Leveraging the Academic Artificial Intelligence Silecosystem to Advance the Community Oncology Enterprise
Abstract
:1. Introduction
2. The AI Silecosystem as Kuhnian Paradigm
3. Origins of the AI Silecosystem: A Chronicle of an Emergent Paradigm
3.1. Inception: Articulation Anticipates Actualization
3.2. Intermission: Expectations Exceed Experience
3.3. Invigoration: Innovation Invites Implementation and Investment
3.3.1. Advances in Computer Hardware: The Engines That Power the AI Silecosystem
Quantum Computing
Artificial-Intelligence-Boosted Internet of Things (AIoT)
Distributive Edge Computing
Cloud Computing
Neuromorphic Computing
Analog Neural Networks
Monolithic-3D AI Systems
The Graphics Processing Unit
Analog, Non-Volatile Memory Devices
3.3.2. Advances in Data
Synthetic Data
Facilitating Culturally Representative AI Data Sets
Optimizing Data Deposition and Engineering
3.3.3. Advances in Software Algorithms: Piloting the AI Ecosystem
Generative AI
Virtual and Augmented Reality
Explainable Machine Learning
Generative Adversarial Networks
Neuro-Vector-Symbolic Architecture
The Democratization of Resources/Open-Source AI Software
4. Tribulations of the AI Silecosystem: Impending AI Winter or Early Twilight of a Paradigm in Demise?
5. The Academic Origins and Catalysis of the AI Silecosystem
6. Harnessing of the Academic Oncology AI Silecosystem to Advance Community Oncology Practice: The City of Hope Experience
6.1. Hardware Resources: High-Performance Computer Cluster
6.2. Data Resources
6.3. Software Resources
6.4. COH AI Silecosystem Engagement with the Community Oncology Network
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Poola, I. How artificial intelligence in impacting real life everyday. Int. J. Adv. Res. Dev. 2017, 2, 96–100. [Google Scholar]
- Lee, R.S.T. Artificial Intelligence in Daily Life; Springer: Singapore, 2020. [Google Scholar]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A new era: Artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.J.; Houssami, N.; Pozzi, G.; Séroussi, B. Artificial intelligence (AI) in breast cancer care–Leveraging multidisciplinary skills to improve care. Breast 2021, 56, 110–113. [Google Scholar] [CrossRef]
- Bhalla, S.; Laganà, A. Artificial intelligence for precision oncology. In Computational Methods for Precision Oncology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 249–268. [Google Scholar]
- Dlamini, Z.; Francies, F.Z.; Hull, R.; Marima, R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct. Biotechnol. J. 2020, 18, 2300–2311. [Google Scholar] [CrossRef]
- Rompianesi, G.; Pegoraro, F.; Ceresa, C.D.; Montalti, R.; Troisi, R.I. Artificial intelligence for precision oncology: Beyond patient stratification. NPJ Precis. Oncol. 2019, 3, 6. [Google Scholar]
- Rompianesi, G.; Pegoraro, F.; Ceresa, C.D.; Montalti, R.; Troisi, R.I. Artificial intelligence in the diagnosis and management of colorectal cancer liver metastases. World J. Gastroenterol. 2022, 28, 108. [Google Scholar] [CrossRef]
- Christie, J.R.; Lang, P.; Zelko, L.M.; Palma, D.A.; Abdelrazek, M.; Mattonen, S.A. Artificial intelligence in lung cancer: Bridging the gap between computational power and clinical decision-making. Can. Assoc. Radiol. J. 2021, 72, 86–97. [Google Scholar] [CrossRef]
- Derbal, Y. Can artificial intelligence improve cancer treatments? Health Inform. J. 2022, 28, 14604582221102314. [Google Scholar] [CrossRef]
- Ibrahim, A.; Gamble, P.; Jaroensri, R.; Abdelsamea, M.M.; Mermel, C.H.; Chen, P.-H.C.; Rakha, E.A. Artificial intelligence in digital breast pathology: Techniques and applications. Breast 2020, 49, 267–273. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Yang, M.; Wang, S.; Li, X.; Sun, Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020, 40, 154–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswanathan, V.S.; Toro, P.; Corredor, G.; Mukhopadhyay, S.; Madabhushi, A. The state of the art for artificial intelligence in lung digital pathology. J. Pathol. 2022, 257, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Försch, S.; Klauschen, F.; Hufnagl, P.; Roth, W. Artificial intelligence in pathology. Dtsch. Ärzteblatt Int. 2021, 118, 199. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Tran, W.T.; Sadeghi-Naini, A.; Lu, F.-I.; Gandhi, S.; Meti, N.; Brackstone, M.; Rakovitch, E.; Curpen, B. Computational radiology in breast cancer screening and diagnosis using artificial intelligence. Can. Assoc. Radiol. J. 2021, 72, 98–108. [Google Scholar] [CrossRef]
- Chassagnon, G.; Vakalopoulou, M.; Paragios, N.; Revel, M.-P. Artificial intelligence applications for thoracic imaging. Eur. J. Radiol. 2020, 123, 108774. [Google Scholar] [CrossRef] [Green Version]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Frownfelter, J.; Blau, S.; Page, R.D.; Showalter, J.; Miller, K.; Kish, J.; Valley, A.W.; Nabhan, C. Artificial intelligence (AI) to improve patient outcomes in community oncology practices. J. Clin. Oncol. 2019, 37, e18098. [Google Scholar] [CrossRef]
- Kappel, C.; Rushton-Marovac, M.; Leong, D.; Dent, S. Pursuing Connectivity in Cardio-Oncology Care—The Future of Telemedicine and Artificial Intelligence in Providing Equity and Access to Rural Communities. Front. Cardiovasc. Med. 2022, 9, 927769. [Google Scholar] [CrossRef]
- Ye, P.; Butler, B.; Vo, D.; He, B.; Turnwald, B.; Hoverman, J.R.; Indurlal, P.; Garey, J.S.; Hoang, S.N. The initial outcome of deploying a mortality prediction tool at community oncology practices. J. Clin. Oncol. 2022, 40, 1521. [Google Scholar] [CrossRef]
- Kuhn, T.S. The Structure of Scientific Revolutions; University of Chicago Press: Chicago, IL, USA, 1962; Volume XV, p. 172. [Google Scholar]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biol. 1990, 52, 99–115. [Google Scholar] [CrossRef]
- Rosenblatt, F. The Perceptron, a Perceiving and Recognizing Automaton Project Para; Cornell Aeronautical Laboratory: Buffalo, NY, USA, 1957. [Google Scholar]
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef] [Green Version]
- Turing, A.M. Computing machinery and intelligence. Mind 1950, 59, 433–460. [Google Scholar] [CrossRef]
- McCarthy, J.; Minsky, M.; Rochester, N.; Shannon, C.E. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955. AI Mag. 2006, 27, 12–14. [Google Scholar]
- Pierce, J.R.; Carroll, J.B. Language and Machines: Computers in Translation and Linguistics; National Academies Press: Washington, DC, USA, 1966. [Google Scholar]
- Science Research Council. Artificial Intelligence; a Paper Symposium; Science Research Council: London, UK, 1973; p. iv. 45p. [Google Scholar]
- ICOT. Shin-Sedai-Konpyūta-Gijutsu-Kaihatsu-Kikō, FGCS’92. Fifth Generation Computer Systems; IOS Press: Amsterdam, The Netherlands, 1992; Volume 1. [Google Scholar]
- Mack, C.A. Fifty years of Moore’s law. IEEE Trans. Semicond. Manuf. 2011, 24, 202–207. [Google Scholar] [CrossRef]
- Gepner, P.; Kowalik, M.K. Multi-core processors: New way to achieve high system performance. In Proceedings of the International Symposium on Parallel Computing in Electrical Engineering (PARELEC’06), Bialystok, Poland, 13–17 September 2006. [Google Scholar]
- Goda, K.; Kitsuregawa, M. The history of storage systems. Proc. IEEE 2012, 100, 1433–1440. [Google Scholar] [CrossRef]
- Arute, F.; Arya, K.; Babbush, R.; Bacon, D.; Bardin, J.C.; Barends, R.; Biswas, R.; Boixo, S.; Brandao, F.G.; Buell, D.A. Quantum supremacy using a programmable superconducting processor. Nature 2019, 574, 505–510. [Google Scholar] [CrossRef] [Green Version]
- Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural products for drug discovery in the 21st century: Innovations for novel drug discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Ziauddin, J.; Leonchyk, P.; Yenkanchi, S.; Geraci, J. Quantum and classical machine learning for the classification of non-small-cell lung cancer patients. SN Appl. Sci. 2020, 2, 1088. [Google Scholar] [CrossRef]
- Davids, J.; Lidströmer, N.; Ashrafian, H. Artificial Intelligence in Medicine Using Quantum Computing in the Future of Healthcare. In Artificial Intelligence in Medicine; Springer: Berlin/Heidelberg, Germany, 2022; pp. 423–446. [Google Scholar]
- Niraula, D.; Jamaluddin, J.; Matuszak, M.M.; Haken, R.K.T.; Naqa, I.E. Quantum deep reinforcement learning for clinical decision support in oncology: Application to adaptive radiotherapy. Sci. Rep. 2021, 11, 23545. [Google Scholar] [CrossRef]
- Majumdar, R.; Baral, B.; Bhalgamiya, B.; Roy, T.D. Histopathological Cancer Detection Using Hybrid Quantum Computing. arXiv 2023, arXiv:2302.04633. [Google Scholar]
- Madakam, S.; Lake, V.; Lake, V.; Lake, V. Internet of Things (IoT): A literature review. J. Comput. Commun. 2015, 3, 164. [Google Scholar] [CrossRef] [Green Version]
- Čolaković, A.; Hadžialić, M. Internet of Things (IoT): A review of enabling technologies, challenges, and open research issues. Comput. Netw. 2018, 144, 17–39. [Google Scholar] [CrossRef]
- Valsalan, P.; Baomar, T.A.B.; Baabood, A.H.O. IoT based health monitoring system. J. Crit. Rev. 2020, 7, 739–743. [Google Scholar]
- Yuehong, Y.; Zeng, Y.; Chen, X.; Fan, Y. The internet of things in healthcare: An overview. J. Ind. Inf. Integr. 2016, 1, 3–13. [Google Scholar]
- Saloni, S.; Hegde, A. WiFi-aware as a connectivity solution for IoT pairing IoT with WiFi aware technology: Enabling new proximity based services. In Proceedings of the 2016 International Conference on Internet of Things and Applications (IOTA), Pune, India, 22–24 January 2016. [Google Scholar]
- Aldhyani, T.H.; Khan, M.A.; Almaiah, M.A.; Alnazzawi, N.; Hwaitat, A.K.A.; Elhag, A.; Shehab, R.T.; Alshebami, A.S. A Secure internet of medical things Framework for Breast Cancer Detection in Sustainable Smart Cities. Electronics 2023, 12, 858. [Google Scholar] [CrossRef]
- Jabarulla, M.Y.; Lee, H.-N. A blockchain and artificial intelligence-based, patient-centric healthcare system for combating the COVID-19 pandemic: Opportunities and applications. Healthcare 2021, 9, 1019. [Google Scholar] [CrossRef]
- Srinivasu, P.N.; Ijaz, M.F.; Shafi, J.; Woźniak, M.; Sujatha, R. 6G driven fast computational networking framework for healthcare applications. IEEE Access 2022, 10, 94235–94248. [Google Scholar] [CrossRef]
- Prayitno; Shyu, C.R.; Putra, K.T.; Chen, H.C.; Tsai, Y.Y.; Hossain, K.T.; Jiang, W.; Shae, Z.Y. A systematic review of federated learning in the healthcare area: From the perspective of data properties and applications. Appl. Sci. 2021, 11, 11191. [Google Scholar] [CrossRef]
- Sung, T.-W.; Tsai, P.-W.; Gaber, T.; Lee, C.-Y. Artificial Intelligence of Things (AIoT) technologies and applications. Wirel. Commun. Mob. Comput. 2021, 2021, 9781271. [Google Scholar] [CrossRef]
- Krasniqi, X.; Hajrizi, E. Use of IoT technology to drive the automotive industry from connected to full autonomous vehicles. IFAC-Pap. 2016, 49, 269–274. [Google Scholar] [CrossRef]
- Jia, W.; Wang, S.; Xie, Y.; Chen, Z.; Gong, K. Disruptive technology identification of intelligent logistics robots in AIoT industry: Based on attributes and functions analysis. Syst. Res. Behav. Sci. 2022, 39, 557–568. [Google Scholar] [CrossRef]
- Wazid, M.; Das, A.K.; Park, Y. Blockchain-Envisioned Secure Authentication Approach in AIoT: Applications, Challenges, and Future Research. Wirel. Commun. Mob. Comput. 2021, 2021, 3866006. [Google Scholar] [CrossRef]
- Perez, F.; Nolde, M.; Crane, T.E.; Kebria, M.; Chan, K.; Dellinger, T.; Sun, V. Integrative review of remote patient monitoring in gynecologic and urologic surgical oncology. J. Surg. Oncol. 2023, 127, 1054–1061. [Google Scholar] [CrossRef]
- Chen, J.; Ran, X. Deep learning with edge computing: A review. Proc. IEEE 2019, 107, 1655–1674. [Google Scholar] [CrossRef]
- Uddin, M.Z. A wearable sensor-based activity prediction system to facilitate edge computing in smart healthcare system. J. Parallel Distrib. Comput. 2019, 123, 46–53. [Google Scholar] [CrossRef]
- Verma, P.; Fatima, S. Smart Healthcare Applications and Real-Tme Analytics through Edge Computing. In Internet of Things Use Cases for the Healthcare Industry; Springer: Cham, Switzerland, 2020; pp. 241–270. [Google Scholar]
- Cao, B.; Zhang, L.; Li, Y.; Feng, D.; Cao, W. Intelligent offloading in multi-access edge computing: A state-of-the-art review and framework. IEEE Commun. Mag. 2019, 57, 56–62. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, X.; Li, E.; Zeng, L.; Luo, K.; Zhang, J. Edge intelligence: Paving the last mile of artificial intelligence with edge computing. Proc. IEEE 2019, 107, 1738–1762. [Google Scholar] [CrossRef] [Green Version]
- Deng, S.; Zhao, H.; Fang, W.; Yin, J.; Dustdar, S.; Zomaya, A.Y. Edge intelligence: The confluence of edge computing and artificial intelligence. IEEE Internet Things J. 2020, 7, 7457–7469. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, A.; Kassem, H.; Padoy, N.; Umeton, R.; Karargyris, A. A Review of Medical Federated Learning: Applications in Oncology and Cancer Research. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, proceedings of the 7th International Workshop, BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, 27 September 2021; Springer: Cham, Switzerland, 2022; Part I. [Google Scholar]
- Rodríguez, C. AIoT for Achieving Sustainable Development Goals. In Proceedings of the 4th International Conference on Recent Trends in Advanced Computing, VIT, Chennai, India, 11–12 November 2021. [Google Scholar]
- Rahimi, M.; Navimipour, N.J.; Hosseinzadeh, M.; Moattar, M.H.; Darwesh, A. A systematic review on cloud computing. J. Supercomput. 2014, 68, 1321–1346. [Google Scholar]
- Dang, L.M.; Piran, M.J.; Han, D.; Min, K.; Moon, H. Cloud healthcare services: A comprehensive and systematic literature review. Trans. Emerg. Telecommun. Technol. 2022, 33, e4473. [Google Scholar]
- Raza, K.; Qazi, S.; Sahu, A.; Verma, S. Computational Intelligence in Oncology: Past, Present, and Future. In Computational Intelligence in Oncology: Applications in Diagnosis, Prognosis and Therapeutics of Cancers; Springer: Berlin/Heidelberg, Germany, 2022; pp. 3–18. [Google Scholar]
- Liu, X.; Luo, X.; Jiang, C.; Zhao, H. Difficulties and challenges in the development of precision medicine. Clin. Genet. 2019, 95, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Schuman, C.D.; Kulkarni, S.R.; Parsa, M.; Mitchell, J.P.; Date, P.; Kay, B. Opportunities for neuromorphic computing algorithms and applications. Nat. Comput. Sci. 2022, 2, 10–19. [Google Scholar] [CrossRef]
- Mead, C. Neuromorphic electronic systems. Proc. IEEE 1990, 78, 1629–1636. [Google Scholar] [CrossRef]
- Indiveri, G.; Linares-Barranco, B.; Hamilton, T.J.; Schaik, A.V.; Etienne-Cummings, R.; Delbruck, T.; Liu, S.-C.; Dudek, P.; Häfliger, P.; Renaud, S. Neuromorphic silicon neuron circuits. Front. Neurosci. 2011, 5, 73. [Google Scholar] [CrossRef] [Green Version]
- Thakur, C.S.; Molin, J.L.; Cauwenberghs, G.; Indiveri, G.; Kumar, K.; Qiao, N.; Schemmel, J.; Wang, R.; Chicca, E.; Olson Hasler, J. Large-scale neuromorphic spiking array processors: A quest to mimic the brain. Front. Neurosci. 2018, 12, 891. [Google Scholar] [CrossRef]
- Bulárka, S.; Gontean, A. Brain-computer interface review. In Proceedings of the 2016 12th IEEE International Symposium on Electronics and Telecommunications (ISETC), Timisoara, Romania, 27–28 October 2016. [Google Scholar]
- Yu, Z.; Zahid, A.; Ansari, S.; Abbas, H.; Abdulghani, A.M.; Heidari, H.; Imran, M.A.; Abbasi, Q.H. Hardware-based hopfield neuromorphic computing for fall detection. Sensors 2020, 20, 7226. [Google Scholar] [CrossRef] [PubMed]
- Ceolini, E.; Frenkel, C.; Shrestha, S.B.; Taverni, G.; Khacef, L.; Payvand, M.; Donati, E. Hand-gesture recognition based on EMG and event-based camera sensor fusion: A benchmark in neuromorphic computing. Front. Neurosci. 2020, 14, 637. [Google Scholar] [CrossRef]
- Aitsam, M.; Davies, S.; Di Nuovo, A. Neuromorphic Computing for Interactive Robotics: A Systematic Review. IEEE Access 2022, 10, 122261–122279. [Google Scholar] [CrossRef]
- Yu, Z.; Abdulghani, A.M.; Zahid, A.; Heidari, H.; Imran, M.A.; Abbasi, Q.H. An overview of neuromorphic computing for artificial intelligence enabled hardware-based hopfield neural network. IEEE Access 2020, 8, 67085–67099. [Google Scholar] [CrossRef]
- Sun, B.; Guo, T.; Zhou, G.; Ranjan, S.; Jiao, Y.; Wei, L.; Zhou, Y.N.; Wu, Y.A. Synaptic devices based neuromorphic computing applications in artificial intelligence. Mater. Today Phys. 2021, 18, 100393. [Google Scholar] [CrossRef]
- Roy, K.; Jaiswal, A.; Panda, P. Towards spike-based machine intelligence with neuromorphic computing. Nature 2019, 575, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Pierangeli, D.; Palmieri, V.; Marcucci, G.; Moriconi, C.; Perini, G.; De Spirito, M.; Papi, M.; Conti, C. Optical neural network by disordered tumor spheroids. In Proceedings of the 2019 Conference on Lasers and Electro-Optics Europe & European Quantum Electronics Conference (CLEO/Europe-EQEC), Munich, Germany, 23–27 June 2019. [Google Scholar]
- Rong, G.; Mendez, A.; Assi, E.B.; Zhao, B.; Sawan, M. Artificial intelligence in healthcare: Review and prediction case studies. Engineering 2020, 6, 291–301. [Google Scholar] [CrossRef]
- Pierangeli, D.; Palmieri, V.; Marcucci, G.; Moriconi, C.; Perini, G.; De Spirito, M.; Papi, M.; Conti, C. Optical Neural Network for Cancer Morphodynamics Sensing. In Nonlinear Optics; Optica Publishing Group: Washington, DC, USA, 2019. [Google Scholar]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- DasGupta, B.; Schnitger, G. Analog versus discrete neural networks. Neural Comput. 1996, 8, 805–818. [Google Scholar] [CrossRef]
- Kakkar, V. Comparative study on analog and digital neural networks. Int. J. Comput. Sci. Netw. Secur. 2009, 9, 14–21. [Google Scholar]
- Xiao, T.P.; Bennett, C.H.; Feinberg, B.; Agarwal, S.; Marinella, M.J. Analog architectures for neural network acceleration based on non-volatile memory. Appl. Phys. Rev. 2020, 7, 031301. [Google Scholar] [CrossRef]
- Cramer, B.; Billaudelle, S.; Kanya, S.; Leibfried, A.; Grübl, A.; Karasenko, V.; Pehle, C.; Schreiber, K.; Stradmann, Y.; Weis, J.; et al. Surrogate gradients for analog neuromorphic computing. Proc. Natl. Acad. Sci. USA 2022, 119, e2109194119. [Google Scholar] [CrossRef]
- Chandrasekaran, S.T.; Jayaraj, A.; Karnam, V.E.G.; Banerjee, I.; Sanyal, A. Fully integrated analog machine learning classifier using custom activation function for low resolution image classification. IEEE Trans. Circuits Syst. I Regul. Pap. 2021, 68, 1023–1033. [Google Scholar] [CrossRef]
- Pan, C.-H.; Hsieh, H.-Y.; Tang, K.-T. An analog multilayer perceptron neural network for a portable electronic nose. Sensors 2012, 13, 193–207. [Google Scholar] [CrossRef]
- Odame, K.; Nyamukuru, M.; Shahghasemi, M.; Bi, S.; Kotz, D. Analog Gated Recurrent Unit Neural Network for Detecting Chewing Events. IEEE Trans. Biomed. Circuits Syst. 2022, 16, 1106–1115. [Google Scholar] [CrossRef]
- Perfetti, R.; Ricci, E. Analog neural network for support vector machine learning. IEEE Trans. Neural Netw. 2006, 17, 1085–1091. [Google Scholar] [CrossRef]
- Krestinskaya, O.; AJames, P.; Chua, L.O. Neuromemristive circuits for edge computing: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 31, 4–23. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.; Shin, K.; Jeon, D. Enhancing reliability of analog neural network processors. IEEE Trans. Very Large Scale Integr. (VLSI) Syst. 2019, 27, 1455–1459. [Google Scholar] [CrossRef]
- Geske, G.; Stupmann, F.; Wego, A. High speed color recognition with an analog neural network chi. In Proceedings of the IEEE International Conference on Industrial Technology, Maribor, Slovenia, 10–12 December 2003. [Google Scholar]
- Kieffer, C.; Genot, A.J.; Rondelez, Y.; Gines, G. Molecular Computation for Molecular Classification. Adv. Biol. 2023, 7, 2200203. [Google Scholar] [CrossRef]
- Pattichis, C.; Schnorrenberg, F.; Schizas, C.; Pattichis, M.; Kyriacou, K. A Modular Neural Network System for the Analysis of Nuclei in Histopathological Sections. In Computational Intelligence Processing in Medical Diagnosis; Physica: Heidelberg, Germany, 2002; pp. 291–322. [Google Scholar]
- Morro, A.; Canals, V.; Oliver, A.; Alomar, M.L.; Galan-Prado, F.; Ballester, P.J.; Rossello, J.L. A stochastic spiking neural network for virtual screening. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 1371–1375. [Google Scholar] [CrossRef]
- Jiang, J.; Parto, K.; Cao, W.; Banerjee, K. Ultimate monolithic-3D integration with 2D materials: Rationale, prospects, and challenges. IEEE J. Electron Devices Soc. 2019, 7, 878–887. [Google Scholar] [CrossRef]
- Wong, S.; El-Gamal, A.; Griffin, P.; Nishi, Y.; Pease, F.; Plummer, J. Monolithic 3D integrated circuits. In Proceedings of the 2007 International Symposium on VLSI Technology, Systems and Applications (VLSI-TSA), Hsinchu, Taiwan, 23–25 April 2007. [Google Scholar]
- Torres-Mapa, M.L.; Singh, M.; Simon, O.; Mapa, J.L.; Machida, M.; Günther, A.; Roth, B.; Heinemann, D.; Terakawa, M.; Heisterkamp, A. Fabrication of a monolithic lab-on-a-chip platform with integrated hydrogel waveguides for chemical sensing. Sensors 2019, 19, 4333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dematté, L.; Prandi, D. GPU computing for systems biology. Brief. Bioinform. 2010, 11, 323–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaki, G.; Plishker, W.; Li, W.; Lee, J.; Quon, H.; Wong, J.; Shekhar, R. The utility of cloud computing in analyzing GPU-accelerated deformable image registration of CT and CBCT images in head and neck cancer radiation therapy. IEEE J. Transl. Eng. Health Med. 2016, 4, 4300311. [Google Scholar] [CrossRef]
- Chakrabarty, S.; Abidi, S.A.; Mousa, M.; Mokkarala, M.; Hren, I.; Yadav, D.; Kelsey, M.; LaMontagne, P.; Wood, J.; Adams, M. Integrative Imaging Informatics for Cancer Research: Workflow Automation for Neuro-Oncology (I3CR-WANO). JCO Clin. Cancer Inform. 2023, 7, e2200177. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.I.; Keshavarzi, A.; Datta, S. The future of ferroelectric field-effect transistor technology. Nat. Electron. 2020, 3, 588–597. [Google Scholar] [CrossRef]
- Ajayan, J.; Mohankumar, P.; Nirmal, D.; Joseph, L.L.; Bhattacharya, S.; Sreejith, S.; Kollem, S.; Rebelli, S.; Tayal, S.; Mounika, B. Ferroelectric Field Effect Transistors (FeFETs): Advancements, Challenges and Exciting Prospects for Next Generation Non-Volatile Memory (NVM) Applications. Mater. Today Commun. 2023, 35, 105591. [Google Scholar] [CrossRef]
- Pan, F.; Gao, S.; Chen, C.; Song, C.; Zeng, F. Recent progress in resistive random access memories: Materials, switching mechanisms, and performance. Mater. Sci. Eng. R Rep. 2014, 83, 1–59. [Google Scholar] [CrossRef]
- Gupta, V.; Kapur, S.; Saurabh, S.; Grover, A. Resistive random access memory: A review of device challenges. IETE Tech. Rev. 2020, 37, 377–390. [Google Scholar] [CrossRef]
- Wu, H.; Wang, X.H.; Gao, B.; Deng, N.; Lu, Z.; Haukness, B.; Bronner, G.; Qian, H. Resistive random access memory for future information processing system. Proc. IEEE 2017, 105, 1770–1789. [Google Scholar] [CrossRef]
- Girard, P.; Cheng, Y.; Virazel, A.; Zhao, W.; Bishnoi, R.; Tahoori, M.B. A survey of test and reliability solutions for magnetic random access memories. Proc. IEEE 2020, 109, 149–169. [Google Scholar] [CrossRef]
- Sethu, K.K.V.; Ghosh, S.; Couet, S.; Swerts, J.; Sorée, B.; De Boeck, J.; Kar, G.S.; Garello, K. Optimization of Tungsten β-phase window for spin-orbit-torque magnetic random-access memory. Phys. Rev. Appl. 2021, 16, 064009. [Google Scholar] [CrossRef]
- Chen, A. A review of emerging non-volatile memory (NVM) technologies and applications. Solid-State Electron. 2016, 125, 25–38. [Google Scholar] [CrossRef]
- Si, M.; Cheng, H.-Y.; Ando, T.; Hu, G.; Ye, P.D. Overview and outlook of emerging non-volatile memories. MRS Bull. 2021, 46, 946–958. [Google Scholar] [CrossRef]
- Noé, P.; Vallée, C.; Hippert, F.; Fillot, F.; Raty, J.-Y. Phase-change materials for non-volatile memory devices: From technological challenges to materials science issues. Semicond. Sci. Technol. 2018, 33, 013002. [Google Scholar] [CrossRef]
- Ambrogio, S.; Narayanan, P.; Tsai, H.; Mackin, C.; Spoon, K.; Chen, A.; Fasoli, A.; Friz, A.; Burr, G.W. Accelerating deep neural networks with analog memory devices. In Proceedings of the 2020 2nd IEEE International Conference on Artificial Intelligence Circuits and Systems (AICAS), Genova, Italy, 31 August 2020–2 September 2020. [Google Scholar]
- Abunahla, H.; Halawani, Y.; Alazzam, A.; Mohammad, B. NeuroMem: Analog graphene-based resistive memory for artificial neural networks. Sci. Rep. 2020, 10, 9473. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zarcone, R.V.; Levy, A.; Khwa, W.-S.; Raina, P.; Olshausen, B.A.; Wong, H.P. High-density analog image storage in an analog-valued non-volatile memory array. Neuromorphic Comput. Eng. 2022, 2, 044018. [Google Scholar] [CrossRef]
- Byun, S.-J.; Kim, D.-G.; Park, K.-D.; Choi, Y.-J.; Kumar, P.; Ali, I.; Kim, D.-G.; Yoo, J.-M.; Huh, H.-K.; Jung, Y.-J.; et al. A Low-Power Analog Processor-in-Memory-Based Convolutional Neural Network for Biosensor Applications. Sensors 2022, 22, 4555. [Google Scholar] [CrossRef]
- Tzouvadaki, I.; Gkoupidenis, P.; Vassanelli, S.; Wang, S.; Prodromakis, T. Interfacing Biology and Electronics with Memristive Materials. Adv. Mater. 2023, e2210035, early view. [Google Scholar] [CrossRef]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef]
- Sayood, K. Introduction to Data Compression; Morgan Kaufmann: Burlington, MA, USA, 2017. [Google Scholar]
- Dirik, C.; Jacob, B. The performance of PC solid-state disks (SSDs) as a function of bandwidth, concurrency, device architecture, and system organization. ACM SIGARCH Comput. Archit. News 2009, 37, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Meena, J.S.; Sze, S.M.; Chand, U.; Tseng, T.-Y. Overview of emerging nonvolatile memory technologies. Nanoscale Res. Lett. 2014, 9, 526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. A review of feature selection methods on synthetic data. Knowl. Inf. Syst. 2013, 34, 483–519. [Google Scholar] [CrossRef]
- Raghunathan, T.E. Synthetic data. Annu. Rev. Stat. Its Appl. 2021, 8, 129–140. [Google Scholar] [CrossRef]
- Arora, A.; Arora, A. Generative adversarial networks and synthetic patient data: Current challenges and future perspectives. Future Healthc. J. 2022, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Rajotte, J.-F.; Bergen, R.; Buckeridge, D.L.; El Emam, K.; Ng, R.; Strome, E. Synthetic data as an enabler for machine learning applications in medicine. Iscience 2022, 25, 105331. [Google Scholar] [CrossRef] [PubMed]
- Delaney, A.M.; Brophy, E.; Ward, T.E. Synthesis of realistic ECG using generative adversarial networks. arXiv 2019, arXiv:1909.09150. [Google Scholar]
- Ramesh, V.; Vatanparvar, K.; Nemati, E.; Nathan, V.; Rahman, M.M.; Kuang, J. Coughgan: Generating synthetic coughs that improve respiratory disease classification. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar]
- Braddon, A.E.; Robinson, S.; Alati, R.; Betts, K.S. Exploring the utility of synthetic data to extract more value from sensitive health data assets: A focused example in perinatal epidemiology. Paediatr. Perinat. Epidemiol. 2022, 37, 292–300. [Google Scholar] [CrossRef]
- Thomas, J.A.; Foraker, R.E.; Zamstein, N.; Morrow, J.D.; Payne, P.R.; Wilcox, A.B. Demonstrating an approach for evaluating synthetic geospatial and temporal epidemiologic data utility: Results from analyzing >1.8 million SARS-CoV-2 tests in the United States National COVID Cohort Collaborative (N3C). J. Am. Med. Inform. Assoc. 2022, 29, 1350–1365. [Google Scholar] [CrossRef]
- Hernandez, M.; Epelde, G.; Beristain, A.; Álvarez, R.; Molina, C.; Larrea, X.; Alberdi, A.; Timoleon, M.; Bamidis, P.; Konstantinidis, E. Incorporation of synthetic data generation techniques within a controlled data processing workflow in the health and wellbeing domain. Electronics 2022, 11, 812. [Google Scholar] [CrossRef]
- Gonzales, A.; Guruswamy, G.; Smith, S.R. Synthetic data in health care: A narrative review. PLoS Digit. Health 2023, 2, e0000082. [Google Scholar] [CrossRef]
- D’Amico, S.; Dall’Olio, D.; Sala, C.; Dall’Olio, L.; Sauta, E.; Zampini, M.; Asti, G.; Lanino, L.; Maggioni, G.; Campagna, A. Synthetic data generation by artificial intelligence to accelerate research and precision medicine in hematology. JCO Clin. Cancer Inform. 2023, 7, e2300021. [Google Scholar] [CrossRef]
- Hahn, W.; Schütte, K.; Schultz, K.; Wolkenhauer, O.; Sedlmayr, M.; Schuler, U.; Eichler, M.; Bej, S.; Wolfien, M. Contribution of Synthetic Data Generation towards an Improved Patient Stratification in Palliative Care. J. Pers. Med. 2022, 12, 1278. [Google Scholar] [CrossRef] [PubMed]
- Elias, A.; Paradies, Y. The costs of institutional racism and its ethical implications for healthcare. J. Bioethical Inq. 2021, 18, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J. Racism, Inequality, And Health Care for African Americans. The Century Foundation, 19 December 2019. [Google Scholar]
- Matalon, D.R.; Zepeda-Mendoza, C.J.; Aarabi, M.; Brown, K.; Fullerton, S.M.; Kaur, S.; Quintero-Rivera, F.; Vatta, M.; Social, E.A.; Issues, C.L.; et al. Clinical, technical, and environmental biases influencing equitable access to clinical genetics/genomics testing: A points to consider statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2023, 25, 100812. [Google Scholar] [CrossRef]
- Dankwa-Mullan, I.; Weeraratne, D. Artificial intelligence and machine learning technologies in cancer care: Addressing disparities, bias, and data diversity. Cancer Discov. 2022, 12, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.V.; Chen, H.; Edwards, M.A.; Faber, L.; Freischlag, J.A. A new look at an old problem: Improving diversity, equity, and inclusion in scientific research. Am. Surg. 2021, 87, 1722–1726. [Google Scholar] [CrossRef]
- Eubanks, V. Automating Inequality: How High-Tech Tools Profile, Police, and Punish the Poor; St. Martin’s Press: New York, NY, USA, 2018. [Google Scholar]
- Holstein, K.; Vaughan, J.W.; Daumé, H.; Dudik, M.; Wallach, H. Improving Fairness in Machine Learning Systems: What Do Industry Practitioners Need? Association for Computing Machinery: New York, NY, USA, 2019; pp. 1–16. [Google Scholar]
- Cachat-Rosset, G.; Klarsfeld, A. Diversity, Equity, and Inclusion in Artificial Intelligence: An Evaluation of Guidelines. Appl. Artif. Intell. 2023, 37, 2176618. [Google Scholar] [CrossRef]
- Washington, V.; Franklin, J.B.; Huang, E.S.; Mega, J.L.; Abernethy, A.P. Diversity, equity, and inclusion in clinical research: A path toward precision health for everyone. Clin. Pharmacol. Ther. 2023, 113, 575–584. [Google Scholar] [CrossRef]
- Al-Jarrah, O.Y.; Yoo, P.D.; Muhaidat, S.; Karagiannidis, G.K.; Taha, K. Efficient machine learning for big data: A review. Big Data Res. 2015, 2, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Anh, T.T.; Luong, N.C.; Niyato, D.; Kim, D.I.; Wang, L.-C. Efficient training management for mobile crowd-machine learning: A deep reinforcement learning approach. IEEE Wirel. Commun. Lett. 2019, 8, 1345–1348. [Google Scholar] [CrossRef] [Green Version]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M. Applications of machine learning in drug discovery and development. Nature reviews. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Ng, K.; Steinhubl, S.R.; DeFilippi, C.; Dey, S.; Stewart, W.F. Early detection of heart failure using electronic health records: Practical implications for time before diagnosis, data diversity, data quantity, and data density. Circ. Cardiovasc. Qual. Outcomes 2016, 9, 649–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, H.-H.; Wei, L.; Cui, S.; Luo, Y.; Haken, R.K.T.; El Naqa, I. Machine learning and imaging informatics in oncology. Oncol. 2020, 98, 344–362. [Google Scholar] [CrossRef] [PubMed]
- Corrie, B.D.; Marthandan, N.; Zimonja, B.; Jaglale, J.; Zhou, Y.; Barr, E.; Knoetze, N.; Breden, F.M.; Christley, S.; Scott, J.K. iReceptor: A platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev. 2018, 284, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Shakhovska, N.; Bolubash, Y.J.; Veres, O. Big data federated repository model. In Proceedings of the Experience of Designing and Application of CAD Systems in Microelectronics, Lviv, Ukraine, 24–27 February 2015. [Google Scholar]
- Barnes, C.; Bajracharya, B.; Cannalte, M.; Gowani, Z.; Haley, W.; Kass-Hout, T.; Hernandez, K.; Ingram, M.; Juvvala, H.P.; Kuffel, G. The Biomedical Research Hub: A federated platform for patient research data. J. Am. Med. Inform. Assoc. 2022, 29, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Crabtree, J.; Dillo, I.; Downs, R.R.; Edmunds, R.; Giaretta, D.; De Giusti, M.; L’Hours, H.; Hugo, W.; Jenkyns, R. The TRUST Principles for digital repositories. Sci. Data 2020, 7, 144. [Google Scholar] [CrossRef]
- Yozwiak, N.L.; Schaffner, S.F.; Sabeti, P.C. Data sharing: Make outbreak research open access. Nature 2015, 518, 477–479. [Google Scholar] [CrossRef] [Green Version]
- Romero, O.; Wrembel, R. Data engineering for data science: Two sides of the same coin. In Proceedings of the Big Data Analytics and Knowledge Discovery: 22nd International Conference, DaWaK 2020, Bratislava, Slovakia, 14–17 September 2020; p. 22. [Google Scholar]
- Tamburri, D.; van den Heuvel, W.-J. Big Data Engineering, in Data Science for Entrepreneurship: Principles and Methods for Data Engineering. In Analytics, Entrepreneurship, and the Society; Springer: Berlin/Heidelberg, Germany, 2023; pp. 25–35. [Google Scholar]
- Schelter, S.; Stoyanovich, J. Taming technical bias in machine learning pipelines. Bull. Tech. Comm. Data Eng. 2020, 43, 1926250. [Google Scholar]
- Gudivada, V.; Apon, A.; Ding, J. Data quality considerations for big data and machine learning: Going beyond data cleaning and transformations. Int. J. Adv. Softw. 2017, 10, 1–20. [Google Scholar]
- Chu, X.; Ilyas, I.F.; Krishnan, S.; Wang, J. Data cleaning: Overview and emerging challenges. In Proceedings of the 2016 International Conference on Management of Data, San Francisco, CA, USA, 26 June–1 July 2016. [Google Scholar]
- Martinez, D.; Malyska, N.; Streilein, B.; Caceres, R.; Campbell, W.; Dagli, C.; Gadepally, V.; Greenfield, K.; Hall, R.; King, A. Artificial Intelligence: Short History, Present Developments, and Future Outlook; MIT Lincoln Laboratory: Lexington, MA, USA, 2019. [Google Scholar]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, H.; Nakayama, K.I. Artificial intelligence in oncology. Cancer Sci. 2020, 111, 1452–1460. [Google Scholar] [CrossRef] [Green Version]
- Khodabandehlou, S.; Zivari Rahman, M. Comparison of supervised machine learning techniques for customer churn prediction based on analysis of customer behavior. J. Syst. Inf. Technol. 2017, 19, 65–93. [Google Scholar] [CrossRef]
- Hambarde, K.; Silahtaroğlu, G.; Khamitkar, S.; Bhalchandra, P.; Shaikh, H.; Kulkarni, G.; Tamsekar, P.; Samale, P. Data Analytics Implemented over E-Commerce Data to Evaluate Performance of Supervised Learning Approaches in Relation to Customer Behavior. In Soft Computing for Problem Solving: SocProS 2018; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1. [Google Scholar]
- Liu, M.; Ylanko, J.; Weekman, E.; Beckett, T.; Andrews, D.; McLaurin, J. Utilizing supervised machine learning to identify microglia and astrocytes in situ: Implications for large-scale image analysis and quantification. J. Neurosci. Methods 2019, 328, 108424. [Google Scholar] [CrossRef]
- Janssens, T.; Antanas, L.; Derde, S.; Vanhorebeek, I.; Van den Berghe, G.; Grandas, F.G. CHARISMA: An integrated approach to automatic H&E-stained skeletal muscle cell segmentation using supervised learning and novel robust clump splitting. Med. Image Anal. 2013, 17, 1206–1219. [Google Scholar] [PubMed] [Green Version]
- Wani, M.A.; Bhat, F.A.; Afzal, S.; Khan, A.I.; Wani, M.A.; Bhat, F.A.; Afzal, S.; Khan, A.I. Supervised deep learning in face recognition. In Advances in Deep Learning. Studies in Big Data; Springer: Singapore, 2020; pp. 95–110. [Google Scholar]
- Nagaraj, P.; Banala, R.; Prasad, A.K. Real Time Face Recognition using Effective Supervised Machine Learning Algorithms. J. Physics Conf. Ser. 2021, 1998, 012007. [Google Scholar] [CrossRef]
- Han, T.; Yao, H.; Sun, X.; Zhao, S.; Zhang, Y. Unsupervised discovery of crowd activities by saliency-based clustering. Neurocomputing 2016, 171, 347–361. [Google Scholar] [CrossRef]
- Xu, S.; Ho, E.S.; Aslam, N.; Shum, H.P. Unsupervised abnormal behaviour detection with overhead crowd video. In Proceedings of the 2017 11th International Conference on Software, Knowledge, Information Management and Applications (SKIMA), Malabe, Sri Lanka, 6–8 December 2017. [Google Scholar]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Rane, M. Convolutional neural networks: An overview and Its applications in pattern recognition. In Information and Communication Technology for Intelligent Systems: Proceedings of ICTIS 2020; Springer: Singapore, 2020; Volume 1, pp. 21–30. [Google Scholar]
- Graves, A.; Liwicki, M.; Bunke, H.; Schmidhuber, J.; Fernández, S. Unconstrained on-line handwriting recognition with recurrent neural networks. Adv. Neural Inf. Process. Syst. 2007, 20, 1–8. [Google Scholar]
- Behnke, S. Hierarchical Neural Networks for Image Interpretation; Springer: Berlin/Heidelberg, Germany, 2003; Volume 2766. [Google Scholar]
- Clark, E.; Ross, A.S.; Tan, C.; Ji, Y.; Smith, N.A. Creative writing with a machine in the loop: Case studies on slogans and stories. In Proceedings of the 23rd International Conference on Intelligent User Interfaces, Tokyo, Japan, 7–11 March 2018. [Google Scholar]
- Hadjeres, G.; Pachet, F.; Nielsen, F. Deepbach: A steerable model for bach chorales generation. In Proceedings of the 34th International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017. [Google Scholar]
- Guzdial, M.; Liao, N.; Chen, J.; Chen, S.-Y.; Shah, S.; Shah, V.; Reno, J.; Smith, G.; Riedl, M.O. Friend, collaborator, student, manager: How design of an ai-driven game level editor affects creators. In Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems, Glasgow, UK, 4–9 May 2019. [Google Scholar]
- Introducing ChatGPT. Available online: https://openai.com/blog/chatgpt (accessed on 10 June 2023).
- Bard, an Experiment by Google. Available online: https://bard.google.com/ (accessed on 10 June 2023).
- Teubner, T.; Flath, C.M.; Weinhardt, C.; van der Aalst, W.; Hinz, O. Welcome to the era of chatgpt et al. The prospects of large language models. Bus. Inf. Syst. Eng. 2023, 65, 95–101. [Google Scholar] [CrossRef]
- Mondal, S.; Das, S.; Vrana, V.G. How to Bell the Cat? A Theoretical Review of Generative Artificial Intelligence towards Digital Disruption in All Walks of Life. Technologies 2023, 11, 44. [Google Scholar] [CrossRef]
- Piccolo, S.R.; Denny, P.; Luxton-Reilly, A.; Payne, S.; Ridge, P.G. Many bioinformatics programming tasks can be automated with ChatGPT. arXiv 2023, arXiv:2303.13528. [Google Scholar]
- Surameery, N.M.S.; Shakor, M.Y. Use chat GPT to solve programming bugs. Int. J. Inf. Technol. Comput. Eng. 2023, 3, 17–22. [Google Scholar] [CrossRef]
- van Dis, E.A.; Bollen, J.; Zuidema, W.; van Rooij, R.; Bockting, C.L. ChatGPT: Five priorities for research. Nature 2023, 614, 224–226. [Google Scholar] [CrossRef]
- Naser, M.; Ross, B.; Ogle, J.; Kodur, V.; Hawileh, R.; Abdalla, J.; Thai, H.-T. Can AI Chatbots Pass the Fundamentals of Engineering (FE) and Principles and Practice of Engineering (PE) Structural Exams? arXiv 2023, arXiv:2303.18149. [Google Scholar]
- Geerling, W.; Mateer, G.D.; Wooten, J.; Damodaran, N. Is ChatGPT Smarter than a Student in Principles of Economics? 2023. Available online: https://ssrn.com/abstract=4356034 (accessed on 10 June 2023).
- The Brilliance and Weirdness of ChatGPT. Available online: https://www.nytimes.com/2022/12/05/technology/chatgpt-ai-twitter.html (accessed on 10 June 2023).
- Ram, B.; Pratima Verma, P.V. Artificial intelligence AI-based Chatbot Study of ChatGPT, Google AI Bard and Baidu AI. World J. Adv. Eng. Technol. Sci. 2023, 8, 258–261. [Google Scholar]
- Cascella, M.; Montomoli, J.; Bellini, V.; Bignami, E. Evaluating the feasibility of ChatGPT in healthcare: An analysis of multiple clinical and research scenarios. J. Med. Syst. 2023, 47, 33. [Google Scholar] [CrossRef] [PubMed]
- Kung, T.H.; Cheatham, M.; Medenilla, A.; Sillos, C.; De Leon, L.; Elepaño, C.; Madriaga, M.; Aggabao, R.; Diaz-Candido, G.; Maningo, J. Performance of ChatGPT on USMLE: Potential for AI-assisted medical education using large language models. PLoS Digit. Health 2023, 2, e0000198. [Google Scholar] [CrossRef]
- Vert, J.-P. How will generative AI disrupt data science in drug discovery? Nat. Biotechnol. 2023, 41, 750–751. [Google Scholar] [CrossRef]
- Uprety, D.; Zhu, D.; West, H. ChatGPT—A promising generative AI tool and its implications for cancer care. Cancer 2023, 129, 2284–2289. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Furukawa, T.; Lami, K.; Pham, H.H.N.; Uegami, W.; Kuroda, K.; Kawai, M.; Sakanashi, H.; Cooper, L.A.D.; Bychkov, A. A narrative review of digital pathology and artificial intelligence: Focusing on lung cancer. Transl. Lung Cancer Res. 2020, 9, 2255. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Chan, K.; Gibson, I. Virtual reality. IEEE Potentials 1998, 17, 20–23. [Google Scholar] [CrossRef]
- Carmigniani, J.; Furht, B. Augmented reality: An overview. Handbook of Augmented Reality; Springer: New York, NY, USA, 2011; pp. 3–46. [Google Scholar]
- Berryman, D.R. Augmented reality: A review. Med. Ref. Serv. Q. 2012, 31, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Fuchsova, M.; Korenova, L. Visualisation in Basic Science and Engineering Education of Future Primary School Teachers in Human Biology Education Using Augmented Reality. Eur. J. Contemp. Educ. 2019, 8, 92–102. [Google Scholar]
- Paembonan, T.L.; Ikhsan, J. Supporting Students’ Basic Science Process S kills by Augmented Reality Learning Media. J. Educ. Sci. Technol. 2021, 7, 188–196. [Google Scholar]
- Chen, S.-Y.; Liu, S.-Y. Using augmented reality to experiment with elements in a chemistry course. Comput. Hum. Behav. 2020, 111, 106418. [Google Scholar] [CrossRef]
- Li, L.; Yu, F.; Shi, D.; Shi, J.; Tian, Z.; Yang, J.; Wang, X.; Jiang, Q. Application of virtual reality technology in clinical medicine. Am. J. Transl. Res. 2017, 9, 3867. [Google Scholar]
- Pottle, J. Virtual reality and the transformation of medical education. Future Healthc. J. 2019, 6, 181. [Google Scholar] [CrossRef] [Green Version]
- Ayoub, A.; Pulijala, Y. The application of virtual reality and augmented reality in Oral & Maxillofacial Surgery. BMC Oral Health 2019, 19, 238. [Google Scholar]
- McKnight, R.R.; Pean, C.A.; Buck, J.S.; Hwang, J.S.; Hsu, J.R.; Pierrie, S.N. Virtual reality and augmented reality—Translating surgical training into surgical technique. Curr. Rev. Musculoskelet. Med. 2020, 13, 663–674. [Google Scholar] [CrossRef]
- Casari, F.A.; Navab, N.; Hruby, L.A.; Kriechling, P.; Nakamura, R.; Tori, R.; de Lourdes dos Santos Nunes, F.; Queiroz, M.C.; Fürnstahl, P.; Farshad, M. Augmented reality in orthopedic surgery is emerging from proof of concept towards clinical studies: A literature review explaining the technology and current state of the art. Curr. Rev. Musculoskelet. Med. 2021, 14, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Carl, B.; Bopp, M.; Saß, B.; Voellger, B.; Nimsky, C. Implementation of augmented reality support in spine surgery. Eur. Spine J. 2019, 28, 1697–1711. [Google Scholar] [CrossRef]
- Georgescu, R.; Fodor, L.A.; Dobrean, A.; Cristea, I.A. Psychological interventions using virtual reality for pain associated with medical procedures: A systematic review and meta-analysis. Psychol. Med. 2020, 50, 1795–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittara, M.; Matsangidou, M.; Stylianides, K.; Petkov, N.; Pattichis, C.S. Virtual reality for pain management in cancer: A comprehensive review. IEEE Access 2020, 8, 225475–225489. [Google Scholar] [CrossRef]
- Sharifpour, S.; Manshaee, G.R.; Sajjadian, I. Effects of virtual reality therapy on perceived pain intensity, anxiety, catastrophising and self-efficacy among adolescents with cancer. Couns. Psychother. Res. 2021, 21, 218–226. [Google Scholar] [CrossRef]
- Cipresso, P.; Giglioli, I.A.C.; Raya, M.A.; Riva, G. The past, present, and future of virtual and augmented reality research: A network and cluster analysis of the literature. Front. Psychol. 2018, 9, 2086. [Google Scholar] [CrossRef] [Green Version]
- Peddie, J. Augmented Reality: Where We Will All Live; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Riva, G.; Baños, R.M.; Botella, C.; Mantovani, F.; Gaggioli, A. Transforming experience: The potential of augmented reality and virtual reality for enhancing personal and clinical change. Front. Psychiatry 2016, 7, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcke, J.; Roscher, R. Explainable Machine Learning. Mach. Learn. Knowl. Extr. 2023, 5, 169–170. [Google Scholar] [CrossRef]
- Roscher, R.; Bohn, B.; Duarte, M.F.; Garcke, J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access 2019, 8, 42200–42216. [Google Scholar] [CrossRef]
- Rasheed, K.; Qayyum, A.; Ghaly, M.; Al-Fuqaha, A.I.; Razi, A.; Qadir, J. Explainable, trustworthy, and ethical machine learning for healthcare: A survey. Comput. Biol. Med. 2021, 149, 106043. [Google Scholar] [CrossRef]
- Ratti, E.; Graves, M. Explainable machine learning practices: Opening another black box for reliable medical AI. AI Ethics 2022, 2, 801–814. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S.B. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef]
- Wells, L.; Bednarz, T. Explainable AI and Reinforcement Learning—A Systematic Review of Current Approaches and Trends. Front. Artif. Intell. 2021, 4, 550030. [Google Scholar] [CrossRef]
- Nauta, M.; Trienes, J.; Pathak, S.; Nguyen, E.; Peters, M.; Schmitt, Y.; Schlötterer, J.; Keulen, M.V.; Seifert, C. From Anecdotal Evidence to Quantitative Evaluation Methods: A Systematic Review on Evaluating Explainable AI. ACM Comput. Surv. 2022, 55, 1–42. [Google Scholar] [CrossRef]
- Jin, W.; Li, X.; Hamarneh, G. Evaluating explainable AI on a multi-modal medical imaging task: Can existing algorithms fulfill clinical requirements? Proc. AAAI Conf. Artif. Intelligence. 2022, 36, 11945–11953. [Google Scholar] [CrossRef]
- Dwivedi, R.; Dave, D.; Naik, H.; Singhal, S.; Omer, R.; Patel, P.; Qian, B.; Wen, Z.; Shah, T.; Morgan, G. Explainable AI (XAI): Core ideas, techniques, and solutions. ACM Comput. Surv. 2023, 55, 194. [Google Scholar] [CrossRef]
- Jiménez-Luna, J.; Grisoni, F.; Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2020, 2, 573–584. [Google Scholar] [CrossRef]
- de Souza, L.A., Jr.; Mendel, R.; Strasser, S.; Ebigbo, A.; Probst, A.; Messmann, H.; Papa, J.P.; Palm, C. Convolutional Neural Networks for the evaluation of cancer in Barrett’s esophagus: Explainable AI to lighten up the black-box. Comput. Biol. Med. 2021, 135, 104578. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014. [Google Scholar]
- Zafar, M.R.; Khan, N.M. DLIME: A deterministic local interpretable model-agnostic explanations approach for computer-aided diagnosis systems. arXiv 2019, arXiv:1906.10263. [Google Scholar]
- Palatnik de Sousa, I.; Vellasco, M.M.B.R.; da Silva, E.C. Local interpretable model-agnostic explanations for classification of lymph node metastases. Sensors 2019, 19, 2969. [Google Scholar] [CrossRef] [Green Version]
- Kumarakulasinghe, N.B.; Blomberg, T.; Liu, J.; Leao, A.S.; Papapetrou, P. Evaluating local interpretable model-agnostic explanations on clinical machine learning classification models. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020. [Google Scholar]
- Binder, A.; Bockmayr, M.; Hägele, M.; Wienert, S.; Heim, D.; Hellweg, K.; Ishii, M.; Stenzinger, A.; Hocke, A.; Denkert, C. Morphological and molecular breast cancer profiling through explainable machine learning. Nat. Mach. Intell. 2021, 3, 355–366. [Google Scholar] [CrossRef]
- Alsinglawi, B.; Alshari, O.; Alorjani, M.; Mubin, O.; Alnajjar, F.; Novoa, M.; Darwish, O. An explainable machine learning framework for lung cancer hospital length of stay prediction. Sci. Rep. 2022, 12, 607. [Google Scholar] [CrossRef]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative adversarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, A.; Mittal, M.; Battineni, G. Generative adversarial network: An overview of theory and applications. Int. J. Inf. Manag. Data Insights 2021, 1, 100004. [Google Scholar] [CrossRef]
- Arjovsky, M.; Bottou, L. Towards principled methods for training generative adversarial networks. arXiv 2017, arXiv:1701.04862. [Google Scholar]
- Gui, J.; Sun, Z.; Wen, Y.; Tao, D.; Ye, J. A review on generative adversarial networks: Algorithms, theory, and applications. IEEE Trans. Knowl. Data Eng. 2021, 35, 3313–3332. [Google Scholar] [CrossRef]
- Nam, S.; Kim, Y.; Kim, S.J. Text-adaptive generative adversarial networks: Manipulating images with natural language. Adv. Neural Inf. Process. Syst. 2018, 31, 1–10. [Google Scholar]
- Xu, J.; Ren, X.; Lin, J.; Sun, X. Diversity-promoting GAN: A cross-entropy based generative adversarial network for diversified text generation. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018. [Google Scholar]
- Lai, C.-T.; Hong, Y.-T.; Chen, H.-Y.; Lu, C.-J.; Lin, S.-D. Multiple text style transfer by using word-level conditional generative adversarial network with two-phase training. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China, 3–7 November 2019. [Google Scholar]
- Yinka-Banjo, C.; Ugot, O.-A. A review of generative adversarial networks and its application in cybersecurity. Artif. Intell. Rev. 2020, 53, 1721–1736. [Google Scholar] [CrossRef]
- Toshpulatov, M.; Lee, W.; Lee, S. Generative adversarial networks and their application to 3D face generation: A survey. Image Vis. Comput. 2021, 108, 104119. [Google Scholar] [CrossRef]
- Kusiak, A. Convolutional and generative adversarial neural networks in manufacturing. Int. J. Prod. Res. 2020, 58, 1594–1604. [Google Scholar] [CrossRef]
- Singh, R.; Garg, R.; Patel, N.S.; Braun, M.W. Generative adversarial networks for synthetic defect generation in assembly and test manufacturing. In Proceedings of the 2020 31st Annual SEMI Advanced Semiconductor Manufacturing Conference (ASMC), Saratoga Springs, NY, USA, 24–26 August 2020. [Google Scholar]
- Wang, J.; Yang, Z.; Zhang, J.; Zhang, Q.; Chien, W.-T.K. AdaBalGAN: An improved generative adversarial network with imbalanced learning for wafer defective pattern recognition. IEEE Trans. Semicond. Manuf. 2019, 32, 310–319. [Google Scholar] [CrossRef]
- Meng, F.-j.; Li, Y.-q.; Shao, F.-m.; Yuan, G.-h.; Dai, J.-y. Visual-simulation region proposal and generative adversarial network based ground military target recognition. Def. Technol. 2022, 18, 2083–2096. [Google Scholar] [CrossRef]
- Thompson, S.; Teixeira-Dias, F.; Paulino, M.; Hamilton, A. Predictions on multi-class terminal ballistics datasets using conditional Generative Adversarial Networks. Neural Netw. 2022, 154, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Achuthan, S.; Chatterjee, R.; Kotnala, S.; Mohanty, A.; Bhattacharya, S.; Salgia, R.; Kulkarni, P. Leveraging deep learning algorithms for synthetic data generation to design and analyze biological networks. J. Biosci. 2022, 47, 43. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yang, X.-H.; Wei, Z.; Heidari, A.A.; Zheng, N.; Li, Z.; Chen, H.; Hu, H.; Zhou, Q.; Guan, Q. Generative adversarial networks in medical image augmentation: A review. Comput. Biol. Med. 2022, 144, 105382. [Google Scholar] [CrossRef]
- von Werra, L.; Schöngens, M.; Uzun, E.D.G.; Eickhoff, C. Generative adversarial networks in precision oncology. In Proceedings of the 2019 ACM SIGIR International Conference on Theory of Information Retrieval, Santa Clara, CA, USA, 2–5 October 2019. [Google Scholar]
- Zhan, B.; Xiao, J.; Cao, C.; Peng, X.; Zu, C.; Zhou, J.; Wang, Y. Multi-constraint generative adversarial network for dose prediction in radiotherapy. Med. Image Anal. 2022, 77, 102339. [Google Scholar] [CrossRef] [PubMed]
- Heilemann, G.; Zimmermann, L.; Matthewman, M. Investigating the Potential of Generative Adversarial Networks (GANs) for Autosegmentation in Radiation Oncology. Ph.D. Thesis, Technische Universitä, Viena, Austria, 2021. [Google Scholar]
- Nakamura, M.; Nakao, M.; Imanishi, K.; Hirashima, H.; Tsuruta, Y. Geometric and dosimetric impact of 3D generative adversarial network-based metal artifact reduction algorithm on VMAT and IMPT for the head and neck region. Radiat. Oncol. 2021, 16, 96. [Google Scholar] [CrossRef]
- Hersche, M.; Zeqiri, M.; Benini, L.; Sebastian, A.; Rahimi, A. A neuro-vector-symbolic architecture for solving Raven’s progressive matrices. Nat. Mach. Intell. 2023, 5, 363–375. [Google Scholar] [CrossRef]
- Serre, T. Deep learning: The good, the bad, and the ugly. Annu. Rev. Vis. Sci. 2019, 5, 399–426. [Google Scholar] [CrossRef]
- Shrestha, A.; Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Kleyko, D.; Rachkovskij, D.A.; Osipov, E.; Rahimi, A. A Survey on Hyperdimensional Computing aka Vector Symbolic Architectures, Part I: Models and Data Transformations. ACM Comput. Surv. 2021, 55, 1–40. [Google Scholar] [CrossRef]
- Kleyko, D.; Rachkovskij, D.A.; Osipov, E.; Rahimi, A. A Survey on Hyperdimensional Computing aka Vector Symbolic Architectures, Part II: Applications, Cognitive Models, and Challenges. ACM Comput. Surv. 2021, 55, 1–52. [Google Scholar] [CrossRef]
- This AI Could Likely Beat You at an IQ Test. Available online: https://research.ibm.com/blog/neuro-vector-symbolic-architecture-IQ-test (accessed on 10 June 2023).
- Widdows, D.; Cohen, T. Reasoning with vectors: A continuous model for fast robust inference. Log. J. IGPL 2015, 23, 141–173. [Google Scholar] [CrossRef]
- Abhijith, M.; Nair, D.R. Neuromorphic High Dimensional Computing Architecture for Classification Applications. In Proceedings of the 2021 IEEE International Conference on Nanoelectronics, Nanophotonics, Nanomaterials, Nanobioscience & Nanotechnology (5NANO), Kottayam, Kerala, India, 29–30 April 2021. [Google Scholar]
- Fortunato, L.; Galassi, M. The case for free and open source software in research and scholarship. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2021, 379, 20200079. [Google Scholar] [CrossRef]
- Sahay, S. Free and open source software as global public goods? What are the distortions and how do we address them? Electron. J. Inf. Syst. Dev. Ctries. 2019, 85, e12080. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Manure, A.; Singh, P.; Manure, A. Introduction to Tensorflow 2.0. In Learn TensorFlow 2.0: Implement Machine Learning and Deep Learning Models with Python; Apress: New York, NY, USA, 2020; pp. 1–24. [Google Scholar]
- Stevens, E.; Antiga, L.; Viehmann, T. Deep Learning with PyTorch; Manning Publications: Shelter Island, NY, USA, 2020. [Google Scholar]
- Pang, B.; Nijkamp, E.; Wu, Y.N. Deep learning with tensorflow: A review. J. Educ. Behav. Stat. 2020, 45, 227–248. [Google Scholar] [CrossRef]
- Sarang, P. Artificial Neural Networks with TensorFlow 2; Apress: Berkeley, CA, USA, 2021. [Google Scholar]
- Rao, D.; McMahan, B. Natural Language Processing with PyTorch: Build Intelligent Language Applications Using Deep Learning; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2019. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M. Huggingface’s transformers: State-of-the-art natural language processing. arXiv 2019, arXiv:1910.03771. [Google Scholar]
- Ayyadevara, V.K.; Reddy, Y. Modern Computer Vision with Pytorch: Explore Deep Learning Concepts and Implement Over 50 Real-World Image Applications; Packt Publishing Ltd.: Birmingham, UK, 2020. [Google Scholar]
- Anderson, B.M.; Wahid, K.A.; Brock, K.K. Simple python module for conversions between DICOM images and radiation therapy structures, masks, and prediction arrays. Pract. Radiat. Oncol. 2021, 11, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Norton, I.; Essayed, W.I.; Zhang, F.; Pujol, S.; Yarmarkovich, A.; Golby, A.J.; Kindlmann, G.; Wassermann, D.; Estepar, R.S.J.; Rathi, Y. SlicerDMRI: Open source diffusion MRI software for brain cancer research. Cancer Res. 2017, 77, e101–e103. [Google Scholar] [CrossRef] [Green Version]
- Gutman, D.A.; Khalilia, M.; Lee, S.; Nalisnik, M.; Mullen, Z.; Beezley, J.; Chittajallu, D.R.; Manthey, D.; Cooper, L.A. The digital slide archive: A software platform for management, integration, and analysis of histology for cancer research. Cancer Res. 2017, 77, e75–e78. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Qiu, P.; Ji, Y. TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nat. Methods 2014, 11, 599–600. [Google Scholar] [CrossRef] [PubMed]
- Jo, A. The Promise and Peril of Generative AI. Nature 2023, 614, 214–216. [Google Scholar]
- Bucknall, B.S.; Dori-Hacohen, S. Current and near-term AI as a potential existential risk factor. In Proceedings of the 2022 AAAI/ACM Conference on AI, Ethics, and Society, Oxford, UK, 19–21 May 2022. [Google Scholar]
- Roose, K.A.I. Poses ‘Risk of Extinction,’ Industry Leaders Warn. New York Times, 30 May 2023. [Google Scholar]
- Letters, F.O. Pause giant AI Experiments: An Open Letter. Future of Life Institution. 2023. Available online: https://futureoflife.org/open-letter/pause-giant-ai-experiments (accessed on 10 June 2023).
- Kann, B.H.; Hosny, A.; Aerts, H. Artificial intelligence for clinical oncology. Cancer Cell 2021, 39, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Gha, A.E.; Takov, P.; Shang, N. The Superhuman Born Out of Artificial Intelligence and Genetic Engineering: The Destruction of Human Ontological Dignity. Horiz. J. Humanit. Artif. Intell. 2023, 2, 56–65. [Google Scholar]
- Zhang, J.; Zhang, Z.-M. Ethics and governance of trustworthy medical artificial intelligence. BMC Med. Inform. Decis. Mak. 2023, 23, 7. [Google Scholar] [CrossRef]
- Langlotz, C.P. Will artificial intelligence replace radiologists? Radiol. Soc. North America. 2019, 1, e190058. [Google Scholar] [CrossRef]
- Goldhahn, J.; Rampton, V.; Spinas, G.A. Could artificial intelligence make doctors obsolete? BMJ 2018, 363, k4563. [Google Scholar] [CrossRef] [Green Version]
- Razzaki, S.; Baker, A.; Perov, Y.; Middleton, K.; Baxter, J.; Mullarkey, D.; Sangar, D.; Taliercio, M.; Butt, M.; Majeed, A. A comparative study of artificial intelligence and human doctors for the purpose of triage and diagnosis. arXiv 2018, arXiv:1806.10698. [Google Scholar]
- Botha, J.; Pieterse, H. Fake news and deepfakes: A dangerous threat for 21st century information security. In Proceedings of the ICCWS 2020 15th International Conference on Cyber Warfare and Security, Norfolk, VA, USA, 12–13 March 2020. [Google Scholar]
- Pantserev, K.A. The Malicious Use of AI-Based Deepfake Technology as the New Threat to Psychological Security and Political Stability. In Cyber Defence in the Age of AI, Smart Societies and Augmented Humanity; Springer: Cham, Switzerland, 2020; pp. 37–55. [Google Scholar]
- Borji, A. A categorical archive of ChatGPT failures. arXiv 2023, arXiv:2302.03494. [Google Scholar]
- Brainard, J. Journals take up arms against AI-written text. Science 2023, 379, 740–741. [Google Scholar] [CrossRef]
- Vaishya; Misra, A.; Vaish, A. ChatGPT: Is this version good for healthcare and research? Diabetes Metab. Syndr. Clin. Res. Rev. 2023, 17, 102744. [Google Scholar] [CrossRef]
- DeGrave, A.; Janizek, J.; Lee, S. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell. 2021, 3, 610–619. [Google Scholar] [CrossRef]
- Khullar, D.; Casalino, L.P.; Qian, Y.; Lu, Y.; Krumholz, H.M.; Aneja, S. Perspectives of patients about artificial intelligence in health care. JAMA Netw. Open 2022, 5, e2210309. [Google Scholar] [CrossRef]
- Seyyed-Kalantari, L.; Zhang, H.; McDermott, M.B.; Chen, I.Y.; Ghassemi, M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat. Med. 2021, 27, 2176–2182. [Google Scholar] [CrossRef]
- Kim, E.J.; Woo, H.S.; Cho, J.H.; Sym, S.J.; Baek, J.-H.; Lee, W.-S.; Kwon, K.A.; Kim, K.O.; Chung, J.-W.; Park, D.K. Early experience with Watson for oncology in Korean patients with colorectal cancer. PLoS ONE 2019, 14, e0213640. [Google Scholar] [CrossRef]
- Richardson, J.P.; Smith, C.; Curtis, S.; Watson, S.; Zhu, X.; Barry, B.; Sharp, R.R. Patient apprehensions about the use of artificial intelligence in healthcare. NPJ Digit. Med. 2021, 4, 140. [Google Scholar] [CrossRef]
- Lim, A.K.; Thuemmler, C. Opportunities and challenges of internet-based health interventions in the future internet. In Proceedings of the 2015 12th International Conference on Information Technology-New Generations, Las Vegas, NV, USA, 13–15 April 2015. [Google Scholar]
- Hatherley, J.J. Limits of trust in medical AI. J. Med. Ethics 2020, 46, 478–481. [Google Scholar] [CrossRef] [PubMed]
- DeCamp, M.; Tilburt, J.C. Why we cannot trust artificial intelligence in medicine. Lancet Digit. Health 2019, 1, e390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nundy, S.; Montgomery, T.; Wachter, R.M. Promoting trust between patients and physicians in the era of artificial intelligence. JAMA 2019, 322, 497–498. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Albizri, A.; Harfouche, A. Responsible artificial intelligence in healthcare: Predicting and preventing insurance claim denials for economic and social wellbeing. Inf. Syst. Front. 2021, 1–17. [Google Scholar] [CrossRef]
- Lysaght, T.; Lim, H.Y.; Xafis, V.; Ngiam, K.Y. AI-assisted decision-making in healthcare: The application of an ethics framework for big data in health and research. Asian Bioeth. Rev. 2019, 11, 299–314. [Google Scholar] [CrossRef] [Green Version]
- Dueno, T. Racist Robots and the Lack of Legal Remedies in the Use of Artificial Intelligence in Healthcare. Conn. Ins. LJ 2020, 27, 337. [Google Scholar]
- Formosa, P.; Rogers, W.; Griep, Y.; Bankins, S.; Richards, D. Medical AI and human dignity: Contrasting perceptions of human and artificially intelligent (AI) decision making in diagnostic and medical resource allocation contexts. Comput. Hum. Behav. 2022, 133, 107296. [Google Scholar] [CrossRef]
- Muthukrishnan, N.; Maleki, F.; Ovens, K.; Reinhold, C.; Forghani, B.; Forghani, R. Brief history of artificial intelligence. Neuroimaging Clin. 2020, 30, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y. Heading toward artificial intelligence 2.0. Engineering 2016, 2, 409–413. [Google Scholar] [CrossRef]
- Fan, J.; Campbell, M.; Kingsbury, B. Artificial intelligence research at IBM. IBM J. Res. Dev. 2011, 55, 16:1–16:4. [Google Scholar] [CrossRef]
- Bory, P. Deep new: The shifting narratives of artificial intelligence from Deep Blue to AlphaGo. Convergence 2019, 25, 627–642. [Google Scholar] [CrossRef]
- McCorduck, P.; Minsky, M.; Selfridge, O.; Simon, H.A. History of artificial intelligence. In Proceedings of the IJCAI’77: Proceedings of the 5th International Joint Conference on Artificial Intelligence, Cambridge, MA, USA, 22–25 August 1977. [Google Scholar]
- Bernstein, J. Three Degrees above Zero: Bell Laboratories in the Information Age; Cambridge University Press: Cambridge, UK, 1987. [Google Scholar]
- Horowitz, M.C. Artificial intelligence, international competition, and the balance of power. Texas National Security Review 2018, 2018, 22. [Google Scholar]
- Dick, S. Artificial intelligence. Harv. Data Sci. Rev. 2019, 1, 1–9. [Google Scholar]
- Wasilow, S.; Thorpe, J.B. Artificial intelligence, robotics, ethics, and the military: A Canadian perspective. AI Mag. 2019, 40, 37–48. [Google Scholar] [CrossRef]
- Bistron, M.; Piotrowski, Z. Artificial intelligence applications in military systems and their influence on sense of security of citizens. Electronics 2021, 10, 871. [Google Scholar] [CrossRef]
- Lohr, S. MIT Plans College for Artificial Intelligence, Backed by $1 Billion. The New York Times, 15 October 2018; 15. [Google Scholar]
- Kubassova, O.; Shaikh, F.; Melus, C.; Mahler, M. History, current status, and future directions of artificial intelligence. In Precision Medicine and Artificial Intelligence; Academic Press: Cambridge, MA, USA, 2021; pp. 1–38. [Google Scholar]
- Fulbright, R. A Brief History of Artificial Intelligence. In Democratization of Expertise; Routledge: London, UK, 2020. [Google Scholar]
- Marvin, M.; Seymour, A.P. Perceptrons; MIT Press: Cambridge, MA, USA, 1969; pp. 318–362. [Google Scholar]
- McCarthy, J.; Minsky, M.; Rochester, N.; Shannon, C. Dartmouth Artificial Intelligence (AI) Conference; Dartmouth College: Hanover, NH, USA, 1956. [Google Scholar]
- Lim, H.-o.; Takanishi, A. Biped walking robots created at Waseda University: WL and WABIAN family. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2007, 365, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Güzel, M.S. Autonomous vehicle navigation using vision and mapless strategies: A survey. Adv. Mech. Eng. 2013, 5, 234747. [Google Scholar] [CrossRef] [Green Version]
- Fei-Fei, L.; Deng, J.; Li, K. ImageNet: Constructing a large-scale image database. J. Vis. 2009, 9, 1037. [Google Scholar] [CrossRef]
- Chang, A. History of Artificial Intelligence in Medicine. Gastrointest. Endosc. 2020, 92, 807–812. [Google Scholar]
- Beam, A.L.; Drazen, J.M.; Kohane, I.S.; Leong, T.Y.; Manrai, A.K.; Rubin, E.J. Artificial Intelligence in Medicine. N. Engl. J. Med. 2023, 388, 1220–1221. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [Green Version]
- Tang, A.; Tam, R.; Cadrin-Chênevert, A.; Guest, W.; Chong, J.; Barfett, J.; Chepelev, L.; Cairns, R.; Mitchell, J.R.; Cicero, M.D. Canadian Association of Radiologists white paper on artificial intelligence in radiology. Can. Assoc. Radiol. J. 2018, 69, 120–135. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.W.; Soto, J.T.; Glicksberg, B.S.; Shameer, K.; Miotto, R.; Ali, M.; Ashley, E.; Dudley, J.T. Artificial intelligence in cardiology. J. Am. Coll. Cardiol. 2018, 71, 2668–2679. [Google Scholar] [CrossRef]
- Lopez-Jimenez, F.; Attia, Z.; Arruda-Olson, A.M.; Carter, R.; Chareonthaitawee, P.; Jouni, H.; Kapa, S.; Lerman, A.; Luong, C.; Medina-Inojosa, J.R. Artificial intelligence in cardiology: Present and future. Mayo Clin. Proc. 2020, 95, 1015–1039. [Google Scholar] [CrossRef]
- Miyazawa, A.A. Artificial intelligence: The future for cardiology. Heart 2019, 105, 1214. [Google Scholar] [CrossRef]
- Le Berre, C.; Sandborn, W.J.; Aridhi, S.; Devignes, M.-D.; Fournier, L.; Smail-Tabbone, M.; Danese, S.; Peyrin-Biroulet, L. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology 2020, 158, 76–94.e2. [Google Scholar] [CrossRef] [Green Version]
- Christou, C.D.; Tsoulfas, G. Challenges and opportunities in the application of artificial intelligence in gastroenterology and hepatology. World J. Gastroenterol. 2021, 27, 6191. [Google Scholar] [CrossRef]
- Kröner, P.T.; Engels, M.M.; Glicksberg, B.S.; Johnson, K.W.; Mzaik, O.; van Hooft, J.E.; Wallace, M.B.; El-Serag, H.B.; Krittanawong, C. Artificial intelligence in gastroenterology: A state-of-the-art review. World J. Gastroenterol. 2021, 27, 6794. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Cao, H.; FitzGerald, J.M.; Iannotti, N.; Yang, E.; Kocks, J.W.; Kostikas, K.; Price, D.; Reddel, H.K.; Tsiligianni, I. Artificial intelligence/machine learning in respiratory medicine and potential role in asthma and COPD diagnosis. J. Allergy Clin. Immunol. Pract. 2021, 9, 2255–2261. [Google Scholar] [CrossRef] [PubMed]
- Mekov, E.; Miravitlles, M.; Petkov, R. Artificial intelligence and machine learning in respiratory medicine. Expert Rev. Respir. Med. 2020, 14, 559–564. [Google Scholar] [CrossRef]
- Ferrante, G.; Licari, A.; Marseglia, G.L.; La Grutta, S. Artificial intelligence as an emerging diagnostic approach in paediatric pulmonology. Respirology 2020, 25, 1029–1030. [Google Scholar] [CrossRef]
- Hunter, B.; Hindocha, S.; Lee, R.W. The role of artificial intelligence in early cancer diagnosis. Cancers 2022, 14, 1524. [Google Scholar] [CrossRef] [PubMed]
- Kenner, B.; Chari, S.T.; Kelsen, D.; Klimstra, D.S.; Pandol, S.J.; Rosenthal, M.; Rustgi, A.K.; Taylor, J.A.; Yala, A.; Abul-Husn, N. Artificial intelligence and early detection of pancreatic cancer: 2020 summative review. Pancreas 2021, 50, 251. [Google Scholar] [CrossRef]
- Ballester, P.J.; Carmona, J. Artificial intelligence for the next generation of precision oncology. NPJ Precis. Oncol. 2021, 5, 79. [Google Scholar] [CrossRef]
- Windisch, P.; Hertler, C.; Blum, D.; Zwahlen, D.; Förster, R. Leveraging advances in artificial intelligence to improve the quality and timing of palliative care. Cancers 2020, 12, 1149. [Google Scholar] [CrossRef]
- Periyakoil, V.S.; Gunten, C.F.v. Palliative Care Is Proven. J. Palliat. Med. 2023, 26, 2–4. [Google Scholar] [CrossRef]
- Courdy, S.; Hulse, M.; Nadaf, S.; Mao, A.; Pozhitkov, A.; Berger, S.; Chang, J.; Achuthan, S.; Kancharla, C.; Kunz, I. The City of Hope POSEIDON enterprise-wide platform for real-world data and evidence in cancer. J. Clin. Oncol. 2021, 39, e18813. [Google Scholar] [CrossRef]
- Melstrom, L.G.; Rodin, A.S.; Rossi, L.A.; Fu, P., Jr.; Fong, Y.; Sun, V. Patient generated health data and electronic health record integration in oncologic surgery: A call for artificial intelligence and machine learning. J. Surg. Oncol. 2021, 123, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Dadwal, S.; Eftekhari, Z.; Thomas, T.; Johnson, D.; Yang, D.; Mokhtari, S.; Nikolaenko, L.; Munu, J.; Nakamura, R. A Machine-Learning Sepsis Prediction Model for Patients Undergoing Hematopoietic Cell Transplantation. Blood 2018, 132, 711. [Google Scholar] [CrossRef]
- Deng, H.; Eftekhari, Z.; Carlin, C.; Veerapong, J.; Fournier, K.F.; Johnston, F.M.; Dineen, S.P.; Powers, B.D.; Hendrix, R.; Lambert, L.A. Development and Validation of an Explainable Machine Learning Model for Major Complications After Cytoreductive Surgery. JAMA Netw. Open 2022, 5, e2212930. [Google Scholar] [CrossRef]
- Zachariah, F.J.; Rossi, L.A.; Roberts, L.M.; Bosserman, L.D. Prospective comparison of medical oncologists and a machine learning model to predict 3-month mortality in patients with metastatic solid tumors. JAMA Netw. Open 2022, 5, e2214514. [Google Scholar] [CrossRef]
- Rossi, L.A.; Shawber, C.; Munu, J.; Zachariah, F. Evaluation of embeddings of laboratory test codes for patients at a cancer center. arXiv 2019, arXiv:1907.09600. [Google Scholar]
- Achuthan, S.; Chang, M.; Shah, A. SPIRIT-ML: A machine learning platform for deriving knowledge from biomedical datasets. In Proceedings of the Data Integration in the Life Sciences: 11th International Conference, DILS 2015, Los Angeles, CA, USA, 9–10 July 2015. [Google Scholar]
- Karolak, A.; Branciamore, S.; McCune, J.S.; Lee, P.P.; Rodin, A.S.; Rockne, R.C. Concepts and applications of information theory to immuno-oncology. Trends Cancer 2021, 7, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Rosen, S.T. Why precision medicine continues to be the future of health care. Oncol. Times UK 2017, 39, 24. [Google Scholar] [CrossRef]
- Budhraja, K.K.; McDonald, B.R.; Stephens, M.D.; Contente-Cuomo, T.; Markus, H.; Farooq, M.; Favaro, P.F.; Connor, S.; Byron, S.A.; Egan, J.B. Genome-wide analysis of aberrant position and sequence of plasma DNA fragment ends in patients with cancer. Science Transl. Med. 2023, 15, eabm6863. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Germino, E.; Han, C.; Watkins, W.; Amini, A.; Wong, J.; Williams, T. Clinical Validation of Artificial Intelligence Based Auto-Segmentation of Organs-at-Risk in Total Marrow Irradiation Treatment. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, e302–e303. [Google Scholar] [CrossRef]
- Watkins, W.; Du, D.; Qing, K.; Ladbury, C.; Liu, J.; Liu, A. Validation of Automated Segmentation Algorithms. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, e152. [Google Scholar] [CrossRef]
- Watkins, W.; Liu, J.; Hui, S.; Liu, A. Clinical Efficiency Gains with Artificial-Intelligence Auto-Segmentation in the Entire Human Body. Int. J. Radiat. Oncol. Biol. Phys. 2022, 114, e558. [Google Scholar] [CrossRef]
- Jossart, J.; Kenjić, N.; Perry, J. Structural-Based Drug Discovery Targeting PCNA: A Novel Cancer Therapeutic. FASEB J. 2021, 35. [Google Scholar] [CrossRef]
- Djulbegovic, B.; Teh, J.B.; Wong, L.; Hozo, I.; Armenian, S.H. Diagnostic Predictive Model for Diagnosis of Heart Failure after Hematopoietic Cell Transplantation (HCT): Comparison of Traditional Statistical with Machine Learning Modeling. Blood 2019, 134, 5799. [Google Scholar] [CrossRef]
- Ladbury, C.; Amini, A.; Govindarajan, A.; Mambetsariev, I.; Raz, D.J.; Massarelli, E.; Williams, T.; Rodin, A.; Salgia, R. Integration of artificial intelligence in lung cancer: Rise of the machine. Cell Rep. Med. 2023, 4, 100933. [Google Scholar] [CrossRef] [PubMed]
- Kothari, R.; Jones, V.; Mena, D.; Reyes, V.B.; Shon, Y.; Smith, J.P.; Schmolze, D.; Cha, P.D.; Lai, L.; Fong, Y. Raman spectroscopy and artificial intelligence to predict the Bayesian probability of breast cancer. Sci. Rep. 2021, 11, 6482. [Google Scholar] [CrossRef]
- Pozhitkov, A.; Seth, N.; Kidambi, T.D.; Raytis, J.; Achuthan, S.; Lew, M.W. Machine learning algorithm to perform ASA Physical Status Classification. medRxiv 2021. [Google Scholar] [CrossRef]
- Han, C.; Liu, A.; Wong, J. Application of Machine Learning for Prediction of Normal Organ Dose: Feasibility Study in Treatment Planning for Total Marrow Irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, e782–e783. [Google Scholar] [CrossRef]
- Two Studies for Patients with High Risk Prostate Cancer Testing Less Intense Treatment for Patients with a Low Gene Risk Score and Testing a More Intense Treatment for Patients With a High Gene Risk Score, The PREDICT-RT Trial. Available online: https://clinicaltrials.gov/ct2/show/NCT04513717 (accessed on 10 June 2023).
- Ngiam, K.Y.; Khor, W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019, 20, e262–e273. [Google Scholar] [CrossRef]
- Bosserman, L.D.; Cianfrocca, M.; Yuh, B.; Yeon, C.; Chen, H.; Sentovich, S.; Polverini, A.; Zachariah, F.; Deaville, D.; Lee, A.B. Integrating academic and community cancer care and research through multidisciplinary oncology pathways for value-based care: A review and the City of Hope experience. J. Clin. Med. 2021, 10, 188. [Google Scholar] [CrossRef] [PubMed]
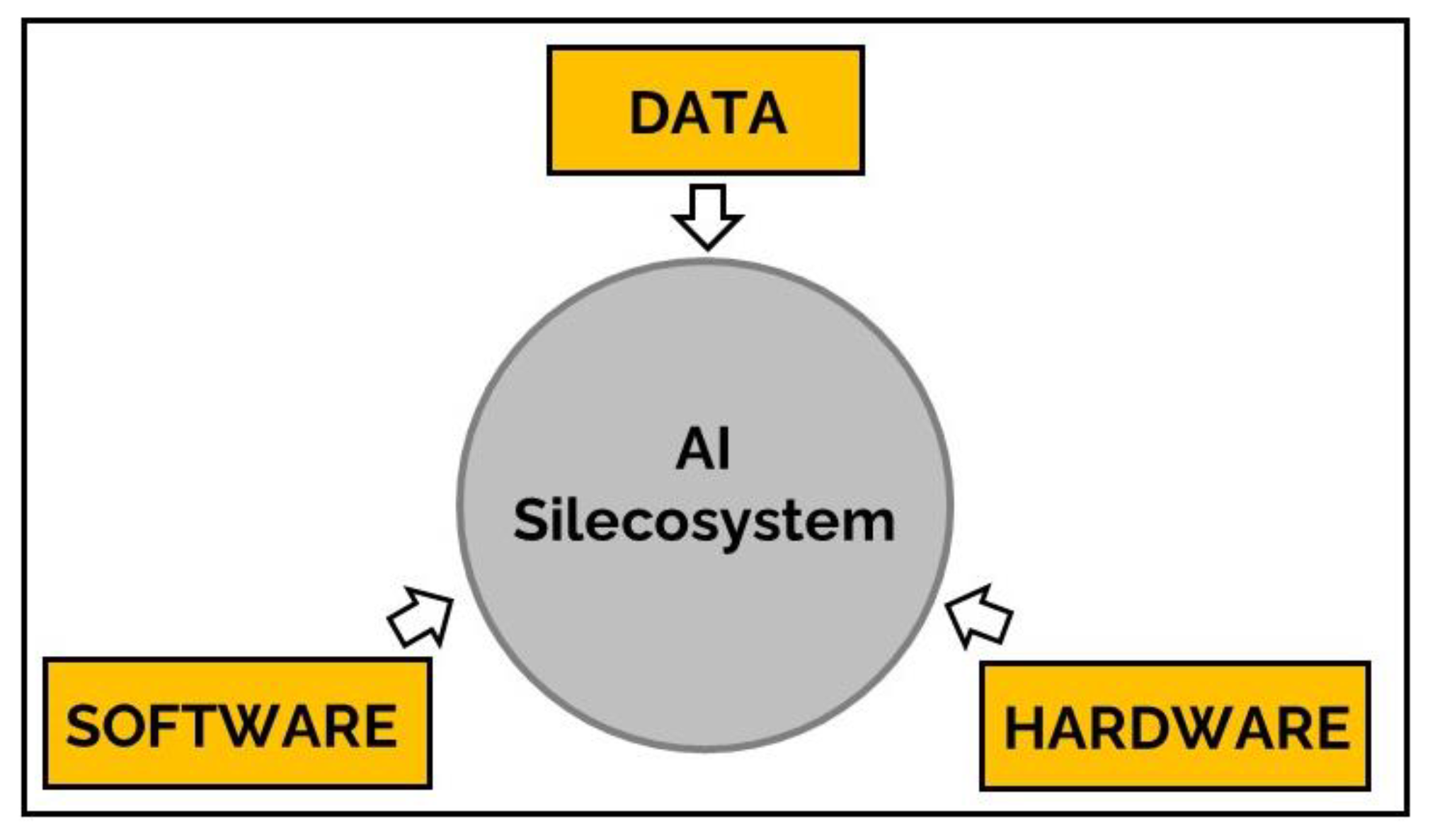


| AI Silecosystem Metric | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Component | Innovation | AI Algorithmic Speed | Efficiency/Cost | Utility | Agility | Accuracy/Validity/Reliability | Security/Safety | Accessibility | ||
| Hardware | Quantum Computing | |||||||||
| AI Internet of Things | ||||||||||
| Distributive Edge Computing | ||||||||||
| Cloud Computing | ||||||||||
| Neuromorphic Computing | ||||||||||
| Analog Neural Networks | ||||||||||
| Monolithic 3D AI Systems | ||||||||||
| Graphics Processing Unit | ||||||||||
| Analog Non-Volatile Memory | ||||||||||
| Data | Synthetic Data | |||||||||
| Culturally Representative Data Sets | ||||||||||
| Data Optimization | ||||||||||
| Software | Generative AI | |||||||||
| Virtual and Augmented Reality | ||||||||||
| Explainable Machine Learning | ||||||||||
| Generative Adversarial Networks | ||||||||||
| Neuro-Vector-Symbolic Architecture | ||||||||||
| Open-Source AI Software | ||||||||||
| Greater Impact | Lesser Impact | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonnell, K.J. Leveraging the Academic Artificial Intelligence Silecosystem to Advance the Community Oncology Enterprise. J. Clin. Med. 2023, 12, 4830. https://doi.org/10.3390/jcm12144830
McDonnell KJ. Leveraging the Academic Artificial Intelligence Silecosystem to Advance the Community Oncology Enterprise. Journal of Clinical Medicine. 2023; 12(14):4830. https://doi.org/10.3390/jcm12144830
Chicago/Turabian StyleMcDonnell, Kevin J. 2023. "Leveraging the Academic Artificial Intelligence Silecosystem to Advance the Community Oncology Enterprise" Journal of Clinical Medicine 12, no. 14: 4830. https://doi.org/10.3390/jcm12144830






