; Herrmann, C.; Stürzl, M. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS ONE 2010, 12, e14246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubeseder-Martellato, C.; Guenzi, E.; Jörg, A.; Töpolt, K.; Naschberger, E.; Kremmer, E.; Zietz, C.; Tschachler, E.; Hutzler, P.; Schwemmle, M.; et al. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am. J. Pathol. 2002, 161, 1749–1759. [Google Scholar] [CrossRef] [Green Version]
- Bour, S.; Daviaud, D.; Gres, S.; Lefort, C.; Prévot, D.; Zorzano, A.; Wabitsch, M.; Saulnier-Blache, J.-S.; Valet, P.; Carpéné, C. Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human adipocytes. Biochimie 2007, 89, 916–925. [Google Scholar] [CrossRef] [PubMed]


| Gene 1 | GenBank Number | Primer Sequence (5′ to 3′) | Product Size |
|---|---|---|---|
| OTUD7A | XM_013177443.1 | F: CAGACTTTGTTCGGTCCACA | 236 |
| R: AGCAATGTCATCCTGCCTCT | |||
| MPAO | XM_013199765.1 | F: TCCGACATGCAGTCCTTGTA | 201 |
| R: AACCTTGTTGCCGTAGTTGG | |||
| IFIT5 | XM_013194152.1 | F: CCTTGAAGAAGTCCCTGCTG | 194 |
| R: ATTTTCCAGGGCTTCCTTGT | |||
| IL23R | XM_013193090.1 | F: CTGATGGGCTCCAACATCTC | 161 |
| R: CTGTAGGGCAGCCTGAAGTC | |||
| TNFSF8 | XM_013182840.1 | F: AGGCTTGGAGCCACTAACAA | 201 |
| R: GGTCGGGACATTCTGTAGGA | |||
| SAMD9 | XM_013187038.1 | F: CTCTCGCACAAATGGCACTA | 157 |
| R: AGCTTTTGCAGCTCCAATGT | |||
| RSAD2 | XM_013172803.1 | F: GAGAGCGGTGGTTCAAGAAG | 238 |
| R: TTGAGTGCCATGATCTGCTC | |||
| MX1 | XM_013181384.1 | F: TGGAGCAAGTAAACGCCTCT | 216 |
| R: TTCAGGCACTGGTAGGCTTT | |||
| GBP1 | XM_013170751.1 | F: GAGCAGGAGAGAGAGGCTGA | 190 |
| R: TGTTCTTCTCGGAGCCACTT | |||
| GAPDH | XM_013199522.1 | F: GCCATCAATGATCCCTTCAT | 155 |
| R: CTGGGGTCACGCTCCTG |
| Name | Description | p-Value | Fold Change |
|---|---|---|---|
| SNCG | Gamma-synuclein isoform | 0.019 | 2.11 |
| GPRIN2 | G protein-regulated inducer of neurite outgrowth 2 isoform | 0.039 | 2.29 |
| NMU | Neuromedin-U isoform | 0.004 | 7.86 |
| OTUD7A | OTU domain-containing protein 7A | <0.001 | 134.69 |
| LOC106034831 | CMRF35-like molecule 3 isoform | 0.018 | 4.68 |
| ARHGAP28 | Rho GTPase-activating protein 28 isoform | 0.040 | 2.10 |
| SH3BP1 | SH3 domain-binding protein 1 | 0.040 | 2.82 |
| MEST | Mesoderm-specific transcript homolog protein, partial | 0.045 | 2.75 |
| PROCR | Endothelial protein C receptor | 0.004 | 2.61 |
| CLMP | CXADR-like membrane protein | 0.012 | 2.40 |
| KLHDC8A | Kelch domain-containing protein 8A | 0.047 | 3.32 |
| MYL10 | Myosin regulatory light chain 2B, cardiac muscle isoform | 0.019 | 2.07 |
| IL23R | Interleukin-23 receptor | 0.017 | 2.14 |
| LOC106045877 | Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like | 0.042 | 2.06 |
| ACE | Angiotensin-converting enzyme | 0.002 | 2.05 |
| TMEM114 | Transmembrane protein 114 | 0.004 | 3.64 |
| PPP1R27 | Protein phosphatase 1 regulatory subunit 27 | 0.05 | 2.99 |
| MPAO | Membrane primary amine oxidase | 0.045 | 2.03 |
| LOC106048707 | Uncharacterized protein LOC106048707 isoform | 0.045 | 4.52 |
| Name | Description | p-Value | Fold Change |
|---|---|---|---|
| IFIT5 | Interferon-induced protein with tetratricopeptide repeats 5 | <0.001 | 0.42 |
| OTOGL | Otogelin-like protein | 0.045 | 0.45 |
| LOC106030637 | Interleukin-7-like | 0.022 | 0.17 |
| RSAD2 | Radical S-adenosyl methionine domain-containing protein 2 | <0.001 | 0.48 |
| DDX60 | Probable ATP-dependent RNA helicase DDX60 isoform | <0.001 | 0.42 |
| MX1 | Interferon-induced GTP-binding protein Mx1 | <0.001 | 0.47 |
| NCAM2 | Neural cell adhesion molecule 2 isoform | 0.007 | 0.27 |
| TNFSF8 | Tumor necrosis factor ligand superfamily member 8 | 0.041 | 0.35 |
| PIH1D3 | Protein PIH1D3 | 0.035 | 0.39 |
| LOC106038693 | Low-affinity vacuolar monovalent cation antiporter | 0.043 | 0.08 |
| SAMD9 | Sterile alpha motif domain-containing protein 9 | <0.001 | 0.45 |
| LOC106040189 | Arylacetamide deacetylase-like 4 | 0.010 | 0.42 |
| LOC106041938 | Placenta-specific gene 8 protein-like isoform | <0.001 | 0.44 |
| B3GNT7 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 | <0.001 | 0.36 |
| GBP1 | Interferon-induced guanylate-binding protein 1 | <0.001 | 0.45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Wen, K.; Fan, X.; Sun, Q.; Jauregui, D.; K. Khogali, M.; Liu, L.; Geng, T.; Gong, D. OTUD7A Regulates Inflammation- and Immune-Related Gene Expression in Goose Fatty Liver. Agriculture 2022, 12, 105. https://doi.org/10.3390/agriculture12010105
Zhao M, Wen K, Fan X, Sun Q, Jauregui D, K. Khogali M, Liu L, Geng T, Gong D. OTUD7A Regulates Inflammation- and Immune-Related Gene Expression in Goose Fatty Liver. Agriculture. 2022; 12(1):105. https://doi.org/10.3390/agriculture12010105
Chicago/Turabian StyleZhao, Minmeng, Kang Wen, Xiang Fan, Qingyun Sun, Diego Jauregui, Mawahib K. Khogali, Long Liu, Tuoyu Geng, and Daoqing Gong. 2022. "OTUD7A Regulates Inflammation- and Immune-Related Gene Expression in Goose Fatty Liver" Agriculture 12, no. 1: 105. https://doi.org/10.3390/agriculture12010105





