Bovine Hemoglobin Enzymatic Hydrolysis by a New Ecoefficient Process—Part I: Feasibility of Electrodialysis with Bipolar Membrane and Production of Neokyotorphin (α137-141)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Electrodialysis Cell Configuration
2.3. Protocol
2.3.1. Conventional Hydrolysis
Stock Solution Preparation
Hydrolysis Process
2.3.2. Hydrolysis by EDBM-MCP and EDBM-AEM
2.4. Analyses
2.4.1. Determination of Hemoglobin Concentration: Drabkin’s Method
2.4.2. pH
2.4.3. Conductivity
2.4.4. EDBM Global Resistance
2.4.5. Membrane Thickness and Electrical Conductivity
2.4.6. Total Solid and Ash Contents
2.4.7. Mineral Concentration Measurement
2.4.8. Determination of the Degree of Hydrolysis of Bovine Hemoglobin
2.4.9. RP-UPLC Analyses
Identification of Hydrolysis Mechanism and α137-141 Peptide
Identification and Characterization of Peptide Population
Quantification of α137-141 in the Hydrolysates by Mass Spectrometry
2.4.10. RP-UPLC Statistical Analyses
3. Results and Discussion
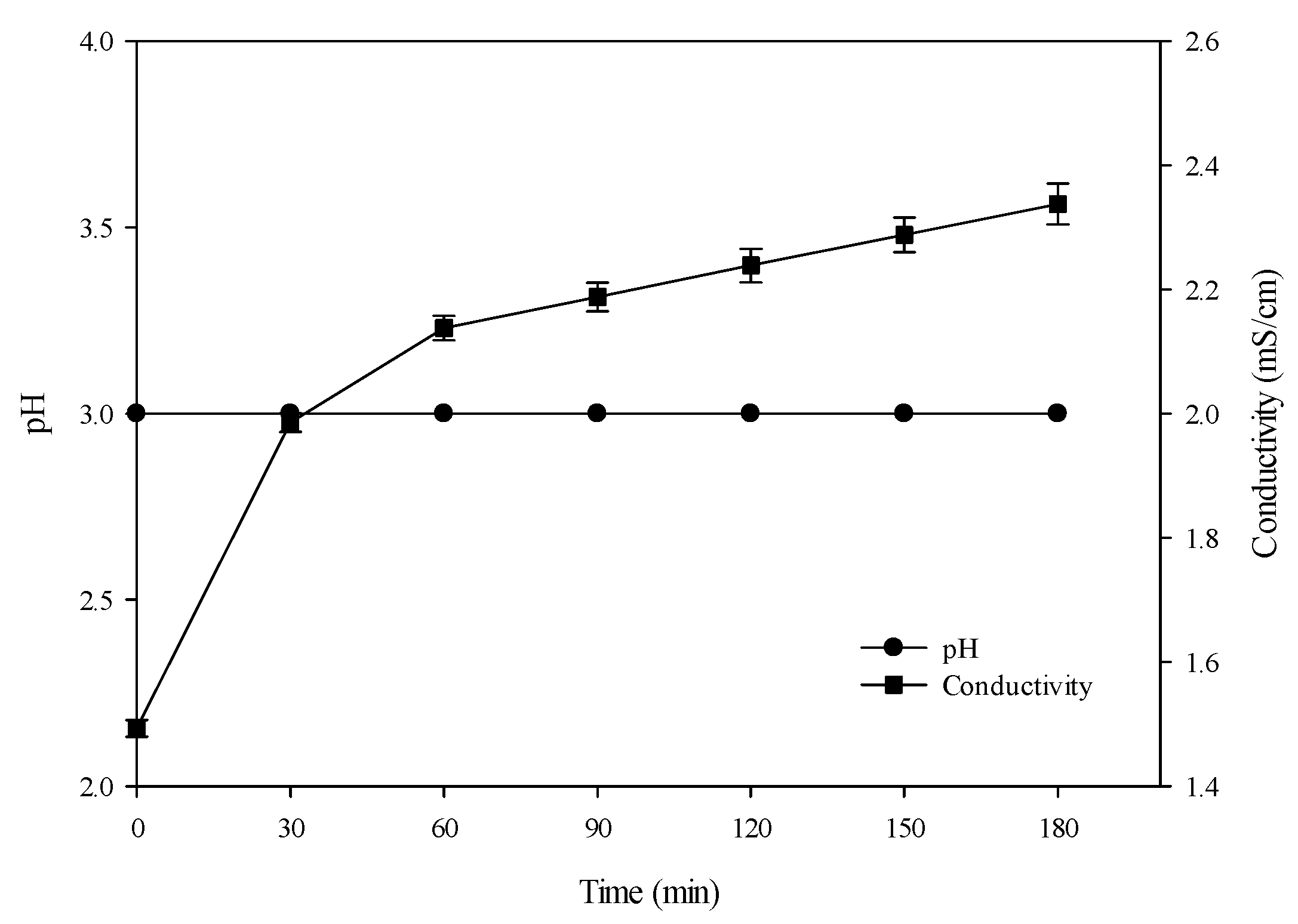
3.1. Conventional Hydrolysis (Control): Evolution of pH and Conductivity
3.2. Hydrolysis in EDBM: Evolution of pH, Conductivity, Global Resistance and Membrane Characterization
3.2.1. pH
3.2.2. Conductivity
3.2.3. ED System Global Resistance
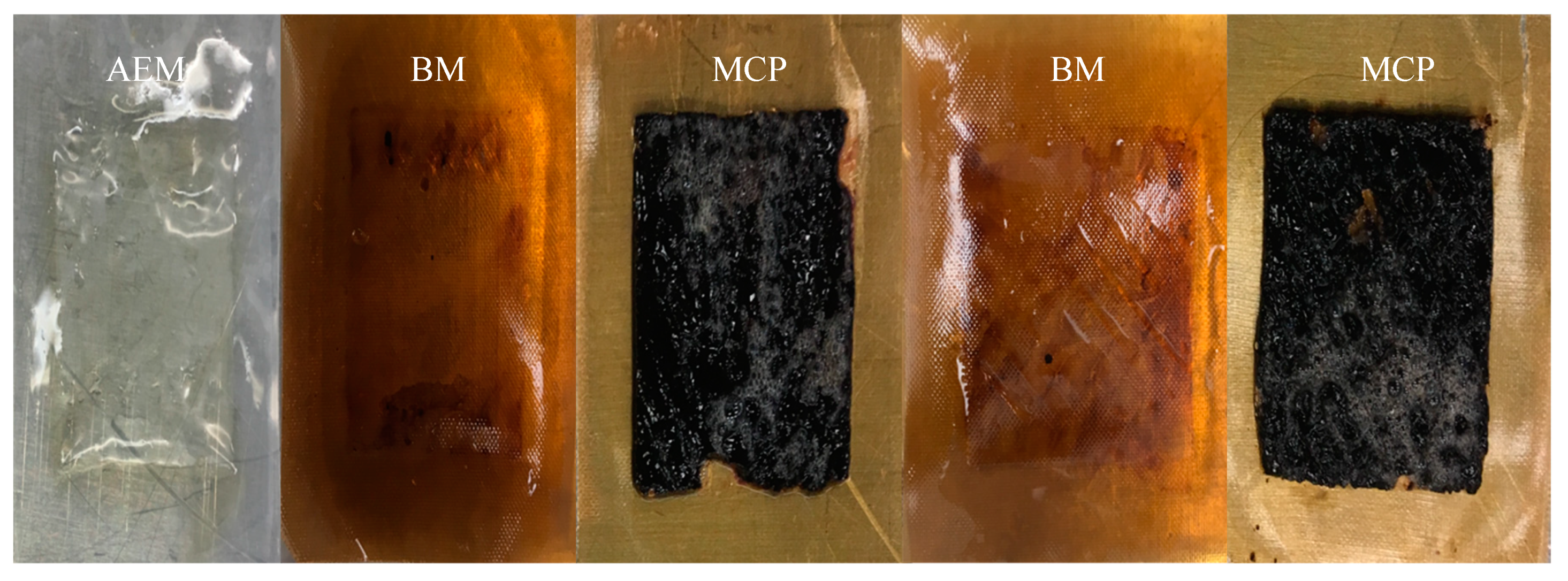
3.2.4. Membrane Characterization
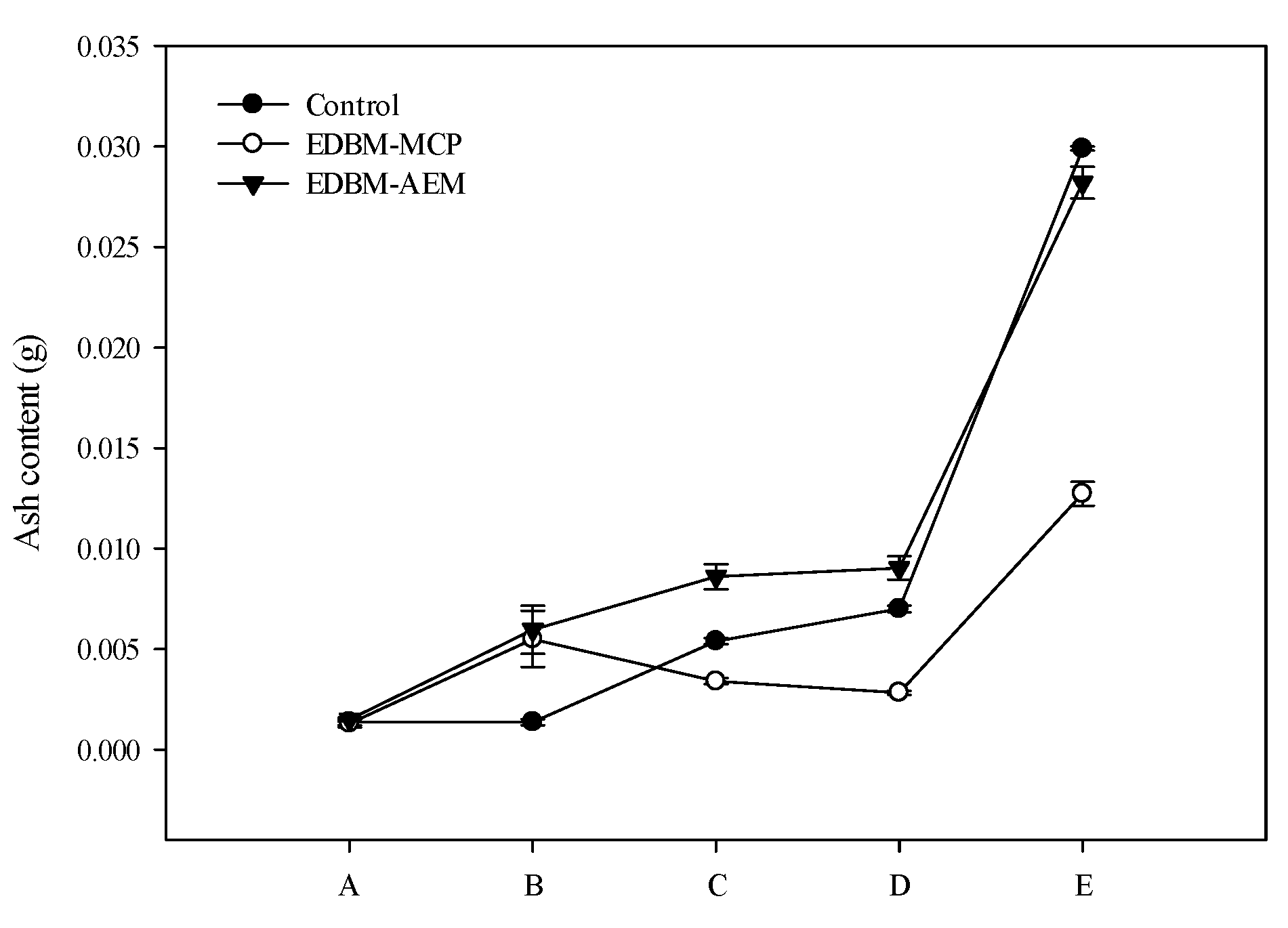
3.3. Ash Content and Mineral Composition
3.3.1. Evolution of Ash Content
3.3.2. Mineral Composition of the 1% HB Solution after Hydrolysis
3.4. Enzymatic Kinetics and Mechanism of Action of the New Process
3.4.1. Determination of the Degree of Hydrolysis
3.4.2. Reaction Mechanism of Bovine Hemoglobin Hydrolysis by Pepsin and Identification of α137-141 Peptide
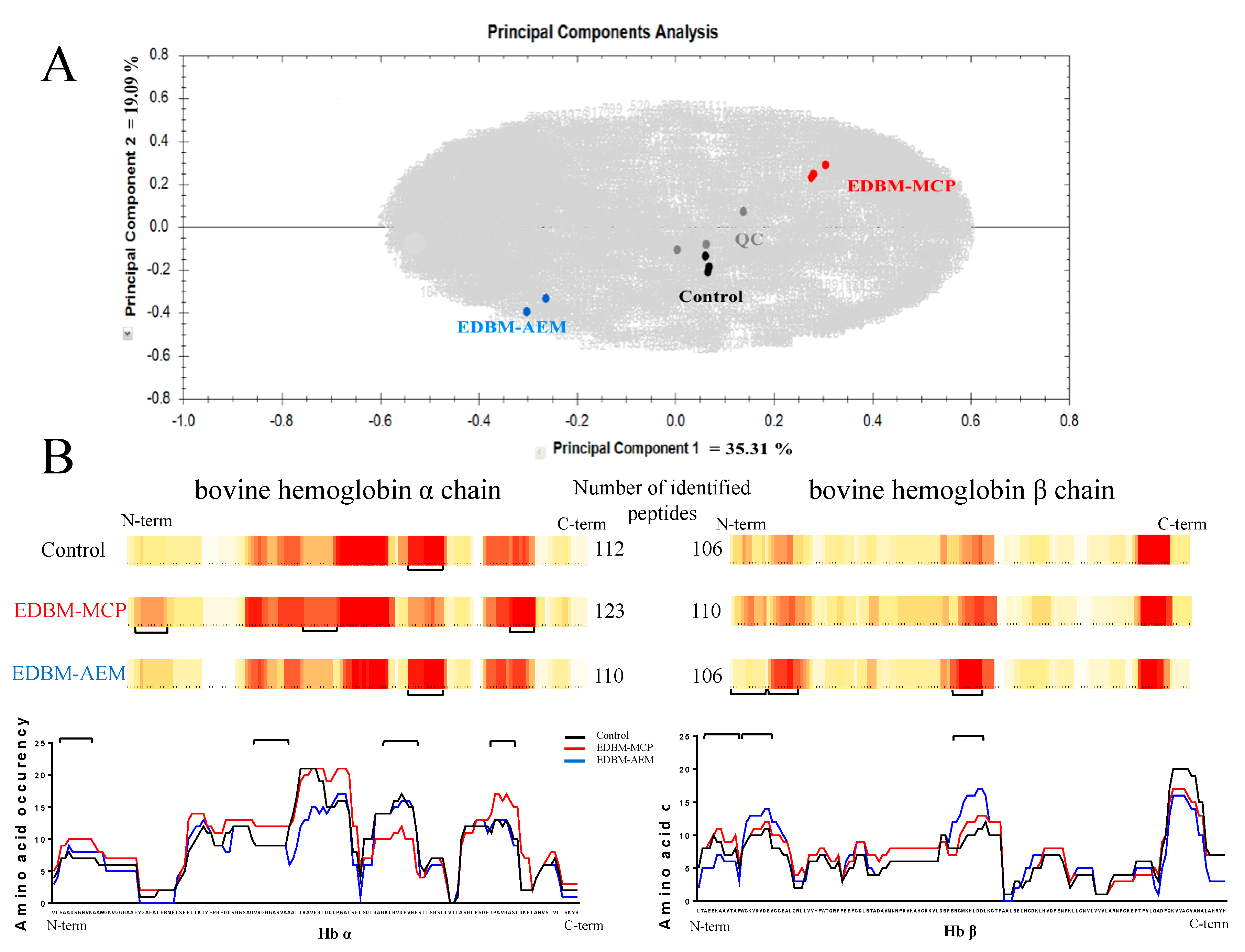
3.4.3. Characterization of the Peptide Populations
Principal Component Analysis (PCA) and Heat Maps
3.4.4. Quantification of α137-141 Derived from the New Process
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gatnau, R.; Polo, J.; Robert, E. Plasma protein antimicrobial substitution at negligible risk. Cah. Opt. Méditerr. 2001, 54, 141–150. [Google Scholar]
- Toldrà, F.; Aristoy, M.C.; Mora, L.; Reig, M. Innovations in value-addition of edible meat by-products. Meat Sci. 2012, 92, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Sasakawa, S. Studies on Hemoglobin. VI. Amino acid composition of the fractionated bovine globin α and β. J. Biochem. 1961, 50, 345–351. [Google Scholar]
- Lignot, B.; Froidevaux, R.; Nedjar-Arroume, N.; Guillochon, D. Solvent effect on kinetics of appearance of neokyotorphin, VVh4 and a bradykinin-potentiating peptide in the course of peptic hydrolysis of bovine hemoglobin. Appl. Biochem. Biotechnol. 1999, 30, 201–207. [Google Scholar]
- Zhao, Q.; Piot, J.M. Neokyotorphin formation and quantitative evolution following human hemoglobin hydrolysis with cathepsin D. Peptides 1997, 19, 759–766. [Google Scholar] [CrossRef]
- Zhao, Q.Y.; Piot, J.M. Investigation of inhibition angiotensin-converting enzyme (ACE) activity and opioid activity of two hemorphins, LVV-hemorphin-5 and VVhemorphin-5, isolated from a defined peptic hydrolysate of bovine hemoglobin. Neuropeptides 1997, 2, 147–153. [Google Scholar]
- Vercaigne-Marko, D.; Kosciarz, E.; Nedjar-Arroume, N.; Guillochon, D. Improvement of Staphylococcus aureus-V8-protease hydrolysis of bovine hemoglobin by its adsorption on to a solid phase in the presence of SDS: Peptide mapping and obtaining of two haemopoietic peptides. Appl. Biochem. Biotechnol. 2000, 31, 127–134. [Google Scholar]
- Adje, E.Y.; Balti, R.; Kouach, M.; Guillochon, D.; Nedjar-Arroume, N. α67-106 of bovine hemoglobin: A new family of antimicrobial and angiotensin I-converting enzyme inhibitory peptides. Eur. Food Res. Technol. 2011, 232, 637–646. [Google Scholar]
- Daoud, R.; Dubois, V.; Bors-Dodita, L.; Nedjar-Arroume, N.; Krier, F.; Chihib, N.-E.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. New antibacterial peptide derived from bovine hemoglobin. Peptides 2005, 26, 713–719. [Google Scholar] [CrossRef]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Adje, E.Y.; Traisnel, J.; Krier, F.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. Bovine hemoglobin: An attractive source of antibacterial peptides. Peptides 2008, 29, 969–977. [Google Scholar] [CrossRef]
- Choisnard, L.; Froidevaux, R.; Nedjar-Arroume, N.; Lignot, B.; Vercaigne-Marko, D.; Krier, F.; Dhulster, P.; Guillochon, D. Kinetic study of the appearance of an anti-bacterial peptide in the course of bovine hemoglobin peptic hydrolysis. Appl. Biochem. Biotechnol. 2002, 36, 187–194. [Google Scholar]
- Przybylski, R.; Firdaous, L.; Châtaigné, G.; Dhulster, P.; Nedjar, N. Production of an antimicrobial peptide derived from slaughterhouse by-product and its potential application on meat as preservative. Food Chem. 2016, 211, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Adje, E.Y.; Balti, R.; Kouach, M.; Dhulster, P.; Guillochon, D.; Nedjar-Arroume, N. Obtaining antimicrobial peptides by controlled peptic hydrolysis of bovine hemoglobin. Int. J. Biol. Macromol. 2011, 49, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.; Dale, C.S.; Casten, K.; Geigner, M.A.; Gozzo, F.C.; Ferro, E.S.; Heimann, A.S.; Lakshmi, A.D. Hemoglobin-derived peptides as novel type of bioactive signaling molecules. AAPS J. 2010, 12, 658–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Miloudi, K.; Daoud, R.; Krier, F.; Kouach, M.; Briand, G.; Guillochon, D. Isolation and characterization of four antibacterial peptides from bovine hemoglobin. Peptides 2006, 27, 2082–2089. [Google Scholar] [CrossRef]
- Mikhaylin, S.; Patouillard, L.; Margni, M.; Bazinet, L. Milk protein production by a more environmentally sustainable process: Bipolar membrane electrodialysis coupled with ultrafiltration. Green Chem. 2018, 20, 449. [Google Scholar] [CrossRef]
- Bazinet, L.; Lamarche, F.; Labrecque, R.; Toupin, R.; Boulet, M.; Ippersiel, D. Electroacidification of soybean proteins for production of isolate. Food Technol. 1997, 51, 52–60. [Google Scholar]
- Lin Teng Shee, F.; Arul, J.; Brunet, S.; Mateescu, M.A.; Bazinet, L. Solubilization of Chitosan by Bipolar Membrane Electroacidification. J. Agric. Food Chem. 2006, 54, 6760–6764. [Google Scholar] [CrossRef]
- Faucher, M.; Serre, E.; Langevin, M.E.; Mikhaylin, S.; Lutin, F.; Bazinet, L. Drastic energy consumption reduction and ecoefficiency improvement of cranberry juice deacidification by electrodialysis with bipolar membranes at semi-industrial scale: Reuse of the recovery solution. J. Membr. Sci. 2018, 555, 105–114. [Google Scholar] [CrossRef]
- Abdu, S.; Martí-Calatayud, M.C.; Wong, J.E.; García-Gabaldón, M.; Wessling, M. Layer-by-layer modification of cation exchange membranes controls ion selectivity and water splitting. ACS Appl. Mater. Interfaces 2014, 6, 1843–1854. [Google Scholar]
- Van der Bruggen, B.; Koninckx, A.; Vandecasteele, C. Separation of monovalent and divalent ions from aqueous solution by electrodialysis and nanofiltration. Water Res. 2004, 38, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Persico, M.; Bazinet, L. Fouling prevention of peptides from a tryptic whey hydrolysate during electromembrane processes by use of monovalent ion permselective membranes. J. Membr. Sci. 2018, 549, 486–494. [Google Scholar] [CrossRef]
- Takagi, H.; Shiomi, H.; Fukui, K.; Hayashi, K.; Kiso, Y.; Kitagawa, K. Isolation of a novel analgesic pentapeptide, neo-kyotorphin, from bovine brain. Life Sci. 1982, 31, 1733–1736. [Google Scholar] [CrossRef]
- Catiau, L.; Traisnel, J.; Delval-Dubois, V.; Chihib, N.-E.; Guillochon, D.; Nedjar- Arroume, N. Minimal antimicrobial peptide sequence from hemoglobin alpha chain: KYR. Peptides 2011, 32, 633–638. [Google Scholar] [PubMed]
- Crosby, W.H.; Mann, J.I.; Furth, F.W. Standardizing a method for clinical hemoglobinometry. U.S. Armed Forces Med. J. 1954, 5, 693–703. [Google Scholar]
- Dunn, B.M. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002, 102, 4431–4458. [Google Scholar]
- Mikhaylin, S.; Nikonenko, V.; Pismenskaya, N.; Pourcelly, G.; Choi, S.; Kwon, H.J.; Han, J.; Bazinet, L. How physico-chemical and surface properties of cation exchange membrane affect membrane scaling and electroconvective vortices: Influence on performance of electrodialysis with pulsed electric field. Desalination 2016, 393, 102–114. [Google Scholar] [CrossRef]
- Lteif, R.; Dammak, L.; Larchet, C.; Auclair, B. Conductivité membranaire: Étude de l’effet de la concentration, de la nature de l’électrolyte et de la structure membranaire. Eur. Polym. J. 1999, 35, 1187–1195. [Google Scholar]
- Lebrun, L.; Da Silva, E.; Pourcelly, G.; Metayer, M. Elaboration and characterisation of ion-exchange films used in the fabrication of bipolar membranes. J. Membr. Sci. 2003, 227, 95–111. [Google Scholar] [CrossRef]
- AOAC International. Method 945.46. In Official Methods of Analysis, 15th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 1990. [Google Scholar]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983, 66, 1219–1227. [Google Scholar] [CrossRef]
- Spellman, D.; McEvoy, E.; O’Cuin, G.; Fitzgerald, R.J. Proteinase and exopeptidase hydrolysis of whey protein: Comparison of the TNBS, OPA and pH stat methods for quantification of degree of hydrolysis. Int. Dairy J. 2003, 13, 447–453. [Google Scholar] [CrossRef]
- Church, F.C.; Porter, D.H.; Catignani, G.L.; Swaisgood, H.E. An o-phthaldialdehyde spectrometric assay for proteinases. Anal. Biochem. 1985, 146, 343–348. [Google Scholar] [PubMed]
- Zhao, Q.Y.; Sannier, F.; Ricart, G.; Piot, J.M. A rapid detection and identification of hemorphins released from bovin hemoglobin enzymatic hydrolysate by use of HPLC coupled with Photodiode Array Detector. J. Liq. Chromatogr. 1995, 18, 93–103. [Google Scholar]
- Persico, M.; Mikhaylin, S.; Doyen, A.; Firdaous, L.; Hammami, R.; Bazinet, L. How peptide physicochemical and structural characteristics affect anion-exchange membranes fouling by a tryptic whey protein hydrolysate. J. Membr. Sci. 2016, 520, 914–923. [Google Scholar] [CrossRef]
- Persico, M.; Mikhaylin, S.; Doyen, A.; Firdaous, L.; Hammami, R.; Chevalier, M.; Flahaut, C.; Dhulster, P.; Bazinet, L. Formation of peptide layers and adsorption mechanisms on a negatively charged cation-exchange membrane. J. Colloid Interface Sci. 2017, 508, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Bazinet, L.; Ippersiel, D.; Gendron, C.; Beaudry, J.; Mahdavi, B.; Amiot, J.; Lamarche, F. Cationic balance in skim milk during bipolar membrane electroacidification. J. Membr. Sci. 2000, 173, 201–209. [Google Scholar] [CrossRef]
- Lin Teng Shee, F.; Angers, P.; Bazinet, L. Precipitation of cheddar cheese whey lipids by electrochemical acidification. J. Agric. Food Chem. 2005, 53, 5635–5639. [Google Scholar] [CrossRef] [PubMed]
- Vera, E.; Ruales, J.; Dornier, M.; Sandeaux, J.; Persin, F.; Pourcelly, G.; Vaillant, F.; Reynes, M. Comparison of different methods for deacidification of clarified passion fruit juice. J. Food Eng. 2003, 59, 361–367. [Google Scholar] [CrossRef]
- Jaime-Ferrer, J.S.; Couallier, E.; Viers, P.; Rakib, M. Two-compartment bipolar membrane electrodialysis for splitting of sodium formate into formic acid and sodium hydroxide: Modelling. J. Membr. Sci. 2009, 328, 75–80. [Google Scholar] [CrossRef] [Green Version]
- Kravtsov, V.; Kulikova, I.; Mikhaylin, S.; Bazinet, L. Alkalinization of acid whey by means of electrodialysis with bipolar membranes and analysis of induced membrane fouling. J. Food Eng. 2020, 277, 109891. [Google Scholar] [CrossRef]
- Simmons, D.A.; Wilson, D.J.; Lajoie, G.A.; Doherty-Kirby, A.; Konermann, L. Subunit disassembly and unfolding kinetics of hemoglobin studied by time-resolved electrospray mass spectrometry. Biochemistry 2004, 43, 14792–14801. [Google Scholar] [CrossRef] [PubMed]
- Boys, B.L.; Kuprowski, M.C.; Konermann, L. Symmetric behavior of hemoglobin α- and β-subunits during acid-induced denaturation observed by electrospray mass spectrometry. Biochemistry 2007, 46, 10675–10684. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, F.; Bazus, A.; Dhulster, P.; Guillochon, D. Solubility of hem in hem-iron enriched bovine hemoglobin hydrolysates. J. Agric. Food Chem. 1998, 46, 5017–5025. [Google Scholar] [CrossRef]
- Langevin, M.E.; Bazinet, L. Ion-exchange membrane fouling by peptides: A phenomenon governed by electrostatic interactions. J. Membr. Sci. 2011, 369, 359–366. [Google Scholar] [CrossRef]
- Lin Teng Shee, F.; Arul, J.; Brunet, S.; Bazinet, L. Chitosan solubilization by bipolar membrane electroacidification: Reduction of membrane fouling. J. Membr. Sci. 2007, 290, 29–35. [Google Scholar] [CrossRef]
- Linderstrom-Lang, K. Les phases initiales de la dégradation des protéines par les enzymes. Bull. Soc. Chim. Biol. 1953, 35, 100–116. [Google Scholar]
- Dubois, V.; Nedjar-Arroume, N.; Guillochon, D. Influence of pH on the appearance of active peptides in the course of peptic hydrolysis of bovine hemoglobin. Prep. Biochem. Biotechnol. 2005, 35, 85–102. [Google Scholar] [CrossRef]
- Bazinet, L.; Geoffroy, T.R. Electrodialytic processes: Market overview, membrane phenomena, recent developments and sustainable strategies. Membranes 2020, 10, 221. [Google Scholar] [CrossRef]





| Protein (%) | ≃98 |
| Ash (%) | ≤2 |
| Water (%) | ≤8 |
| Iron (Fe) (%) | 0.25–0.35 |
| Process | EDBM-MCP | EDBM-AEM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Conductivity (mS/cm) | AEM | MCP-1 | MCP-2 | BM-1 | BM-2 | AEM-1 | AEM-2 | AEM-3 | BM-1 | BM-2 |
| Before | 5.3 ± 0.05 a | 4.3 ± 0.1 a | 4.23 ± 0.05 a | 5.8 ± 0.05 a | 5.7 ± 0.2 a | 5.3 ± 0.15 a | 5 ± 0.2 a | 4.9 ± 0.2 a | 5.9 ± 0.2 a | 5.8 ± 0.15 a |
| After | 5.3 ± 0.15 a | 0.86 ± 0.11 b | 1.06 ± 0.25 b | 5.06 ± 0.05 b | 5.13 ± 0.15 b | 5.1 ± 0.05 a | 4.76 ± 0.25 a | 5.06 ± 0.2 a | 5.33 ± 0.2 b | 5.4 ± 0.11 b |
| Conditions | Cl | K | S | Na |
|---|---|---|---|---|
| Control | 388.3 ± 5.03 a | 3.73 ± 0.34 a | 61.35 ± 1.31 a | 2.85 ± 0.11 a |
| EDBM-MCP | 197.6 ± 4.04 b | 6.48 ± 0.15 a | 41.37 ± 0.98 b | 1.68 ± 0.8 a |
| EDBM-AEM | 235.6 ± 4.16 c | 364.76 ± 3.8 b | 313.69 ± 3.87 c | 23.35 ± 2.06 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abou-Diab, M.; Thibodeau, J.; Deracinois, B.; Flahaut, C.; Fliss, I.; Dhulster, P.; Nedjar, N.; Bazinet, L. Bovine Hemoglobin Enzymatic Hydrolysis by a New Ecoefficient Process—Part I: Feasibility of Electrodialysis with Bipolar Membrane and Production of Neokyotorphin (α137-141). Membranes 2020, 10, 257. https://doi.org/10.3390/membranes10100257
Abou-Diab M, Thibodeau J, Deracinois B, Flahaut C, Fliss I, Dhulster P, Nedjar N, Bazinet L. Bovine Hemoglobin Enzymatic Hydrolysis by a New Ecoefficient Process—Part I: Feasibility of Electrodialysis with Bipolar Membrane and Production of Neokyotorphin (α137-141). Membranes. 2020; 10(10):257. https://doi.org/10.3390/membranes10100257
Chicago/Turabian StyleAbou-Diab, Mira, Jacinthe Thibodeau, Barbara Deracinois, Christophe Flahaut, Ismail Fliss, Pascal Dhulster, Naima Nedjar, and Laurent Bazinet. 2020. "Bovine Hemoglobin Enzymatic Hydrolysis by a New Ecoefficient Process—Part I: Feasibility of Electrodialysis with Bipolar Membrane and Production of Neokyotorphin (α137-141)" Membranes 10, no. 10: 257. https://doi.org/10.3390/membranes10100257







