Longitudinal Changes of Ocular Surface Microbiome in Patients Undergoing Hemopoietic Stem Cell Transplant (HSCT)
Abstract
:1. Introduction
2. Materials & Methods
2.1. Subjects
2.2. Haematological Characteristics
2.3. Ocular Examination and Sampling
2.4. Sampling, DNA Extraction, 16S rRNA Sequencing, and Processing
2.5. Statistical Analysis
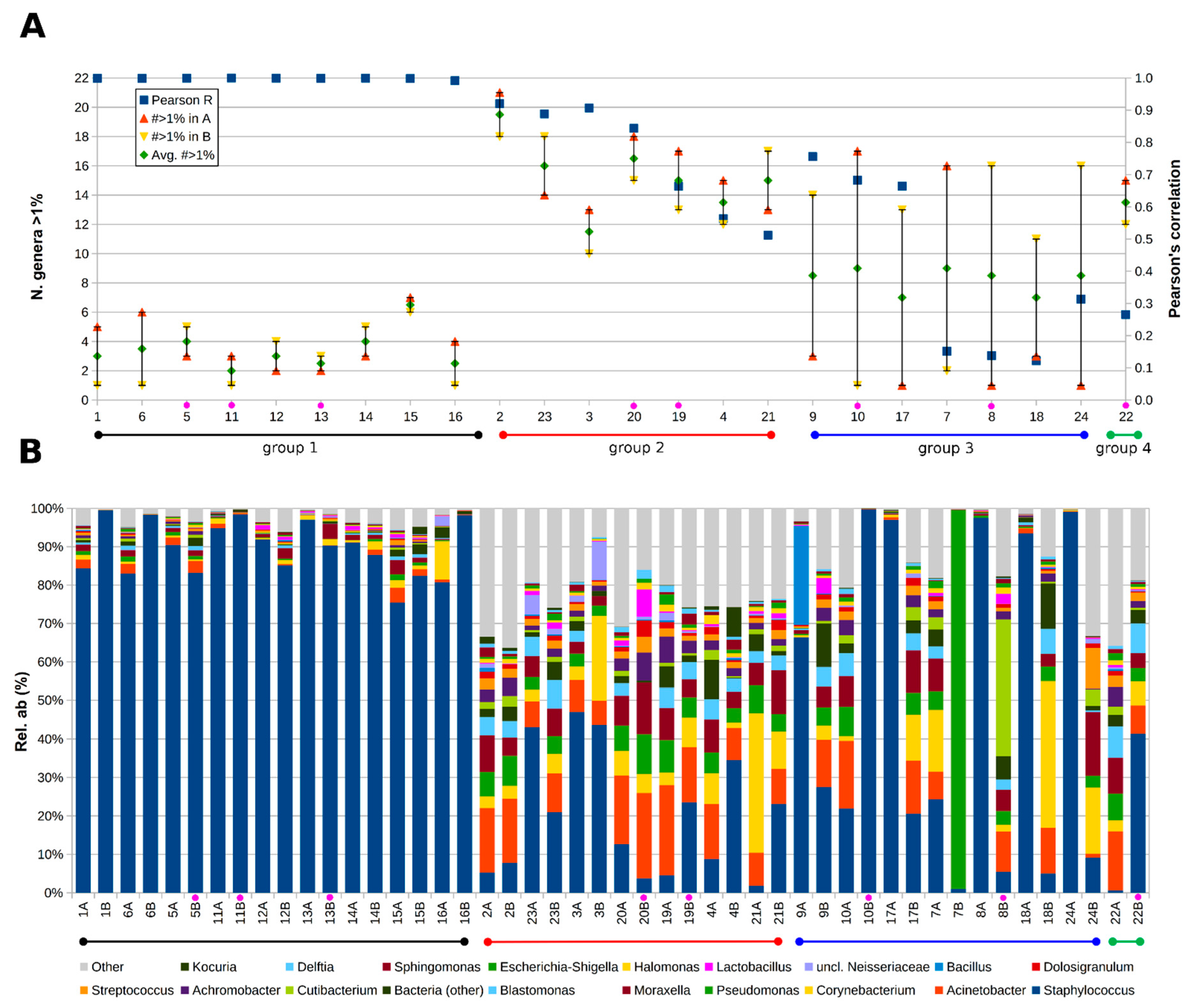
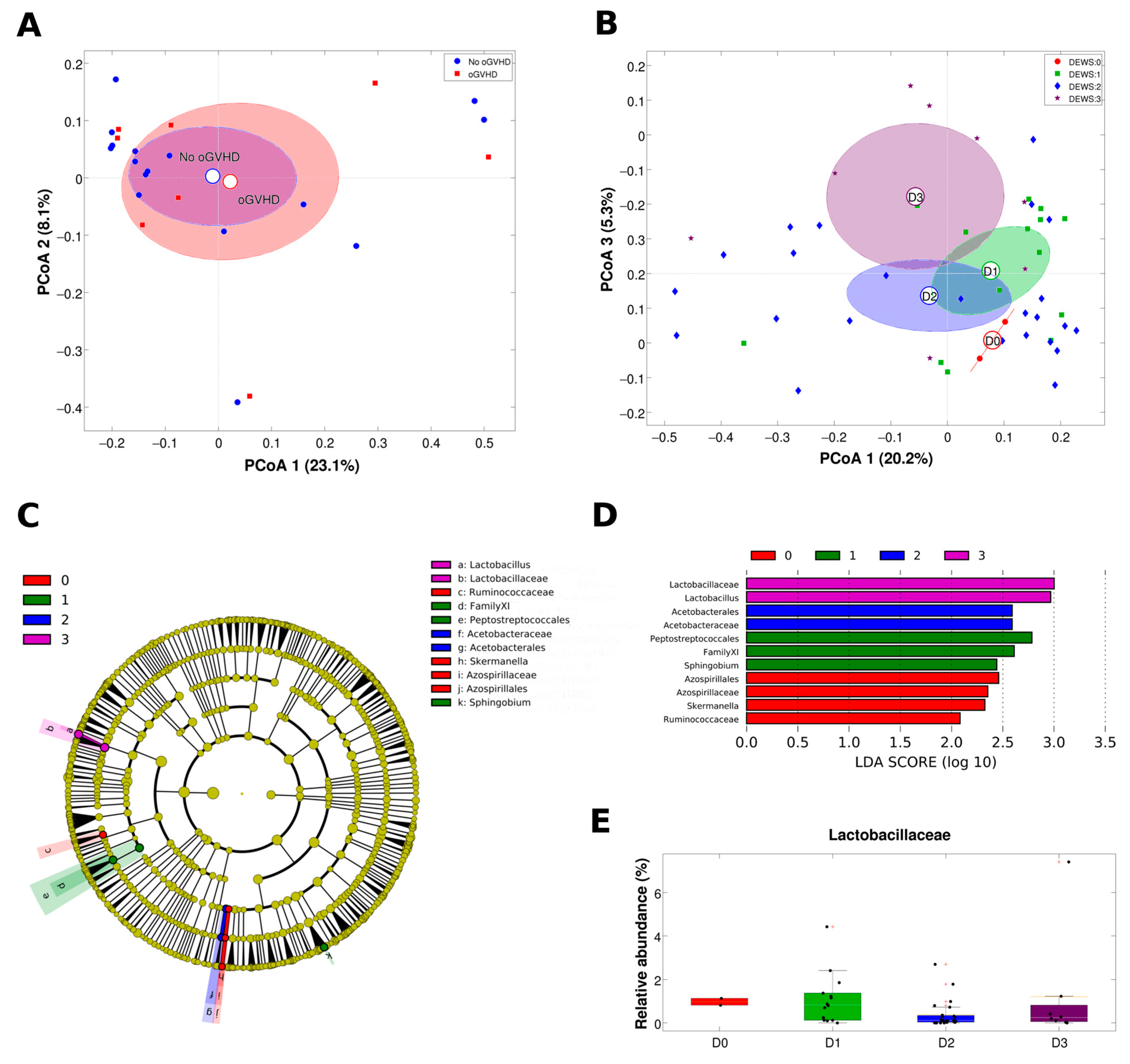
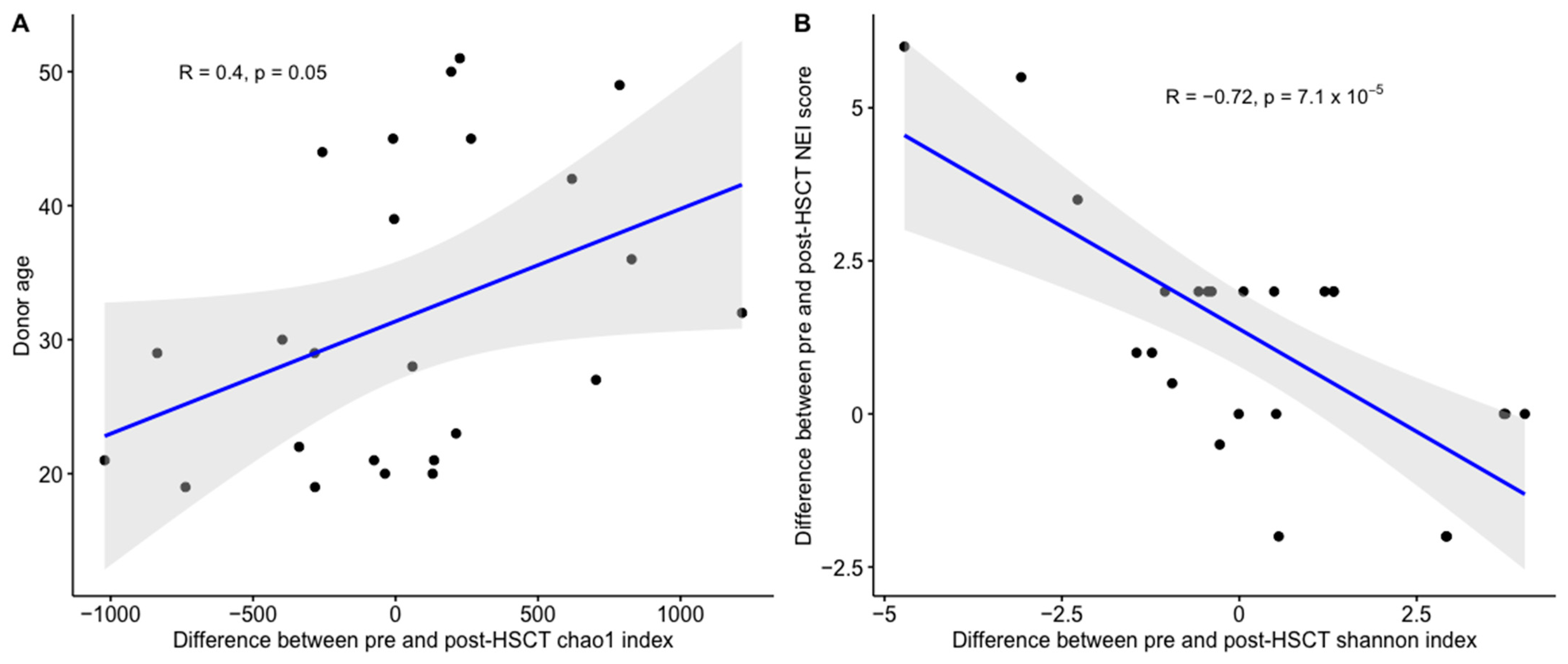
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.J. Classification Systems for Chronic Graft-versus-Host Disease. Blood 2017, 129, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Inamoto, Y.; Valdés-Sanz, N.; Ogawa, Y.; Alves, M.; Berchicci, L.; Galvin, J.; Greinix, H.; Hale, G.A.; Horn, B.; Kelly, D.; et al. Ocular Graft-versus-Host Disease after Hematopoietic Cell Transplantation: Expert Review from the Late Effects and Quality of Life Working Committee of the Center for International Blood and Marrow Transplant Research and Transplant Complications Working Party of the European Society of Blood and Marrow Transplantation. Biol. Blood Marrow Transplant. 2019, 25, e46–e54. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Hu, B.; Li, P.; Zhao, Y.; Xu, Y.; Wu, D. Roles of the Intestinal Microbiota and Microbial Metabolites in Acute GVHD. Exp. Hematol. Oncol. 2021, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Staffas, A.; Burgos da Silva, M.; van den Brink, M.R.M. The Intestinal Microbiota in Allogeneic Hematopoietic Cell Transplant and Graft-versus-Host Disease. Blood 2017, 129, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Hayase, E.; Jenq, R.R. The Role of Microbiota in Allogeneic Hematopoietic Stem Cell Transplantation. Expert. Opin. Biol. Ther. 2021, 21, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.P.; Koh, A.Y. The Gut Microbiota in Transplant Patients. Blood Rev. 2020, 39, 100614. [Google Scholar] [CrossRef]
- Hino, A.; Fukushima, K.; Kusakabe, S.; Ueda, T.; Sudo, T.; Fujita, J.; Motooka, D.; Takeda, A.K.; Shinozaki, N.O.; Watanabe, S.; et al. Prolonged Gut Microbial Alterations in Post-Transplant Survivors of Allogeneic Haematopoietic Stem Cell Transplantation. Br. J. Haematol. 2022, 201, 725–737. [Google Scholar] [CrossRef]
- Jacobs, R.; Tran, U.; Chen, H.; Kassim, A.; Engelhardt, B.G.; Greer, J.P.; Goodman, S.G.; Clifton, C.; Lucid, C.; Vaughan, L.A.; et al. Prevalence and Risk Factors Associated with Development of Ocular GVHD Defined by NIH Consensus Criteria. Bone Marrow Transplant. 2012, 47, 1470–1473. [Google Scholar] [CrossRef]
- Na, K.-S.; Yoo, Y.-S.; Mok, J.W.; Lee, J.W.; Joo, C.-K. Incidence and Risk Factors for Ocular GVHD after Allogeneic Hematopoietic Stem Cell Transplantation. Bone Marrow Transplant. 2015, 50, 1459–1464. [Google Scholar] [CrossRef]
- Pellegrini, M.; Bernabei, F.; Barbato, F.; Arpinati, M.; Giannaccare, G.; Versura, P.; Bonifazi, F. Incidence, Risk Factors and Complications of Ocular Graft-Versus-Host Disease Following Hematopoietic Stem Cell Transplantation. Am. J. Ophthalmol. 2021, 227, 25–34. [Google Scholar] [CrossRef]
- Shimizu, E.; Aketa, N.; Yazu, H.; Uchino, M.; Kamoi, M.; Sato, Y.; Tsubota, K.; Ogawa, Y. Corneal Higher-Order Aberrations in Eyes with Chronic Ocular Graft-versus-Host Disease. Ocul. Surf. 2020, 18, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-C.; Chai, X.; Inamoto, Y.; Pidala, J.; Martin, P.J.; Flowers, M.E.D.; Shen, T.T.; Lee, S.J.; Jagasia, M. Impact of Ocular Chronic Graft-versus-Host Disease on Quality of Life. Biol. Blood Marrow Transplant. 2015, 21, 1687–1691. [Google Scholar] [CrossRef] [PubMed]
- Filipovich, A.H.; Weisdorf, D.; Pavletic, S.; Socie, G.; Wingard, J.R.; Lee, S.J.; Martin, P.; Chien, J.; Przepiorka, D.; Couriel, D.; et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. Diagnosis and Staging Working Group Report. Biol. Blood Marrow Transplant. 2005, 11, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Hessen, M.; Akpek, E.K. Ocular Graft-versus-Host Disease. Curr. Opin. Allergy Clin. Immunol. 2012, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Jagasia, M.H.; Greinix, H.T.; Arora, M.; Williams, K.M.; Wolff, D.; Cowen, E.W.; Palmer, J.; Weisdorf, D.; Treister, N.S.; Cheng, G.-S.; et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group Report. Biol. Blood Marrow Transplant. 2015, 21, 389–401.e1. [Google Scholar] [CrossRef]
- Campagnoli, L.I.M.; Varesi, A.; Barbieri, A.; Marchesi, N.; Pascale, A. Targeting the Gut-Eye Axis: An Emerging Strategy to Face Ocular Diseases. Int. J. Mol. Sci. 2023, 24, 13338. [Google Scholar] [CrossRef]
- Doan, T.; Akileswaran, L.; Andersen, D.; Johnson, B.; Ko, N.; Shrestha, A.; Shestopalov, V.; Lee, C.S.; Lee, A.Y.; Van Gelder, R.N. Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5116–5126. [Google Scholar] [CrossRef]
- Ozkan, J.; Willcox, M.D. The Ocular Microbiome: Molecular Characterisation of a Unique and Low Microbial Environment. Curr. Eye Res. 2019, 44, 685–694. [Google Scholar] [CrossRef]
- Delbeke, H.; Younas, S.; Casteels, I.; Joossens, M. Current Knowledge on the Human Eye Microbiome: A Systematic Review of Available Amplicon and Metagenomic Sequencing Data. Acta Ophthalmol. 2021, 99, 16–25. [Google Scholar] [CrossRef]
- Matysiak, A.; Kabza, M.; Karolak, J.A.; Jaworska, M.M.; Rydzanicz, M.; Ploski, R.; Szaflik, J.P.; Gajecka, M. Characterization of Ocular Surface Microbial Profiles Revealed Discrepancies between Conjunctival and Corneal Microbiota. Pathogens 2021, 10, 405. [Google Scholar] [CrossRef]
- Kang, Y.; Lin, S.; Ma, X.; Che, Y.; Chen, Y.; Wan, T.; Zhang, D.; Shao, J.; Xu, J.; Xu, Y.; et al. Strain Heterogeneity, Cooccurrence Network, Taxonomic Composition and Functional Profile of the Healthy Ocular Surface Microbiome. Eye Vis. 2021, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Garza, A.; Diaz, G.; Hamdan, M.; Shetty, A.; Hong, B.-Y.; Cervantes, J. Homeostasis and Defense at the Surface of the Eye. The Conjunctival Microbiota. Curr. Eye Res. 2021, 46, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gong, Y.; Chen, S.; Li, S.; Zhang, Y.; Zhong, H.; Wang, Z.; Chen, Y.; Deng, Q.; Jiang, Y.; et al. Comparative Portrayal of Ocular Surface Microbe with and without Dry Eye. J. Microbiol. 2019, 57, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Ham, B.; Hwang, H.B.; Jung, S.H.; Chang, S.; Kang, K.D.; Kwon, M.J. Distribution and Diversity of Ocular Microbial Communities in Diabetic Patients Compared with Healthy Subjects. Curr. Eye Res. 2018, 43, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Xiao, K.; Min, H.; Chen, Z.; Long, Q. Characterization of Conjunctival Sac Microbiome from Patients with Allergic Conjunctivitis. J. Clin. Med. 2022, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, E.; Ogawa, Y.; Saijo, Y.; Yamane, M.; Uchino, M.; Kamoi, M.; Fukui, M.; Yang, F.; He, J.; Mukai, S.; et al. Commensal Microflora in Human Conjunctiva; Characteristics of Microflora in the Patients with Chronic Ocular Graft-versus-Host Disease. Ocul. Surf. 2019, 17, 265–271. [Google Scholar] [CrossRef]
- Li, J.; Liang, Q.; Huang, F.; Liao, Y.; Zhao, W.; Yang, J.; Wen, X.; Li, X.; Chen, T.; Guo, S.; et al. Metagenomic Profiling of the Ocular Surface Microbiome in Patients After Allogeneic Hematopoietic Stem Cell Transplantation. Am. J. Ophthalmol. 2022, 242, 144–155. [Google Scholar] [CrossRef]
- Andersson, J.; Vogt, J.K.; Dalgaard, M.D.; Pedersen, O.; Holmgaard, K.; Heegaard, S. Ocular Surface Microbiota in Patients with Aqueous Tear-Deficient Dry Eye. Ocul. Surf. 2020, 19, 210–217. [Google Scholar] [CrossRef]
- Giannaccare, G.; Bonifazi, F.; Sessa, M.; Dan, E.; Arpinati, M.; Fresina, M.; Bandini, G.; Cavo, M.; Versura, P.; Campos, E.C. Ocular Surface Analysis in Hematological Patients before and after Allogeneic Hematopoietic Stem Cell Transplantation: Implication for Daily Clinical Practice. Eye 2017, 31, 1417–1426. [Google Scholar] [CrossRef]
- Wolffsohn, J.S.; Arita, R.; Chalmers, R.; Djalilian, A.; Dogru, M.; Dumbleton, K.; Gupta, P.K.; Karpecki, P.; Lazreg, S.; Pult, H.; et al. TFOS DEWS II Diagnostic Methodology Report. Ocul. Surf. 2017, 15, 539–574. [Google Scholar] [CrossRef]
- Schiffman, R.M.; Christianson, M.D.; Jacobsen, G.; Hirsch, J.D.; Reis, B.L. Reliability and Validity of the Ocular Surface Disease Index. Arch. Ophthalmol. 2000, 118, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Lemp, M.A. Report of the National Eye Institute/Industry Workshop on Clinical Trials in Dry Eyes. CLAO J. 1995, 21, 221–232. [Google Scholar] [PubMed]
- Ousler, G.W.; Hagberg, K.W.; Schindelar, M.; Welch, D.; Abelson, M.B. The Ocular Protection Index. Cornea 2008, 27, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Versura, P.; Profazio, V.; Fresina, M.; Campos, E.C. A Novel Scraping Cytology Score System (SCSS) Grades Inflammation in Dry Eye Patients. Curr. Eye Res. 2009, 34, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.D.; Whitmer, D.; Nichols, K.K.; Tomlinson, A.; Foulks, G.N.; Geerling, G.; Pepose, J.S.; Kosheleff, V.; Porreco, A.; Lemp, M.A. An Objective Approach to Dry Eye Disease Severity. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6125–6130. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Kim, S.K.; Dana, R.; Clayton, J.; Jain, S.; Rosenblatt, M.I.; Perez, V.L.; Shikari, H.; Riemens, A.; Tsubota, K. International Chronic Ocular Graft-vs-Host-Disease (GVHD) Consensus Group: Proposed Diagnostic Criteria for Chronic GVHD (Part I). Sci. Rep. 2013, 3, 3419. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-End Assembler for Illumina Sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An Effective Distance Metric for Microbial Community Comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Bernabei, F.; Versura, P.; Pellegrini, M.; Moscardelli, F.; Bonifazi, F.; Sessa, M.; Arpinati, M.; Scorcia, V.; Giannaccare, G. Longitudinal Analysis of Infrared Meibography in Patients Undergoing Hematopoietic Stem Cell Transplantation. Cornea 2020, 39, 812. [Google Scholar] [CrossRef] [PubMed]
- Giannaccare, G.; Versura, P.; Bonifazi, F.; Sessa, M.; Campos, E.C. Comparison among Different Diagnostic Criteria for Chronic Ocular Graft-versus-Host Disease Applied with and without Pre-Transplant Ophthalmological Examination. Eye 2019, 33, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, M.; Giannaccare, G.; Bernabei, F.; Moscardelli, F.; Sessa, M.; Arpinati, M.; Bonifazi, F.; Versura, P. Longitudinal Corneal Endothelial Cell Changes in Patients Undergoing Hematopoietic Stem Cell Transplantation. Cornea 2021, 40, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-Based Metagenomics Analysis Reveals Markers for Gut Microbiome Composition and Diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Rothschild, D.; Leviatan, S.; Hanemann, A.; Cohen, Y.; Weissbrod, O.; Segal, E. An Atlas of Robust Microbiome Associations with Phenotypic Traits Based on Large-Scale Cohorts from Two Continents. PLoS ONE 2022, 17, e0265756. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, H.; Hu, M.; Ma, Y.; Chen, P.; Zhao, Z.; Li, J.; Ye, Y.; Zheng, M.; Lou, Y. Alterations in the Ocular Surface Microbiome in Traumatic Corneal Ulcer Patients. Investig. Ophthalmol. Vis. Sci. 2020, 61, 35. [Google Scholar] [CrossRef]
- Widman, A.; Reshef, R. Precision in Donor Selection: Identifying Ideal Stem-Cell Donors through Their T Cells. Exp. Hematol. 2016, 44, 1020–1023. [Google Scholar] [CrossRef]
- Chiang, J.C.B.; Zahari, I.; Markoulli, M.; Krishnan, A.V.; Park, S.B.; Semmler, A.; Goldstein, D.; Edwards, K. The Impact of Anticancer Drugs on the Ocular Surface. Ocul. Surf. 2020, 18, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Giannaccare, G.; Bonifazi, F.; Sessa, M.; Fresina, M.; Arpinati, M.; Bandini, G.; Versura, P. Dry Eye Disease Is Already Present in Hematological Patients Before Hematopoietic Stem Cell Transplantation. Cornea 2016, 35, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Giannaccare, G.; Pellegrini, M.; Bernabei, F.; Scorcia, V.; Campos, E. Ocular Surface System Alterations in Ocular Graft-versus-Host Disease: All the Pieces of the Complex Puzzle. Graefes Arch. Clin. Exp. Ophthalmol. 2019, 257, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.-L.; Sun, Y.-C.; Wu, J.-H.; Hsieh, Y.-T.; Huang, W.-L.; Chen, W.-L. The Ocular Graft-versus-Host Disease: The Path from Current Knowledge to Future Managements. Eye 2022, 37, 1982–1992. [Google Scholar] [CrossRef]
- Zilliox, M.J.; Gange, W.S.; Kuffel, G.; Mores, C.R.; Joyce, C.; de Bustros, P.; Bouchard, C.S. Assessing the Ocular Surface Microbiome in Severe Ocular Surface Diseases. Ocul. Surf. 2020, 18, 706–712. [Google Scholar] [CrossRef]
- Kim, J.; Choi, S.H.; Kim, Y.J.; Jeong, H.J.; Ryu, J.S.; Lee, H.J.; Kim, T.W.; Im, S.-H.; Oh, J.Y.; Kim, M.K. Clinical Effect of IRT-5 Probiotics on Immune Modulation of Autoimmunity or Alloimmunity in the Eye. Nutrients 2017, 9, 1166. [Google Scholar] [CrossRef]
- Wen, X.; Miao, L.; Deng, Y.; Bible, P.W.; Hu, X.; Zou, Y.; Liu, Y.; Guo, S.; Liang, J.; Chen, T.; et al. The Influence of Age and Sex on Ocular Surface Microbiota in Healthy Adults. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6030. [Google Scholar] [CrossRef]
| Characteristics | Value |
|---|---|
| Recipient sex (age) | |
| Females n. | 6 (57.5, 18–65) |
| Males n. | 18 (52, 24–68) |
| Diagnosis | |
| ALL | 4 (16.7%) |
| AML | 12 (50%) |
| APLASTIC ANEMIA | 1 (4.2%) |
| MDS/MPN | 5 (20.8%) |
| NHL | 2 (8.3%) |
| Source of stem cells | |
| Bone marrow | 3 (12.5%) |
| Peripheral blood stem cells | 21 (87.5%) |
| Donor age | 28.5 (19–51) |
| Type of donor | |
| Matched unrelated donor | 17 (70.8%) |
| Matched related donor | 7 (29.2%) |
| Donor–recipient sex matching | |
| Female to male | 11 (45.8%) |
| Others | 13 (54.2%) |
| HLA compatibility | |
| Matched | 12 (50%) |
| Mismatched (≥1 antigen) | 12 (50%) |
| Type of conditioning | |
| Myeloablative | 15 (62.5%) |
| Reduced intensity | 9 (37.5%) |
| ATLG | 21 (87.5%) |
| Parameters | Pre-HSCT | Post-HSCT | p |
|---|---|---|---|
| OSDI score | 11.1 ± 15.7 | 14.5 ± 14.3 | 0.33 |
| VAS | 1.4 ± 2.2 | 2.4 ± 2.3 | 0.03 |
| TFBUT (sec) | 7.2 ± 2.6 | 6.6 ± 3.4 | 0.40 |
| Schirmer test (mm length/5′) | 13.8 ± 7.7 | 17.9 ± 11.2 | 0.08 |
| Conjunctival staining (van Bijsterveldt score) | 2.1 ± 2.4 | 2.8 ± 2.9 | 0.31 |
| Corneal damage (NEI score) | 1.2 ± 0.8 | 2.3 ± 1.9 | 0.01 |
| MGD score (%) | 36.3 ± 10.2 | 33.5 ± 7.3 | 0.32 |
| Scraping cytology score | 3.6 ± 1.8 | 5.5 ± 2.8 | 0.01 |
| OPI | 1.4 ± 1.0 | 1.0 ± 0.6 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clougher, S.; Severgnini, M.; Marangoni, A.; Consolandi, C.; Camboni, T.; Morselli, S.; Arpinati, M.; Bonifazi, F.; Dicataldo, M.; Lazzarotto, T.; et al. Longitudinal Changes of Ocular Surface Microbiome in Patients Undergoing Hemopoietic Stem Cell Transplant (HSCT). J. Clin. Med. 2024, 13, 208. https://doi.org/10.3390/jcm13010208
Clougher S, Severgnini M, Marangoni A, Consolandi C, Camboni T, Morselli S, Arpinati M, Bonifazi F, Dicataldo M, Lazzarotto T, et al. Longitudinal Changes of Ocular Surface Microbiome in Patients Undergoing Hemopoietic Stem Cell Transplant (HSCT). Journal of Clinical Medicine. 2024; 13(1):208. https://doi.org/10.3390/jcm13010208
Chicago/Turabian StyleClougher, Suzanne, Marco Severgnini, Antonella Marangoni, Clarissa Consolandi, Tania Camboni, Sara Morselli, Mario Arpinati, Francesca Bonifazi, Michele Dicataldo, Tiziana Lazzarotto, and et al. 2024. "Longitudinal Changes of Ocular Surface Microbiome in Patients Undergoing Hemopoietic Stem Cell Transplant (HSCT)" Journal of Clinical Medicine 13, no. 1: 208. https://doi.org/10.3390/jcm13010208





