A Universal Influenza Virus Vaccine Candidate Tested in a Pig Vaccination-Infection Model in the Presence of Maternal Antibodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals
2.3. Experimental Design, Vaccination
2.4. Challenge and Sample Collection
2.5. Virus Titration of Nasal Swabs and BALF
2.6. Pathology
2.7. HI Assay
2.8. ELISA
2.9. Immune Cell Isolation
2.10. IFNγ ELISpot
2.11. Intracellular Cytokine Staining (ICS) and Flow Cytometry
2.12. Statistical Analyses
3. Results
3.1. Vaccination Did Not Result in an Increase of Antibodies against H1 HA Head and Stalk Domains
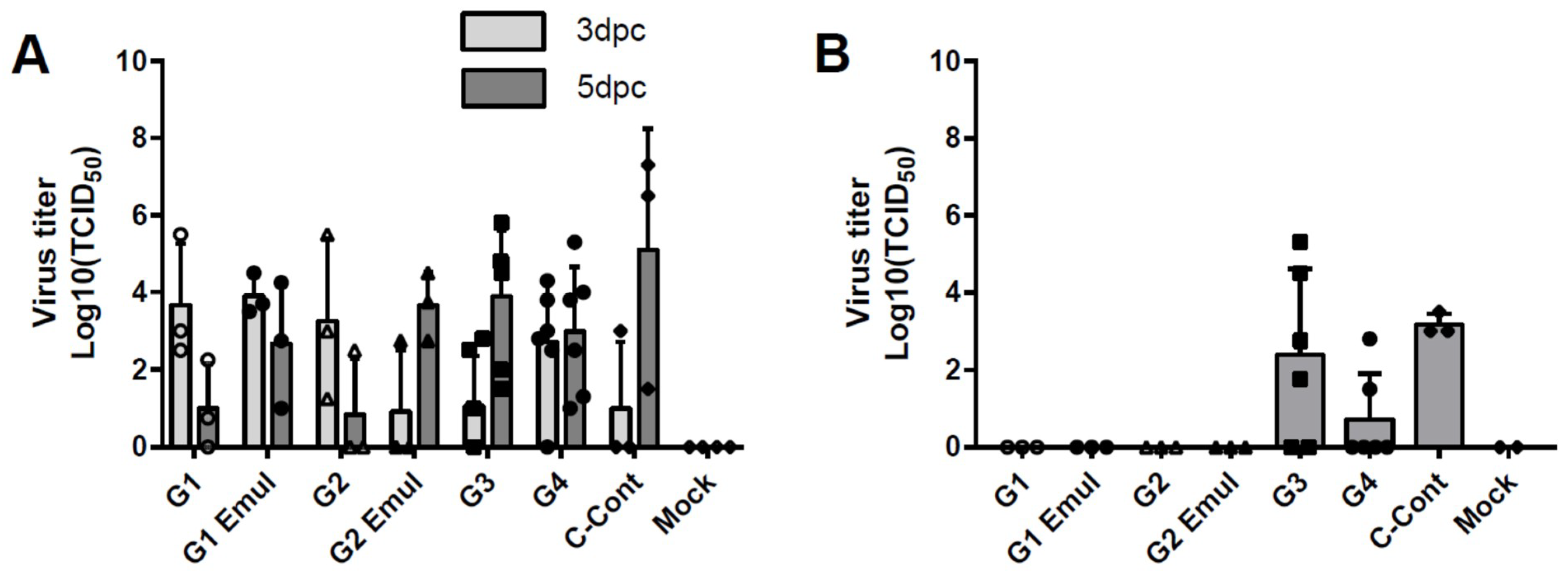
3.2. Virus Replication in the Respiratory Tract
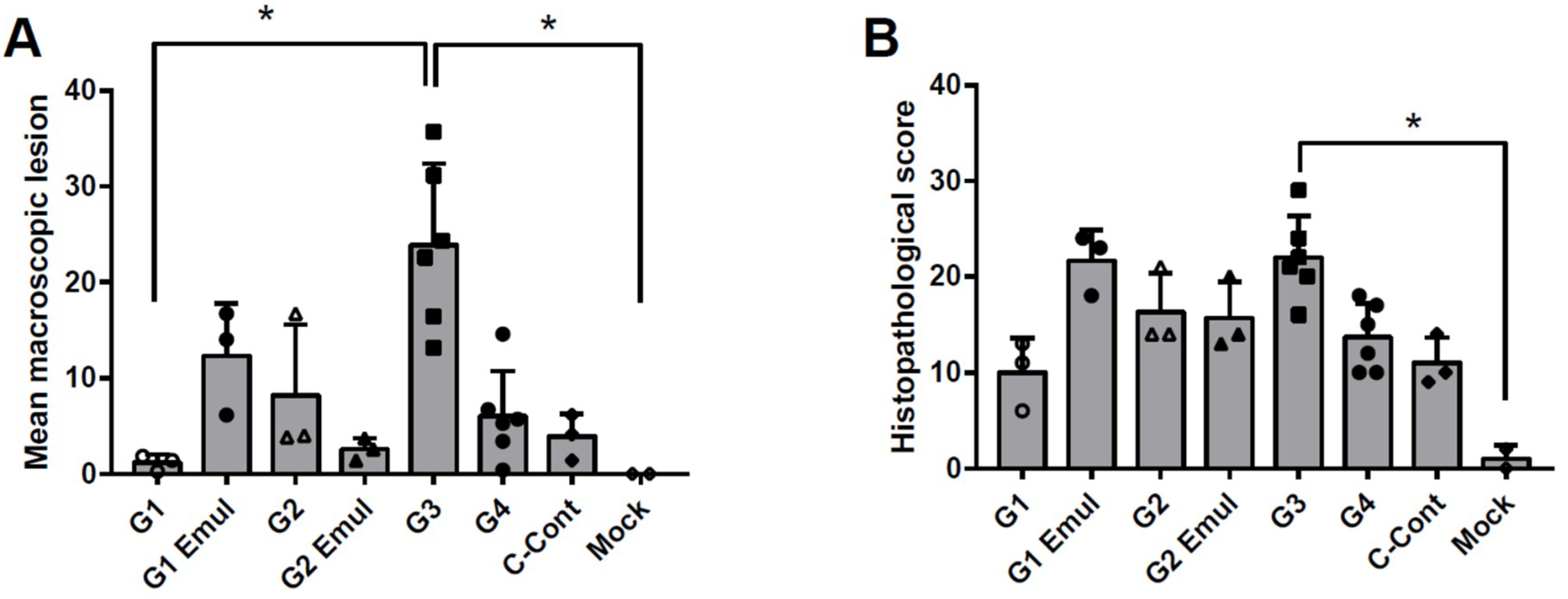
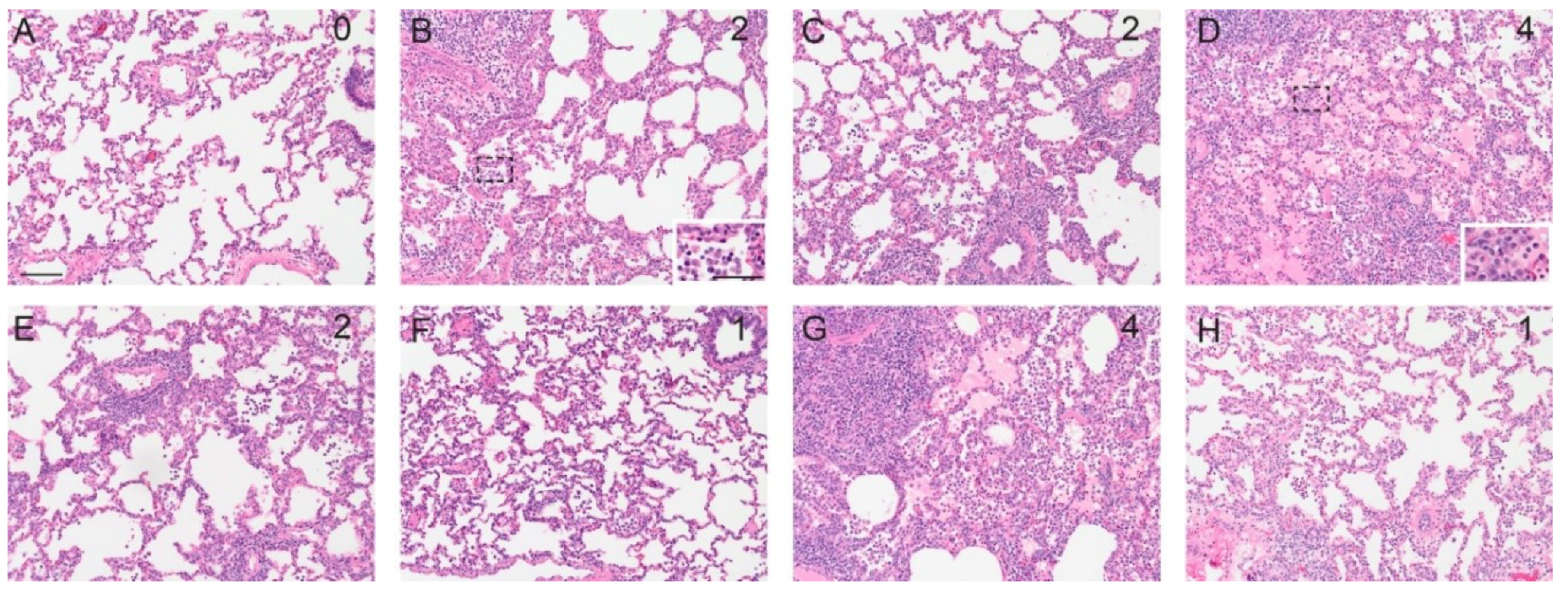
3.3. Pathology
3.4. Cellular Responses Measured after Challenge Reflect Priming by Influenza Virus Vaccination
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BALF | bronchio-alveolar lavage fluid |
| cHA | chimeric hemagglutinin |
| CPE | cytophathic effect |
| DPC | days post challenge |
| Emul | Emulsigen adjuvant |
| H&E staining | hematoxylin and eosin staining |
| HA | hemagglutinin |
| HI | hemagglutination inhibition |
| ICS | intracellular cytokine staining |
| IFN | interferon |
| Ig | immunoglobulin |
| IHC | immunohistochemistry |
| IIV | inactivated whole virus influenza vaccine |
| LAIV | live attenuated influenza virus vaccine |
| MDCK | Madin Darby canine kidney |
| MEM | minimal essential medium |
| NP | nucleoprotein |
| PBMCs | peripheral blood mononuclear cells |
| PFU | Plaque forming units |
| TCID50 | tissue culture infectious dose 50 |
| TIV | trivalent inactivated virus vaccine |
| TPCK | tosyl phenylalanyl chloromethyl ketone |
| VAERD | vaccine-associated enhanced respiratory disease |
References
- WHO. Ask the Expert: Influenza Q&A. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 11 September 2018).
- Molinari, N.A.; Ortega-Sanchez, I.R.; Messonnier, M.L.; Thompson, W.W.; Wortley, P.M.; Weintraub, E.; Bridges, C.B. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine 2007, 25, 5086–5096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanton, L.; Alabi, N.; Mustaquim, D.; Taylor, C.; Kniss, K.; Kramer, N.; Budd, A.; Garg, S.; Cummings, C.N.; Chung, J.; et al. Update: Influenza activity in the United States during the 2016–17 season and composition of the 2017–18 influenza vaccine. Morb. Mortal. Wkly. Rep. 2017, 66, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.F.; Neumann, G.; Kawaoka, Y. Orthomyxoviruses. In Fields Virology, 5th ed.; LWW: Philadelphia, PA, USA, 2006. [Google Scholar]
- Palese, P. Influenza: Old and new threats. Nat. Med. 2004, 10, S82–S87. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015, 14, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Gerdil, C. The annual production cycle for influenza vaccine. Vaccine 2003, 21, 1776–1779. [Google Scholar] [CrossRef]
- Heaton, N.S.; Sachs, D.; Chen, C.J.; Hai, R.; Palese, P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20248–20253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doud, M.B.; Bloom, J.D. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses 2016, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- De Jong, J.C.; Beyer, W.E.; Palache, A.M.; Rimmelzwaan, G.F.; Osterhaus, A.D. Mismatch between the 1997/1998 influenza vaccine and the major epidemic a(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol. 2000, 61, 94–99. [Google Scholar] [CrossRef]
- Xie, H.; Wan, X.F.; Ye, Z.; Plant, E.P.; Zhao, Y.; Xu, Y.; Li, X.; Finch, C.; Zhao, N.; Kawano, T.; et al. H3N2 mismatch of 2014–15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci. Rep. 2015, 5, 15279. [Google Scholar] [CrossRef] [PubMed]
- Berlanda Scorza, F.; Tsvetnitsky, V.; Donnelly, J.J. Universal influenza vaccines: Shifting to better vaccines. Vaccine 2016, 34, 2926–2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachbagauer, R.; Krammer, F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017, 23, 222–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, L.; Compans, R.W.; Wang, B.Z. Universal influenza vaccines, a dream to be realized soon. Viruses 2014, 6, 1974–1991. [Google Scholar] [CrossRef] [PubMed]
- Throsby, M.; Van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; Van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5H1 and H1H1 recovered from human IGM+ memory B cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, H.; Rajendran, M.; Choi, A.; Sjursen, H.; Brokstad, K.A.; Cox, R.J.; Palese, P.; Krammer, F.; Nachbagauer, R. Influenza virus hemagglutinin stalk-specific antibodies in human serum are a surrogate marker for in vivo protection in a serum transfer mouse challenge model. MBio 2017, 8, e01463-17. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Wilson, I.A. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr. Opin. Virol. 2012, 2, 134–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Kinzler, D.; Choi, A.; Hirsh, A.; Beaulieu, E.; Lecrenier, N.; Innis, B.L.; Palese, P.; Mallett, C.P.; Krammer, F. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Ermler, M.E.; Tan, G.S.; Krammer, F.; Palese, P.; Hai, R. Influenza A viruses expressing intra- or intergroup chimeric hemagglutinins. J. Virol. 2016, 90, 3789–3793. [Google Scholar] [CrossRef] [PubMed]
- Hai, R.; Krammer, F.; Tan, G.S.; Pica, N.; Eggink, D.; Maamary, J.; Margine, I.; Albrecht, R.A.; Palese, P. Influenza viruses expressing chimeric hemagglutinins: Globular head and stalk domains derived from different subtypes. J. Virol. 2012, 86, 5774–5781. [Google Scholar] [CrossRef] [PubMed]
- Kahn, R.E.; Ma, W.; Richt, J.A. Swine and influenza: A challenge to one health research. Curr. Top. Microbiol. Immunol. 2014, 385, 205–218. [Google Scholar] [PubMed]
- Vincent, A.L.; Lager, K.M.; Anderson, T.K. A brief introduction to influenza A virus in swine. Methods Mol. Biol. 2014, 1161, 243–258. [Google Scholar] [PubMed]
- Powdrill, T.F.; Nipp, T.L.; Rinderknecht, J.L. One health approach to influenza: Assessment of critical issues and options. Emerg. Infect. Dis. 2010, 16, e1. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; Rijsewijk, F.A.; De Boer-Luijtze, E.A.; Bianchi, A.T. Vaccination of pigs with a DNA construct expressing an influenza virus m2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J. Gen. Virol. 2002, 83, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Lager, K.M.; Janke, B.H.; Gramer, M.R.; Richt, J.A. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet. Microbiol. 2008, 126, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Gauger, P.C.; Vincent, A.L.; Loving, C.L.; Lager, K.M.; Janke, B.H.; Kehrli, M.E.; Roth, J.A. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (δ-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 2011, 29, 2712–2719. [Google Scholar] [CrossRef] [PubMed]
- Rajão, D.S.; Chen, H.; Perez, D.R.; Sandbulte, M.R.; Gauger, P.C.; Loving, C.L.; Shanks, G.D.; Vincent, A. Vaccine-associated enhanced respiratory disease is influenced by haemagglutinin and neuraminidase in whole inactivated influenza virus vaccines. J. Gen. Virol. 2016, 97, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Rajão, D.S.; Loving, C.L.; Gauger, P.C.; Kitikoon, P.; Vincent, A.L. Influenza A virus hemagglutinin protein subunit vaccine elicits vaccine-associated enhanced respiratory disease in pigs. Vaccine 2014, 32, 5170–5176. [Google Scholar] [CrossRef] [PubMed]
- Rajao, D.S.; Sandbulte, M.R.; Gauger, P.C.; Kitikoon, P.; Platt, R.; Roth, J.A.; Perez, D.R.; Loving, C.L.; Vincent, A.L. Heterologous challenge in the presence of maternally-derived antibodies results in vaccine-associated enhanced respiratory disease in weaned piglets. Virology 2016, 491, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.K.; Rajão, D.S.; Loving, C.L.; Gauger, P.C.; Pérez, D.R.; Vincent, A.L. Age at vaccination and timing of infection do not alter vaccine-associated enhanced respiratory disease in influenza A virus-infected pigs. Clin. Vaccine Immunol. 2016, 23, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Loving, C.L.; Manischewitz, J.; King, L.R.; Gauger, P.C.; Henningson, J.; Vincent, A.L.; Golding, H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013, 5, 200ra114. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Liu, W.C.; Choi, A.; Wohlbold, T.J.; Atlas, T.; Rajendran, M.; Solorzano, A.; Berlanda-Scorza, F.; Garcia-Sastre, A.; Palese, P.; et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2017, 2, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachbagauer, R.; Choi, A.; Izikson, R.; Cox, M.M.; Palese, P.; Krammer, F. Age dependence and isotype specificity of influenza virus hemagglutinin stalk-reactive antibodies in humans. MBio 2016, 7, e01996-15. [Google Scholar] [CrossRef] [PubMed]
- Isakova-Sivak, I.; Korenkov, D.; Smolonogina, T.; Kotomina, T.; Donina, S.; Matyushenko, V.; Mezhenskaya, D.; Krammer, F.; Rudenko, L. Broadly protective anti-hemagglutinin stalk antibodies induced by live attenuated influenza vaccine expressing chimeric hemagglutinin. Virology 2018, 518, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, M.; Sun, W.; Comella, P.; Nachbagauer, R.; Wohlbold, T.J.; Amanat, F.; Kirkpatrick, E.; Palese, P.; Krammer, F. An immuno-assay to quantify influenza virus hemagglutinin with correctly folded stalk domains in vaccine preparations. PLoS ONE 2018, 13, e0194830. [Google Scholar] [CrossRef] [PubMed]
- De Vleeschauwer, A.; Atanasova, K.; Van Borm, S.; van den Berg, T.; Rasmussen, T.B.; Uttenthal, A.; Van Reeth, K. Comparative pathogenesis of an avian H5N2 and a swine H1N1 influenza virus in pigs. PLoS ONE 2009, 4, e6662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Poucke, S.G.; Nicholls, J.M.; Nauwynck, H.J.; Van Reeth, K. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol. J. 2010, 7, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Belisle, S.E.; Mosier, D.; Li, X.; Stigger-Rosser, E.; Liu, Q.; Qiao, C.; Elder, J.; Webby, R.; Katze, M.G.; et al. 2009 pandemic H1N1 influenza virus causes disease and upregulation of genes related to inflammatory and immune responses, cell death, and lipid metabolism in pigs. J. Virol. 2011, 85, 11626–11637. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Richt, J.A.; Lager, K.M.; Janke, B.H.; Woods, R.D.; Webster, R.G.; Webby, R.J. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the united states. J. Clin. Microbiol. 2003, 41, 3198–3205. [Google Scholar] [CrossRef] [PubMed]
- Valheim, M.; Gamlem, H.; Gjerset, B.; Germundsson, A.; Lium, B. Pathological findings and distribution of pandemic influenza A (H1N1) 2009 virus in lungs from naturally infected fattening pigs in Norway. Influenza Res. Treat. 2011, 2011, 565787. [Google Scholar] [CrossRef] [PubMed]
- Henningson, J.N.; Rajao, D.S.; Kitikoon, P.; Lorusso, A.; Culhane, M.R.; Lewis, N.S.; Anderson, T.K.; Vincent, A.L. Comparative virulence of wild-type H1N1pdm09 influenza a isolates in swine. Vet. Microbiol. 2015, 176, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Margine, I.; Tan, G.S.; Pica, N.; Krause, J.C.; Palese, P. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS ONE 2012, 7, e43603. [Google Scholar] [CrossRef] [PubMed]
- Margine, I.; Palese, P.; Krammer, F. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp. 2013, 81, 51112. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Miller, M.S.; Hai, R.; Ryder, A.B.; Rose, J.K.; Palese, P.; García-Sastre, A.; Krammer, F.; Albrecht, R.A. Hemagglutinin stalk immunity reduces influenza virus replication and transmission in ferrets. J. Virol. 2015, 90, 3268–3273. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Hai, R.; Yondola, M.; Tan, G.S.; Leyva-Grado, V.H.; Ryder, A.B.; Miller, M.S.; Rose, J.K.; Palese, P.; García-Sastre, A.; et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol. 2014, 88, 3432–3442. [Google Scholar] [CrossRef] [PubMed]
- Ryder, A.B.; Nachbagauer, R.; Buonocore, L.; Palese, P.; Krammer, F.; Rose, J.K. Vaccination with vesicular stomatitis virus-vectored chimeric hemagglutinins protects mice against divergent influenza virus challenge strains. J. Virol. 2015, 90, 2544–2550. [Google Scholar] [CrossRef] [PubMed]
- Larsen, D.L.; Karasin, A.; Zuckermann, F.; Olsen, C.W. Systemic and mucosal immune responses to H1N1 influenza virus infection in pigs. Vet. Microbiol. 2000, 74, 117–131. [Google Scholar] [CrossRef]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Webby, R.J.; García-Sastre, A.; Richt, J.A. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 2007, 25, 7999–8009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, J.T.; Belisle, S.E.; Tchitchek, N.; Tumpey, T.M.; Ma, W.; Richt, J.A.; Safronetz, D.; Feldmann, H.; Katze, M.G. 2009 pandemic H1N1 influenza virus elicits similar clinical course but differential host transcriptional response in mouse, macaque, and swine infection models. BMC Genom. 2012, 13, 627. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Palese, P. Universal influenza virus vaccines: Need for clinical trials. Nat. Immunol. 2014, 15, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. Strategies to induce broadly protective antibody responses to viral glycoproteins. Expert Rev. Vaccines 2017, 16, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. Novel universal influenza virus vaccine approaches. Curr. Opin. Virol. 2016, 17, 95–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.; Van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.J.; Kanekiyo, M.; Kong, W.P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar] [PubMed]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Baranovich, T.; Jones, J.C.; Russier, M.; Vogel, P.; Szretter, K.J.; Sloan, S.E.; Seiler, P.; Trevejo, J.M.; Webby, R.J.; Govorkova, E.A. The hemagglutinin stem-binding monoclonal antibody VIS410 controls influenza virus-induced acute respiratory distress syndrome. Antimicrob. Agents Chemother. 2016, 60, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Tharakaraman, K.; Subramanian, V.; Viswanathan, K.; Sloan, S.; Yen, H.L.; Barnard, D.L.; Leung, Y.H.; Szretter, K.J.; Koch, T.J.; Delaney, J.C.; et al. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc. Natl. Acad Sci. USA 2015, 112, 10890–10895. [Google Scholar] [CrossRef] [PubMed]
- Reuman, P.D.; Ayoub, E.M.; Small, P.A. Effect of passive maternal antibody on influenza illness in children: A prospective study of influenza A in mother-infant pairs. Pediatr. Infect. Dis. J. 1987, 6, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Ohfuji, S.; Deguchi, M.; Tachibana, D.; Koyama, M.; Takagi, T.; Yoshioka, T.; Urae, A.; Ito, K.; Kase, T.; Maeda, A.; et al. Protective effect of maternal influenza vaccination on influenza in their infants: A prospective cohort study. J. Infect. Dis. 2018, 217, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Richt, J.A.; Janke, B.H.; Sandbulte, M.R.; Gauger, P.C.; Loving, C.L.; Webby, R.J.; García-Sastre, A. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J. Virol. 2012, 86, 10597–10605. [Google Scholar] [CrossRef] [PubMed]
- Gauger, P.C.; Loving, C.L.; Khurana, S.; Lorusso, A.; Perez, D.R.; Kehrli, M.E.; Roth, J.A.; Golding, H.; Vincent, A.L. Live attenuated influenza A virus vaccine protects against A(H1N1)pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology 2014, 471–473, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.B.; Hemmink, J.D.; Porter, E.; Harley, R.; Shelton, H.; Aramouni, M.; Everett, H.E.; Brookes, S.M.; Bailey, M.; Townsend, A.M.; et al. Aerosol delivery of a candidate universal influenza vaccine reduces viral load in pigs challenged with pandemic H1N1 virus. J. Immunol. 2016, 196, 5014–5023. [Google Scholar] [CrossRef] [PubMed]
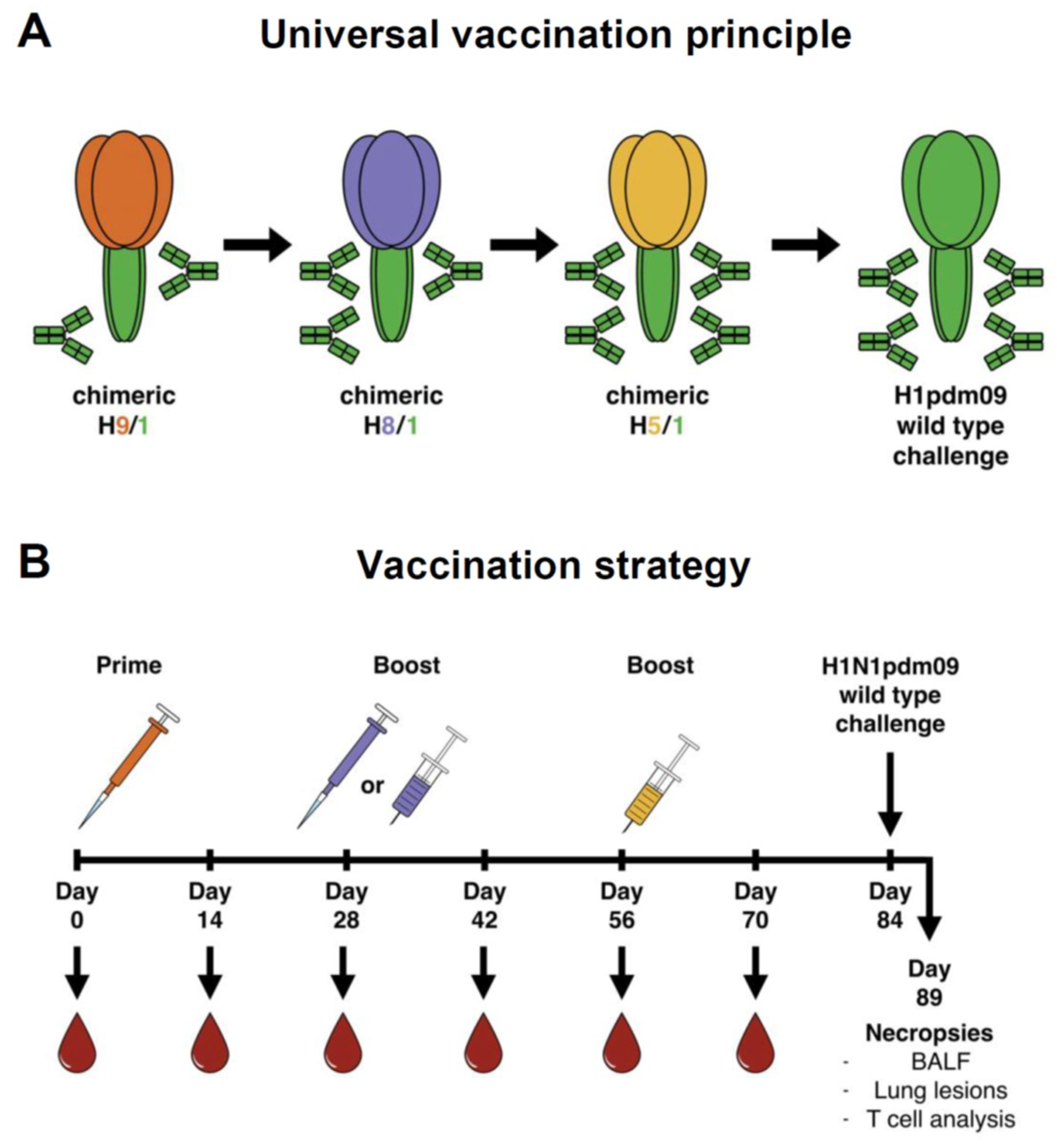


| Group | Vaccine Composition | No. of Pigs | ||
|---|---|---|---|---|
| Prime | Vaccine #1 | Vaccine #2 | ||
| G1 | B-cH9/1 a | IIV cH8/1 b | IIV cH5/1 d | 3 |
| G1 Emul | B-cH9/1 | IIV cH8/1 with adjuvant | IIV cH5/1 with adjuvant | 3 |
| G2 | B-cH9/1 | LAIV cH8/1 c | IIV cH5/1 | 3 |
| G2 Emul | B-cH9/1 | LAIV cH8/1 | IIV cH5/1 with adjuvant | 3 |
| G3 | - | H1N2 inactivated whole virus with adjuvant | 6 | |
| G4 | - | TIV containing H1N1 with adjuvant | 6 | |
| C-Cont | Challenged control | 3 | ||
| Mock | Non-vaccine & non-challenge (Mock) | 2 | ||
| Parameter | HP Score | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| Lung % involvement: Epithelial changes and inflammation | None | 1–25% | 26–50% | 51–75% | Greater than 75% |
| Peribronchiolar cuffing by lymphocytes | None | Mild, loosely formed cuffs of lymphocytes | Moderate, well-formed cuffs of lymphocytes | Prominent thick well-formed cuffs of lymphocytes | |
| Degree of interstitial pneumonia (IP) | None | Mild, focal to multifocal IP | Moderate, locally extensive to multifocal IP | Moderate, multifocal to coalescing IP | Severe, coalescing to diffuse IP |
| Tracheal epithelial changes & Nasal cavity epithelial changes | None | Early epithelial changes with focal to multifocal loss of cilia and epithelial degenerative changes | Mild epithelial flattening with loss of cilia and goblet cells | Moderate epithelial flattening with decreased thickness of respiratory epithelium, loss of cilia and goblet cells | Flattened epithelium with areas of mucosa covered by a single layer of cuboidal epithelium and epithelial loss (necrosis) |
| Degree of rhinitis | None | Mild | Moderate | Severe | |
| Degree of tracheitis | None | Mild | Moderate | Severe | |
| DPC | G1 | G1 Emul | G2 | G2 Emul | G3 | G4 | C-Cont | Mock | |
|---|---|---|---|---|---|---|---|---|---|
| Vaccine Composition | B virus | B virus | B virus | B virus | |||||
| IIV | IIV Emul | LAIV | LAIV | VAERD | TIV | ||||
| IIV | IIV Emul | IIV | IIV Emul | ||||||
| Nasal swab | 0 | 0/3 | 0/3 | 0/3 | 0/3 | 0/6 | 0/6 | 0/3 | 0/2 |
| 1 | 0/3 | 0/3 | 0/3 | 0/3 | 0/6 | 0/6 | 0/3 | 0/2 | |
| 3 | 3/3 | 3/3 | 3/3 | 1/3 | 3/6 | 5/6 | 1/3 | 0/2 | |
| 5 | 2/3 | 3/3 | 1/3 | 3/3 | 6/6 | 6/6 | 3/3 | 0/2 | |
| BALF | 5 | 0/3 | 0/3 | 0/3 | 0/3 | 4/6 | 2/6 | 3/3 | 0/2 |
| Experimental Groups | Immunohistochemistry (IHC): Positive/Total Samples | |||||||
|---|---|---|---|---|---|---|---|---|
| G1 | G1 Emul | G2 | G2 Emul | G3 | G4 | C-Cont | Mock | |
| Lung | 2/3 | 3/3 | 2/3 | 2/3 | 6/6 | 6/6 | 1/3 | 0/2 |
| Trachea | 0/3 | 2/3 | 2/3 | 3/3 | 6/6 | 5/6 | 3/3 | 0/2 |
| Nasal turbinates | 0/3 | 2/3 | 0/3 | 1/3 | 3/6 | 4/6 | 1/3 | 0/2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sunwoo, S.-Y.; Schotsaert, M.; Morozov, I.; Davis, A.S.; Li, Y.; Lee, J.; McDowell, C.; Meade, P.; Nachbagauer, R.; García-Sastre, A.; et al. A Universal Influenza Virus Vaccine Candidate Tested in a Pig Vaccination-Infection Model in the Presence of Maternal Antibodies. Vaccines 2018, 6, 64. https://doi.org/10.3390/vaccines6030064
Sunwoo S-Y, Schotsaert M, Morozov I, Davis AS, Li Y, Lee J, McDowell C, Meade P, Nachbagauer R, García-Sastre A, et al. A Universal Influenza Virus Vaccine Candidate Tested in a Pig Vaccination-Infection Model in the Presence of Maternal Antibodies. Vaccines. 2018; 6(3):64. https://doi.org/10.3390/vaccines6030064
Chicago/Turabian StyleSunwoo, Sun-Young, Michael Schotsaert, Igor Morozov, Anne Sally Davis, Yuhao Li, Jinhwa Lee, Chester McDowell, Philip Meade, Raffael Nachbagauer, Adolfo García-Sastre, and et al. 2018. "A Universal Influenza Virus Vaccine Candidate Tested in a Pig Vaccination-Infection Model in the Presence of Maternal Antibodies" Vaccines 6, no. 3: 64. https://doi.org/10.3390/vaccines6030064





